New Polyene Macrolide Compounds from Mangrove-Derived Strain Streptomyces hiroshimensis GXIMD 06359: Isolation, Antifungal Activity, and Mechanism against Talaromyces marneffei
Abstract
:1. Introduction
2. Results
2.1. Structural Elucidation
2.2. Antifungal Activity of the Compounds
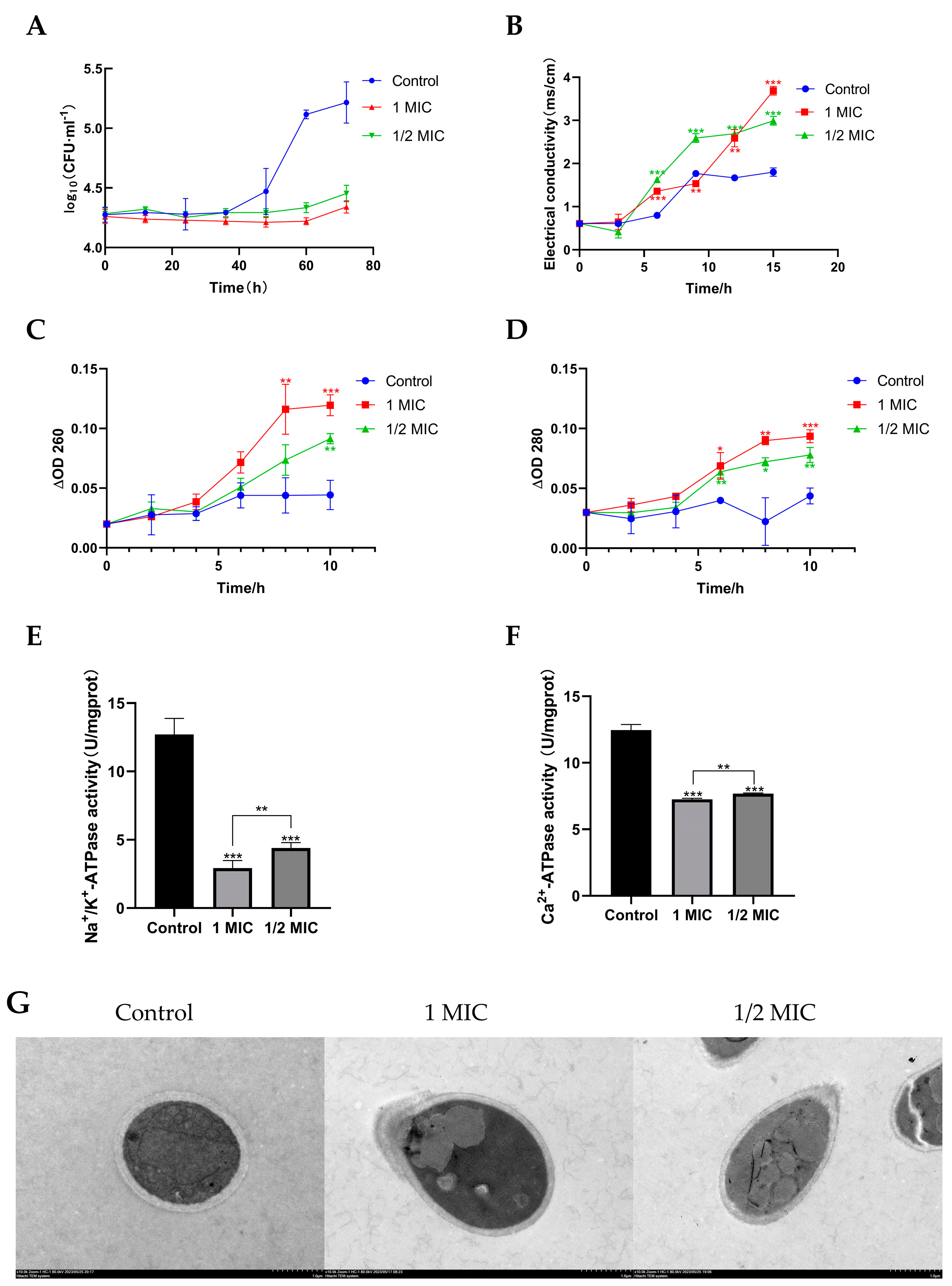
2.3. Compound 1 Inhibited the Growth of T. marneffei
2.4. Compound 1 Disrupted the Cell Membrane of T. marneffei
2.5. Effect of Compound 1 on Morphology of T. marneffe
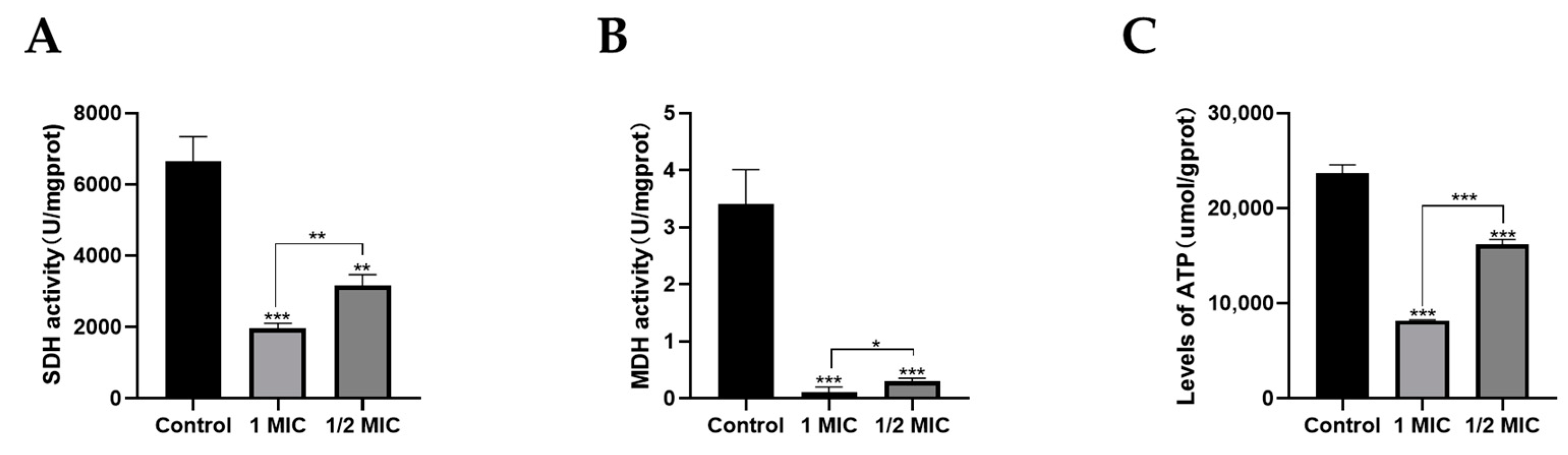
2.6. Effects of Compound 1 on Mitochondrial Function
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Actinomycete Material
4.3. Fermentation, Extraction, and Isolation
4.4. Microbial Strains OriginA and Culture Conditions
4.5. Antifungal Activity
4.6. Determination of Minimal Fungicidal Concentration (MFC)
4.7. Mode of Action of Compound 1
4.7.1. Time-Kill Curve
4.7.2. Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM)
4.7.3. Leakage of Extracellular Conductivity
4.7.4. Leakage of 260 nm and 280 nm Absorbing Material
4.7.5. Detection of Na+/K+-ATPase and Ca2+-ATPase
4.7.6. Measurement of Intracellular ATPase Concentration
4.7.7. Detection of MDH and SDH
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, Y.; Zhang, J.; Li, X.; Yang, Y.; Zhang, Y.; Ma, J.; Xi, L. Penicillium marneffei infection: An emerging disease in mainland China. Mycopathologia 2013, 175, 57–67. [Google Scholar] [CrossRef]
- Imwidthaya, P. Update of Penicillosis marneffei in Thailand. Mycopathologia 1994, 127, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.C.; Lau, S.K.; Woo, P.C. Sixty years from Segretain’s description: What have we learned and should learn about the basic mycology of Talaromyces marneffei? Mycopathologia 2019, 184, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Lo, G.C.; Lam, C.S.; Chow, W.N.; Ngan, A.H.; Wu, A.K. In vitro activity of posaconazole against Talaromyces marneffei by broth microdilution and Etest methods and comparison to itraconazole, voriconazole, and anidulafungin. Antimicrob. Agents Chemother. 2017, 61, e01480-16. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liang, L.; Chen, J. In vitro antifungal drug susceptibilities of Penicillium marneffei from China. J. Infect. Chemother. 2013, 19, 776–778. [Google Scholar] [CrossRef]
- Supparatpinyo, K.; Schlamm, H.T. Voriconazole as therapy for systemic Penicillium marneffei infections in AIDS patients. Am. J. Trop. Med. Hyg. 2007, 77, 350–353. [Google Scholar] [CrossRef]
- Wu, T.C.; Chan, J.W.; Ng, C.K.; Tsang, D.N.; Lee, M.P.; Li, P.C. Clinical presentations and outcomes of Penicillium marneffei infections: A series from 1994 to 2004. Hong Kong Med. J. 2008, 14, 103. [Google Scholar]
- Larsson, M.; Nguyen, L.H.T.; Wertheim, H.F.; Dao, T.T.; Taylor, W.; Horby, P. Clinical characteristics and outcome of Penicillium marneffei infection among HIV-infected patients in northern Vietnam. AIDS Res. Ther. 2012, 9, 24. [Google Scholar] [CrossRef]
- Son, V.T.; Khue, P.M.; Strobel, M. Penicilliosis and AIDS in Haiphong, Vietnam: Evolution and predictive factors of death. Med. Mal. Infect. 2014, 44, 495–501. [Google Scholar] [CrossRef]
- Antinori, S.; Gianelli, E.; Bonaccorso, C.; Ridolfo, A.L.; Croce, F.; Sollima, S.; Parravicini, C. Disseminated Penicillium marneffei infection in an HIV-positive Italian patient and a review of cases reported outside endemic regions. J. Travel Med. 2006, 13, 181–188. [Google Scholar] [CrossRef]
- Castro-Lainez, M.T.; Sierra-Hoffman, M.; LLompart-Zeno, J.; Adams, R.; Howell, A.; Hoffman-Roberts, H. Talaromyces marneffei infection in a non-HIV non-endemic population. IDCases 2018, 12, 21–24. [Google Scholar] [CrossRef]
- Julander, I.; Petrini, B. Penicillium marneffei infection in a Swedish HIV-infected immunodeficient narcotic addict. Scand. J. Infect. Dis. 1997, 29, 320–322. [Google Scholar] [CrossRef]
- De Monte, A.; Risso, K.; Normand, A.C.; Boyer, G.; L’Ollivier, C.; Marty, P.; Gari-Toussaint, M. Chronic pulmonary penicilliosis due to Penicillium marneffei: Late presentation in a french traveler. J. Travel Med. 2014, 21, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Patassi, A.A.; Saka, B.; Landoh, D.E.; Kotosso, A.; Mawu, K.; Halatoko, W.A. First observation in a non-endemic country (Togo) of Penicillium marneffei infection in a human immunodeficiency virus-infected patient: A case report. BMC Res. Notes 2013, 6, 506. [Google Scholar] [CrossRef] [PubMed]
- Pautler, K.B.; Padhye, A.; Ajello, L. Imported Penicilliosis marneffei in the United States: Report of a second human infection. Sabouraudia 1984, 22, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, K.; Dhungana, N.; Jang, Y.; Chaturvedi, V.; Desmond, E. Characterization of clinical isolates of Talaromyces marneffei and related species, California, USA. Emerg. Infect. Dis. 2019, 25, 1765. [Google Scholar] [CrossRef] [PubMed]
- Kalasuba, K.; Miranti, M.; Rahayuningsih, S.R.; Safriansyah, W.; Syamsuri, R.R.P.; Farabi, K.; Oktavia, D.; Alhasnawi, A.N.; Doni, F. Red Mangrove (Rhizophora stylosa Griff.)-A Review of Its Botany, Phytochemistry, Pharmacological Activities, and Prospects. Plants 2023, 12, 2196. [Google Scholar] [CrossRef]
- Ye, J.J.; Zou, R.J.; Zhou, D.D.; Deng, X.L.; Wu, N.L.; Chen, D.D.; Xu, J. Insights into the phylogenetic diversity, biological activities, and biosynthetic potential of mangrove rhizosphere Actinobacteria from Hainan Island. Front. Microbiol. 2023, 14, 1157601. [Google Scholar] [CrossRef]
- Xu, J. Biomolecules produced by mangrove-associated microbes. Curr. Med. Chem. 2011, 18, 5224–5266. [Google Scholar] [CrossRef]
- Xu, J. Bioactive natural products derived from mangrove-associated microbes. RSC Adv. 2015, 5, 841–892. [Google Scholar] [CrossRef]
- Jiang, Z.K.; Tuo, L.; Huang, D.L.; Osterman, I.A.; Tyurin, A.P.; Liu, S.W. Diversity, novelty, and antimicrobial activity of endophytic actinobacteria from mangrove plants in Beilun Estuary National Nature Reserve of Guangxi, China. Front. Microbiol. 2018, 9, 868. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, S.; Pang, X.; Cai, J.; Zhang, X.; Liu, Y. Natural products from mangrove sediments-derived microbes: Structural diversity, bioactivities, biosynthesis, and total synthesis. Eur. J. Med. Chem. 2022, 230, 114117. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zhang, S.; Li, W. Advance in marine actinobacterial research--A review. Wei Sheng Wu Xue Bao = Acta Microbiol. Sin. 2011, 51, 161–169. [Google Scholar]
- Jiang, S.; Li, M.; Hou, S.; Han, M.; Yin, J.; Gao, C. Diversity and anti-aging activity of endophytic actinobacteria from true mangrove plants collected from the west coast of Hainan. Guihaia 2020, 40, 320–326. [Google Scholar]
- Wang, Y.F.; Wei, S.J.; Zhang, Z.P.; Zhan, T.H.; Tu, G.Q. Antifungalmycin, an antifungal macrolide from Streptomyces padanus 702. Nat. Prod. Bioprospecting 2012, 2, 41–45. [Google Scholar] [CrossRef]
- Xiong, Z.Q.; Tu, X.R.; Wei, S.J.; Huang, L.; Li, X.H.; Lu, H.; Tu, G.Q. The mechanism of antifungal action of a new polyene macrolide antibiotic antifungalmycin 702 from Streptomyces padanus JAU4234 on the rice sheath blight pathogen Rhizoctonia solani. PLoS ONE 2013, 8, e73884. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Jiang, X.; Sun, J.; Zhang, C.; Zhang, Y.; Lu, L.; Ju, J. Antibacterial secondary metabolites produced by mangrove-derived Actinomycete Stremptmeces costaricanus SCSIO ZS0073. Nat. Prod. Res. Dev. 2017, 29, 410. [Google Scholar]
- Rochet, P.; Lancelin, J.M. Revised 1H and 13C NMR assignments of the polyene antibiotic filipin III. Magn. Reson. Chem. 1997, 35, 538–542. [Google Scholar] [CrossRef]
- Barreales, E.G.; Rumbero, Á.; Payero, T.D.; de Pedro, A.; Jambrina, E.; Aparicio, J.F. Structural and bioactivity characterization of filipin derivatives from engineered Streptomyces filipinensis strains reveals clues for reduced haemolytic action. Antibiotics 2020, 9, 413. [Google Scholar] [CrossRef]
- Cai, R.; Hu, M.; Zhang, Y.; Niu, C.; Yue, T.; Yuan, Y.; Wang, Z. Antifungal activity and mechanism of citral, limonene and eugenol against Zygosaccharomyces rouxii. LWT 2019, 106, 50–56. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ye, K.P.; Zhang, X.; Pan, D.D.; Sun, Y.Y.; Cao, J.X. Antibacterial activity and mechanism of action of black pepper essential oil on meat-borne Escherichia coli. Front. Microbiol. 2017, 7, 2094. [Google Scholar] [CrossRef] [PubMed]
- Alyousef, A.A. Antifungal Activity and Mechanism of Action of Different Parts of Myrtus communis Growing in Saudi Arabia against Candida Spp. J. Nanomater. 2021, 2021, 3484125. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, X.; Zhang, M.; Bai, B.; Yang, Y.; Fan, S. The antibacterial mechanism of perilla rosmarinic acid. Biotechnol. Appl. Biochem. 2022, 69, 1757–1764. [Google Scholar] [CrossRef]
- Primeau, J.O.; Armanious, G.P.; Fisher, M.L.E.; Young, H.S. The sarcoendoplasmic reticulum calcium ATPase. Membr. Protein Complexes Struct. Funct. 2018, 87, 229–258. [Google Scholar]
- Lau, S.K.P.; Tsang, C.C.; Woo, P.C.Y. Talaromyces marneffei genomic, transcriptomic, proteomic and metabolomic studies reveal mechanisms for environmental adaptations and virulence. Toxins 2017, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhang, C.; Li, C.; Lin, L. Antibacterial mechanism of oregano essential oil. Ind. Crops Prod. 2019, 139, 111498. [Google Scholar] [CrossRef]
- Van Vranken, J.G.; Na, U.; Winge, D.R.; Rutter, J. Protein-mediated assembly of succinate dehydrogenase and its cofactors. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 168–180. [Google Scholar] [CrossRef]
- Mathew, B.P.; Nath, M. Recent approaches to antifungal therapy for invasive mycoses. ChemMedChem Chem. Enabling Drug Discov. 2009, 4, 310–323. [Google Scholar] [CrossRef]
- Zotchev, S.B. Polyene macrolide antibiotics and their applications in human therapy. Curr. Med. Chem. 2003, 10, 211–223. [Google Scholar] [CrossRef]
- Bhatnagar, I.; Kim, S.K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Cai, M.J.; Zhou, L.L.; Wang, H.D. The structure and function of cell membranes studied by atomic force microscopy. Semin. Cell Dev. Biol 2018, 73, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.M.; Wen, X.F.; Xie, Y.T.; Guo, Z.; Zhao, R.B.; Yu, P.; Gong, D.M.; Deng, S.G.; Zeng, Z.L. Antifungal activity and mechanism of monocaprin against food spoilage fungi. Food Control 2018, 84, 561–568. [Google Scholar] [CrossRef]
- Elsadek, L.A.; Matthews, J.H.; Nishimura, S.; Nakatani, T.; Ito, A.; Gu, T. Genomic and targeted approaches unveil the cell membrane as a major target of the antifungal cytotoxin amantelide A. ChemBioChem 2021, 22, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Szomek, M.; Reinholdt, P.; Walther, H.L.; Scheidt, H.A.; Müller, P.; Obermaier, S.; Wüstner, D. Natamycin sequesters ergosterol and interferes with substrate transport by the lysine transporter Lyp1 from yeast. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 184012. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Niu, H.J.; Qu, T.L.; Zhang, X.F.; Du, F.Y. Streptomyces sp. FX13 inhibits fungicide-resistant Botrytis cinerea in vitro and in vivo by producing oligomycin A. Pestic. Biochem. Physiol. 2021, 175, 104834. [Google Scholar] [CrossRef] [PubMed]
- Kamble, M.T.; Rudtanatip, T.; Soowannayan, C.; Nambunruang, B.; Medhe, S.V.; Wongprasert, K. Depolymerized Fractions of Sulfated Galactans Extracted from Gracilaria fisheri and Their Antibacterial Activity against Vibrio parahaemolyticus and Vibrio harveyi. Mar. Drugs 2022, 20, 469. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, J.; Lv, Q.; Han, B. Recent Progress in Research on Mitochondrion-Targeted Antifungal Drugs: A Review. Antimicrob. Agents Chemother. 2023, 67, e00003-23. [Google Scholar] [CrossRef]
- Xin, Z.; OuYang, Q.; Wan, C.; Che, J.; Li, L.; Chen, J.; Tao, N. Isolation of antofine from Cynanchum atratum BUNGE (Asclepiadaceae) and its antifungal activity against Penicillium digitatum. Postharvest Biol. Technol. 2019, 157, 110961. [Google Scholar] [CrossRef]
- Zhao, W.B.; Zhao, Z.M.; Ma, Y.; Li, A.P.; Zhang, Z.J.; Hu, Y.M. Antifungal activity and preliminary mechanism of pristimerin against Sclerotinia sclerotiorum. Ind. Crops Prod. 2022, 185, 115124. [Google Scholar] [CrossRef]
- Kai-Su, P.; Hong, L.; Dong-Yan, Z.; Yan-Qing, Z.; Andrianopoulos, A.; Latgé, J.P.; Cun-Wei, C. Study on the mechanisms of action of berberine combined with fluconazole against fluconazole-resistant strains of Talaromyces marneffei. Front. Microbiol. 2022, 13, 1033211. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kong, F.; Shi, X.; Han, H.; Li, M.; Guan, B. Antibacterial activity and mechanism of lactobionic acid against Pseudomonas fluorescens and Methicillin-resistant Staphylococcus aureus and its application on whole milk. Food Control 2020, 108, 106876. [Google Scholar] [CrossRef]
- Maliehe, T.S.; Nqotheni, M.I.; Shandu, J.S.; Selepe, T.N.; Masoko, P.; Pooe, O.J. Chemical Profile, Antioxidant and Antibacterial Activities, Mechanisms of Action of the Leaf Extract of Aloe arborescens Mill. Plants 2023, 12, 869. [Google Scholar] [CrossRef] [PubMed]
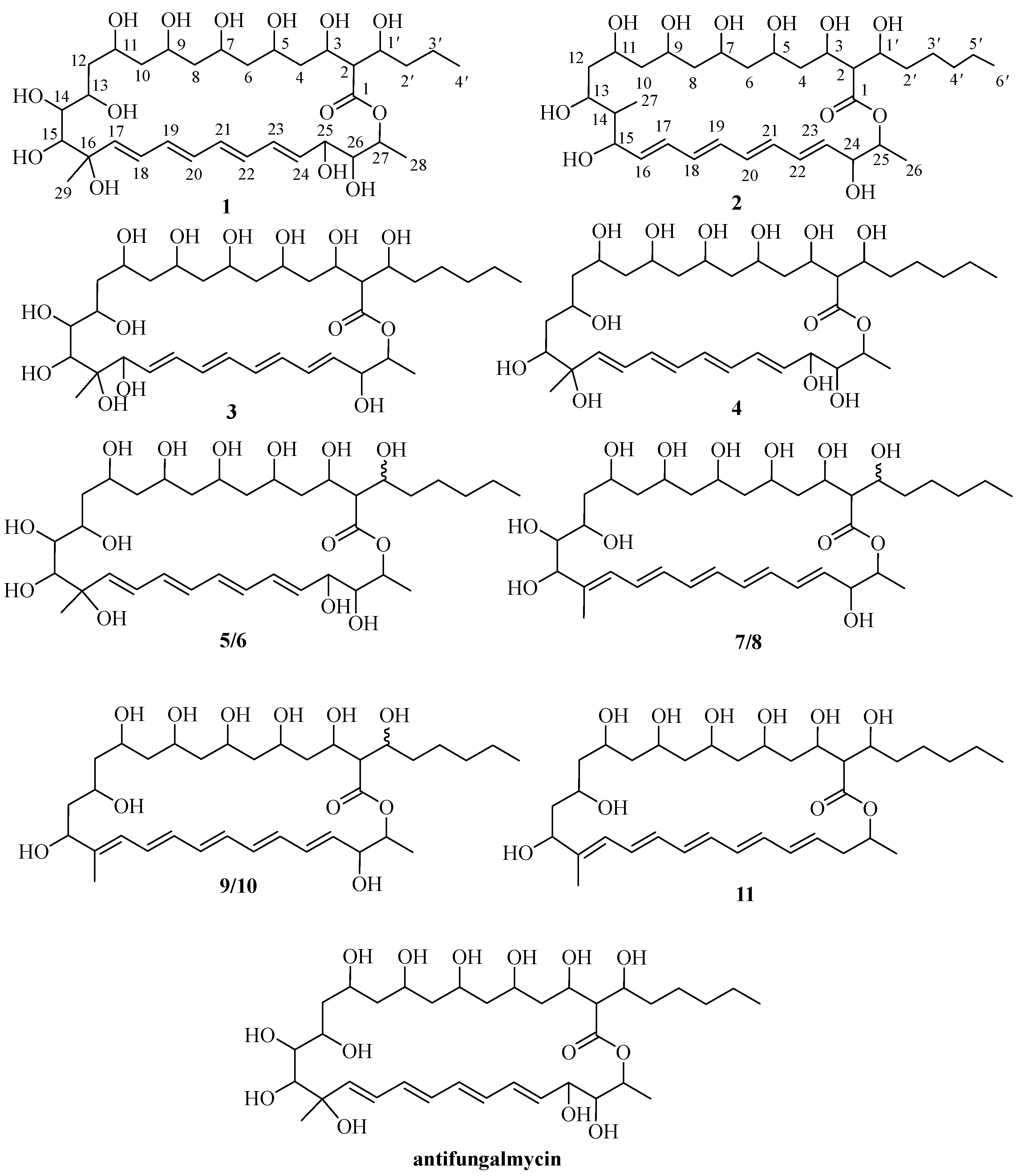
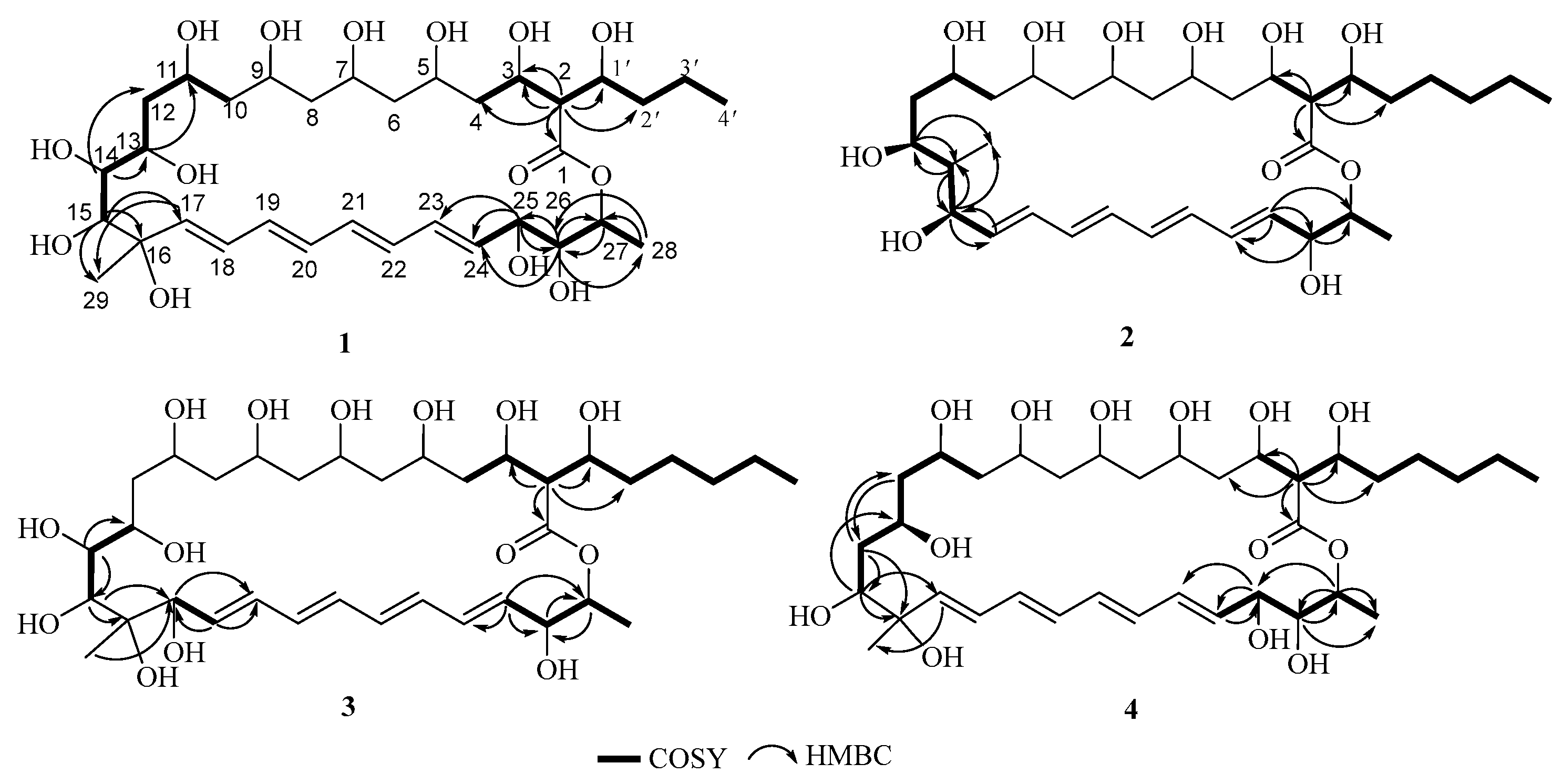
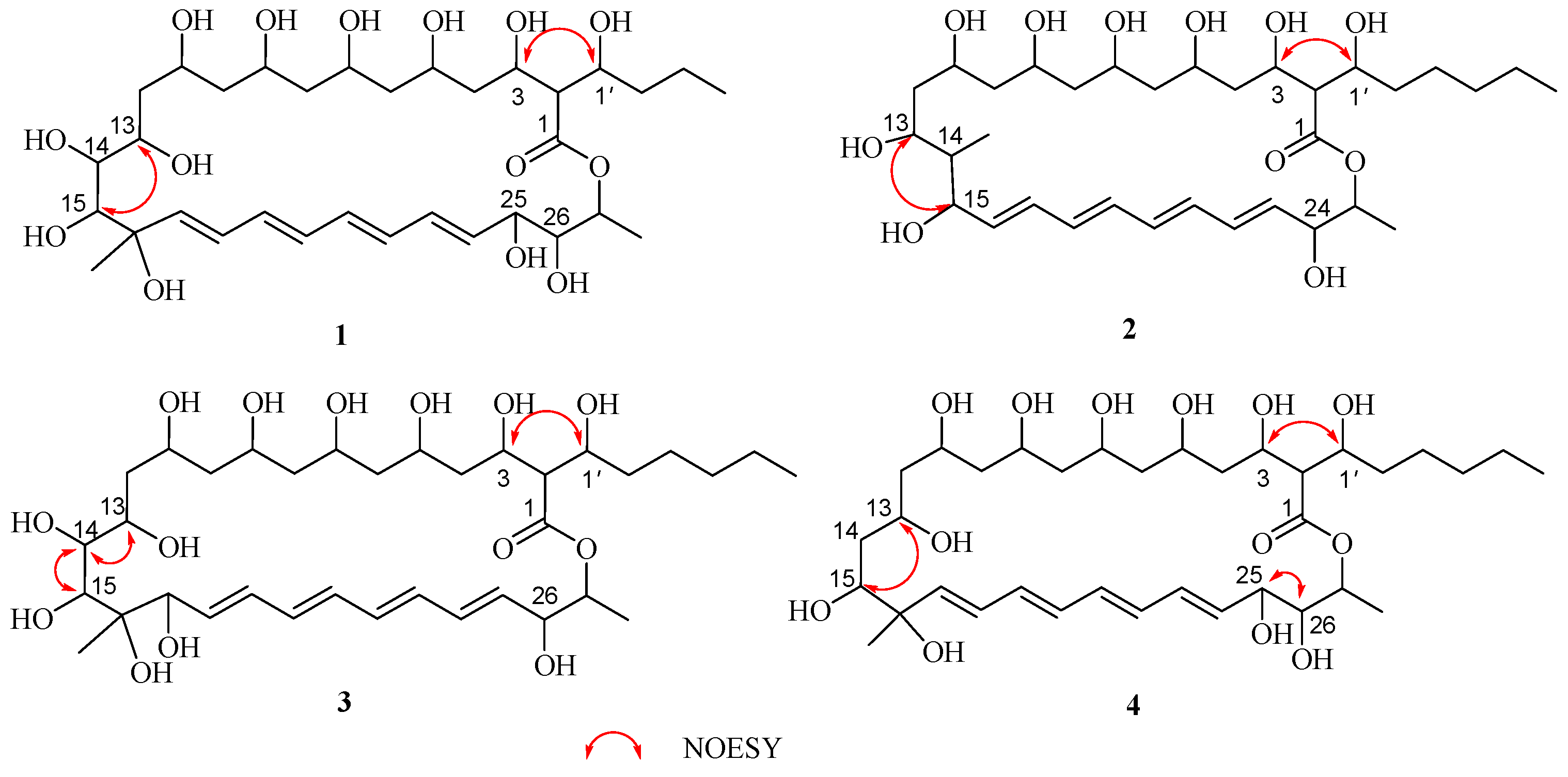

| NO. | 1 a | 2 a | 3 b | 4 b | ||||
|---|---|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | - | 171.6, C | - | 171.2, C | - | 171.3, C | - | 171.7, C |
| 2 | 2.46, dd (8.6, 6.8) | 58.3, CH | 2.38, t (8.0) | 58.5, CH | 2.48, m | 58.2, CH | 2.46, dd (8.8, 6.6) | 58.2, CH |
| 3 | 4.02, m | 69,6, CH | 4.05, m | 69,7, CH | 4.05, m | 70.0, CH | 4.03, m | 69,4, CH |
| 4 | 1.52, m | 40.9, CH2 | 1.38, m | 40.9, CH2 | 1.45, m | 43.3, CH2 | 1.50, m | 40.9, CH2 |
| 5 | 3.90, m | 67.8, CH | 3.93, m | 70.2, CH | 3.93, m | 68.2, CH | 3.92, m | 69.4, CH |
| 6 | 1.54, m | 44.3, CH2 | 1.40, m | 44.0, CH2 | 1.45, m | 45.4, CH2 | 1.50, m | 44.1, CH |
| 7 | 3.90, m | 68.5, CH | 3.93, m | 70.3, CH | 3.98, m | 69.0, CH | 3.92, m | 68.2, CH |
| 8 | 1.47, m | 44.5, CH2 | 1.50, m | 43.7, CH2 | 1.46, m | 46.3, CH2 | 1.50, m | 44.6, CH2 |
| 9 | 3.97, m | 69.2, CH | 3.93, m | 69.8, CH | 3.93, m | 67.0, CH | 3.97, m | 68.2, CH |
| 10 | 1.71, m; 1.25, m | 42.2, CH2 | 1.70, m; 1.25, m | 42.9, CH2 | 1.76, m;1.45, m | 43.6, CH2 | 1.76, m; 1.53, m | 41.4, CH2 |
| 11 | 3.90, m | 67.8, CH | 3.93, m | 66.7, CH | 3.93, m | 66.9, CH | 3.87, m | 67.5, CH |
| 12 | 1.62, m | 41.6, CH2 | 1.92, m; 1.52, m | 38.5, CH2 | 3.33, m | 63.1, CH2 | 1.74, m; 1.50, m | 44.4, CH2 |
| 13 | 3.29, m | 75.2, CH | 3.21, m | 80.5, CH | 3.46, m | 66.0, CH | 3.65, m | 71.2, CH |
| 14 | 3.34, m | 82.3, CH | 2.87, m | 73.0, CH | 3.80, m | 84.6, CH | 2.16, m | 41.9, CH2 |
| 15 | 3.65, d (5.4) | 82.7, CH | 3.63, m | 80.5, CH | 3.71, d | 78.6, CH | 3.90, m | 76.0, CH |
| 16 | - | 82.0, C | 5.89, dd (14.0, 3.1) | 129.5, CH | - | 80.0, C | 84.5, CH | |
| 17 | 5.72, d (15.0) | 138.9, CH | 6.20, m | 133.3, CH | 4.18, d (4.4) | 83.4, CH | 5.65, d (15.0) | 137.8, C |
| 18 | 6.15, dd (15.1, 9.4) | 126.3, CH | 6.18, m | 130.4, CH | 5.72, dd (14.5, 4.3) | 129.1, CH | 6.15, dd (14.5, 6.2) | 127.4, CH |
| 19 | 6.24, m | 131.5, CH | 6.28, m | 131.4, CH | 6.31, m | 131.1, CH | 6.22, m | 130.8, CH |
| 20 | 6.26, m | 133.2, CH | 6.26, m | 130.7, CH | 6.26, m | 130.4, CH | 6.21, m | 131.1, CH |
| 21 | 6.28, m | 131.2, CH | 6.23, m | 131.1, CH | 6.31, m | 131.5, CH | 6.21, m | 133.5, CH |
| 22 | 6.28, m | 133.5, CH | 6.20, m | 133.8, CH | 6.26, m | 131.2, CH | 6.32, m | 134.0, CH |
| 23 | 6.28, m | 130.4, CH | 5.79, dd (14.7, 6.8) | 135.9, CH | 6.23, m | 133.1, CH | 6.25, m | 130.3, CH |
| 24 | 5.67, dd (14.6, 5.8) | 134.1, CH | 3.82, m | 73.3, CH | 6.36, m | 133.9, CH | 5.68, dd (14.6, 6.1) | 134.4, CH |
| 25 | 4.24, dd (5.7, 2.3), | 72.0, CH | 4.62, m | 72.0, CH | 5.99, dd (15.1,5.1) | 136.1, CH | 4.23, m | 72.1, CH |
| 26 | 3.51, dd (7.2, 2.6) | 75.7, CH | 1.19, d (6.3) | 17.8, CH3 | 3.87, m | 72.4, CH | 3.50, m | 75.5, CH |
| 27 | 4.71, m | 70.3, CH | 0.84, m | 15.6, CH3 | 4.57, dd (8.9, 6.3) | 72.5, CH | 4.77, m | 70.2, CH |
| 28 | 1.14, d (6.2) | 17.4, CH3 | - | - | 1.21, d (6.2) | 17.9, CH3 | 1.14, d (6.3) | 17.5, CH3 |
| 29 | 1.06, s (7.1) | 22.9, CH3 | - | - | 1.20, s | 19.6, CH3 | 1.10, s | 22.2, CH3 |
| 1′ | 3.68, m | 69.2, CH | 3.72, m | 69.6, CH | 3.68, m | 69.7, CH | 3.65, m | 69.2, CH |
| 2′ | 1.29, m; 1.24, m | 36.7, CH2 | 1.34, m; 1.23, m | 33.9, CH2 | 1.45, m; 1.25, m | 34.3, CH2 | 1.33, m; 1.24, m | 34.5, CH2 |
| 3′ | 1.44, m; 1.25, m | 18.2, CH2 | 1.42, m; 1.24, m | 24.4, CH2 | 1.45, m; 1.25, m | 24.6, CH2 | 1.42, m; 1.24, m | 24.5, CH2 |
| 4′ | 0.83, t (7.0) | 14.0, CH3 | 1.24, m | 31.3, CH2 | 1.24, m | 31.3, CH2 | 1.24, m | 31.3, CH2 |
| 5′ | - | - | 1.24, m | 22.1, CH2 | 1.25, m | 22.1, CH2 | 1.25, m | 22.1, CH2 |
| 6′ | - | - | 0.84, t (6.9) | 14.0, CH3 | 0.85, t (6.9) | 14.0, CH3 | 0.85, t (7.1) | 14.0, CH3 |
| 3-OH | 5.12, s | - | 5.06, m | - | 5.23, d | - | 5.10, d | - |
| 5-OH | 4.85, m | - | 4.90, d | - | 4.94, d | - | 4.88, m | - |
| 7-OH | 4.85, m | - | 5.04, m | - | 4.82, d | - | 4.88, m | - |
| 9-OH | 4.90, m | - | 5.07, m | - | 4.32, d | - | 4.86, m | - |
| 11-OH | 4.85, m | - | 4.56, d | - | 4.40, d | - | 4.68, d | - |
| 13-OH | - | - | 4.78, d | - | 4.71, m | - | 4.80, m | - |
| 14-OH | - | - | - | - | 4.65, d | - | - | - |
| 15-OH | - | - | 4.78, m | - | 4.92, d | - | 4.88, m | - |
| 16-OH | - | - | - | - | 4.48, d | - | - | - |
| 17-OH | - | - | - | - | 5.00, m | - | - | - |
| 24-OH | - | - | 5.26, d | - | - | - | - | - |
| 25-OH | - | - | - | - | - | - | 4.86, m | - |
| 26-OH | - | - | - | - | 5.29, d | - | 4.91, d | - |
| 1′-OH | 4.85, m | - | 4.82, d | - | 4.85, d | - | 4.80, m | - |
| Compounds | MIC (μg/mL) | MFC (μg/mL) |
|---|---|---|
| 1 | 16 | 64 |
| 2 | >128 | —— |
| 3 | 128 | —— |
| 4 | 32 | —— |
| 5 | 32 | 64 |
| 6 | 128 | —— |
| 7 | 4 | 8 |
| 8 | 16 | 16 |
| 9 | 2 | 4 |
| 10 | 32 | 128 |
| 11 | 32 | —— |
| FLC * | 16 | 64 |
| AMB * | 0.5 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Yin, J.; Bai, M.; Yang, J.; Jiang, C.; Yi, X.; Liu, Y.; Gao, C. New Polyene Macrolide Compounds from Mangrove-Derived Strain Streptomyces hiroshimensis GXIMD 06359: Isolation, Antifungal Activity, and Mechanism against Talaromyces marneffei. Mar. Drugs 2024, 22, 38. https://doi.org/10.3390/md22010038
Wang Z, Yin J, Bai M, Yang J, Jiang C, Yi X, Liu Y, Gao C. New Polyene Macrolide Compounds from Mangrove-Derived Strain Streptomyces hiroshimensis GXIMD 06359: Isolation, Antifungal Activity, and Mechanism against Talaromyces marneffei. Marine Drugs. 2024; 22(1):38. https://doi.org/10.3390/md22010038
Chicago/Turabian StyleWang, Zhou, Jianglin Yin, Meng Bai, Jie Yang, Cuiping Jiang, Xiangxi Yi, Yonghong Liu, and Chenghai Gao. 2024. "New Polyene Macrolide Compounds from Mangrove-Derived Strain Streptomyces hiroshimensis GXIMD 06359: Isolation, Antifungal Activity, and Mechanism against Talaromyces marneffei" Marine Drugs 22, no. 1: 38. https://doi.org/10.3390/md22010038






