Characterization of Unfractionated Polysaccharides in Brown Seaweed by Methylation-GC-MS-Based Linkage Analysis
Abstract
1. Introduction
2. Results
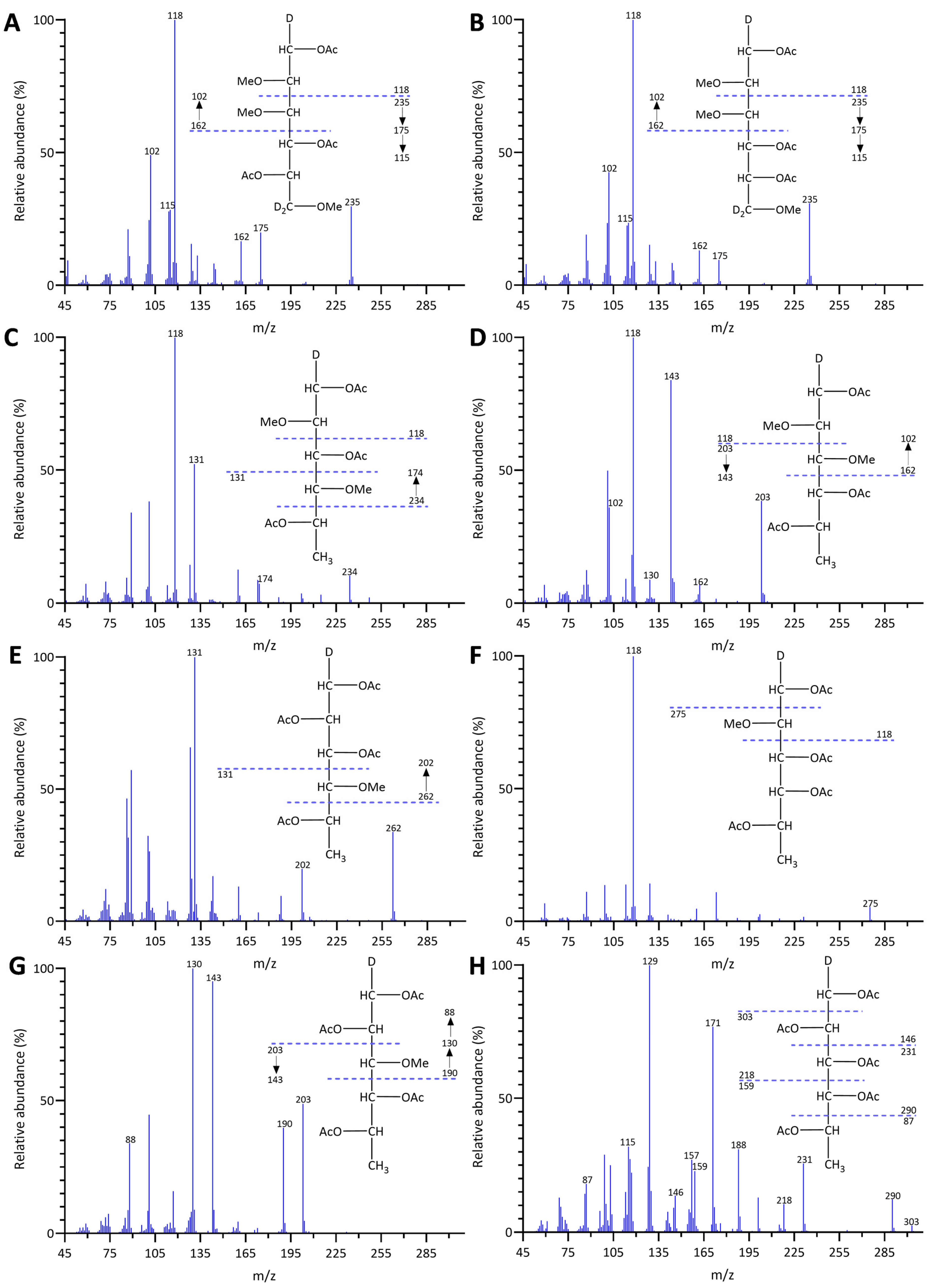
2.1. Unfractionated Polysaccharide Linkage Compositions of Different Brown Seaweeds
2.1.1. Unfractionated Polysaccharide Linkage Compositions of Himanthalia elongata
2.1.2. Unfractionated Polysaccharide Linkage Compositions of Fucus vesiculosus
2.1.3. Unfractionated Polysaccharide Linkage Compositions of Alaria marginata
2.1.4. Unfractionated Polysaccharide Linkage Compositions of Saccharina latissima
2.1.5. Unfractionated Polysaccharide Linkage Compositions of Macrocystis tenuifolia
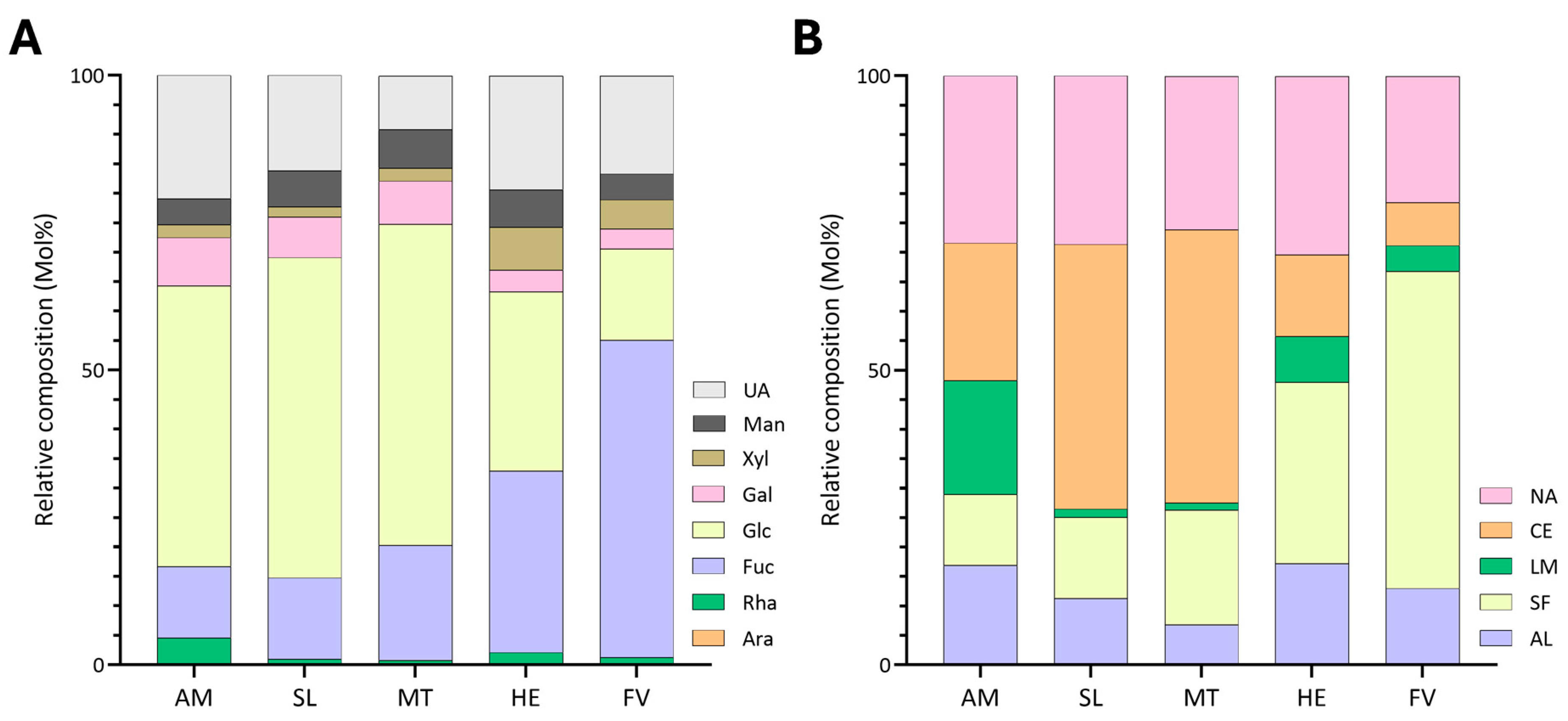
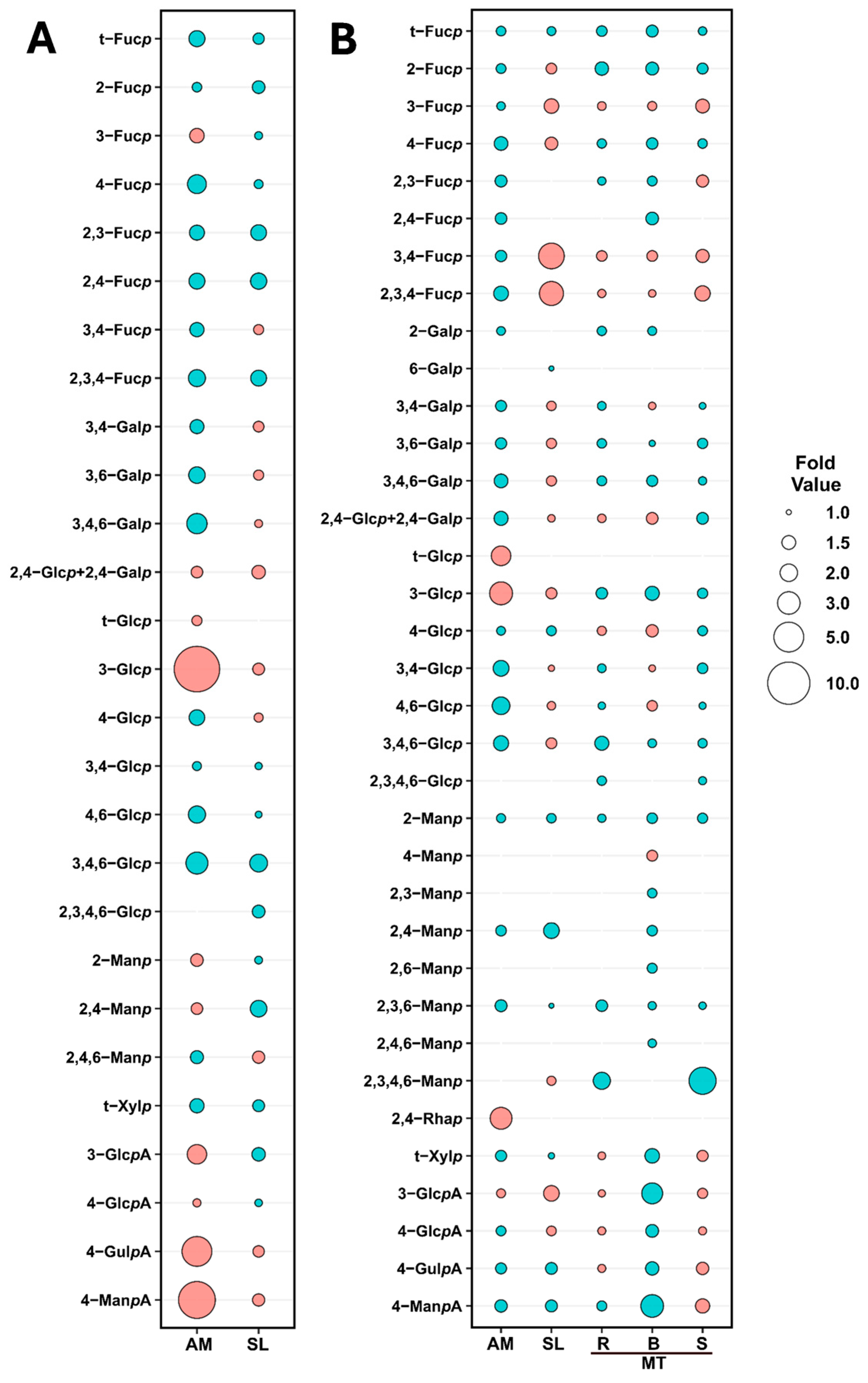
2.2. Variations in Unfractionated Polysaccharide Linkage Compositions Among Brown Seaweed Samples
2.2.1. Variations in Unfractionated Polysaccharide Linkage Compositions Among Tissues of Macrocystis tenuifolia
2.2.2. Annual Variations of Unfractionated Polysaccharide Linkage Compositions of Alaria marginata and Saccharina latissima
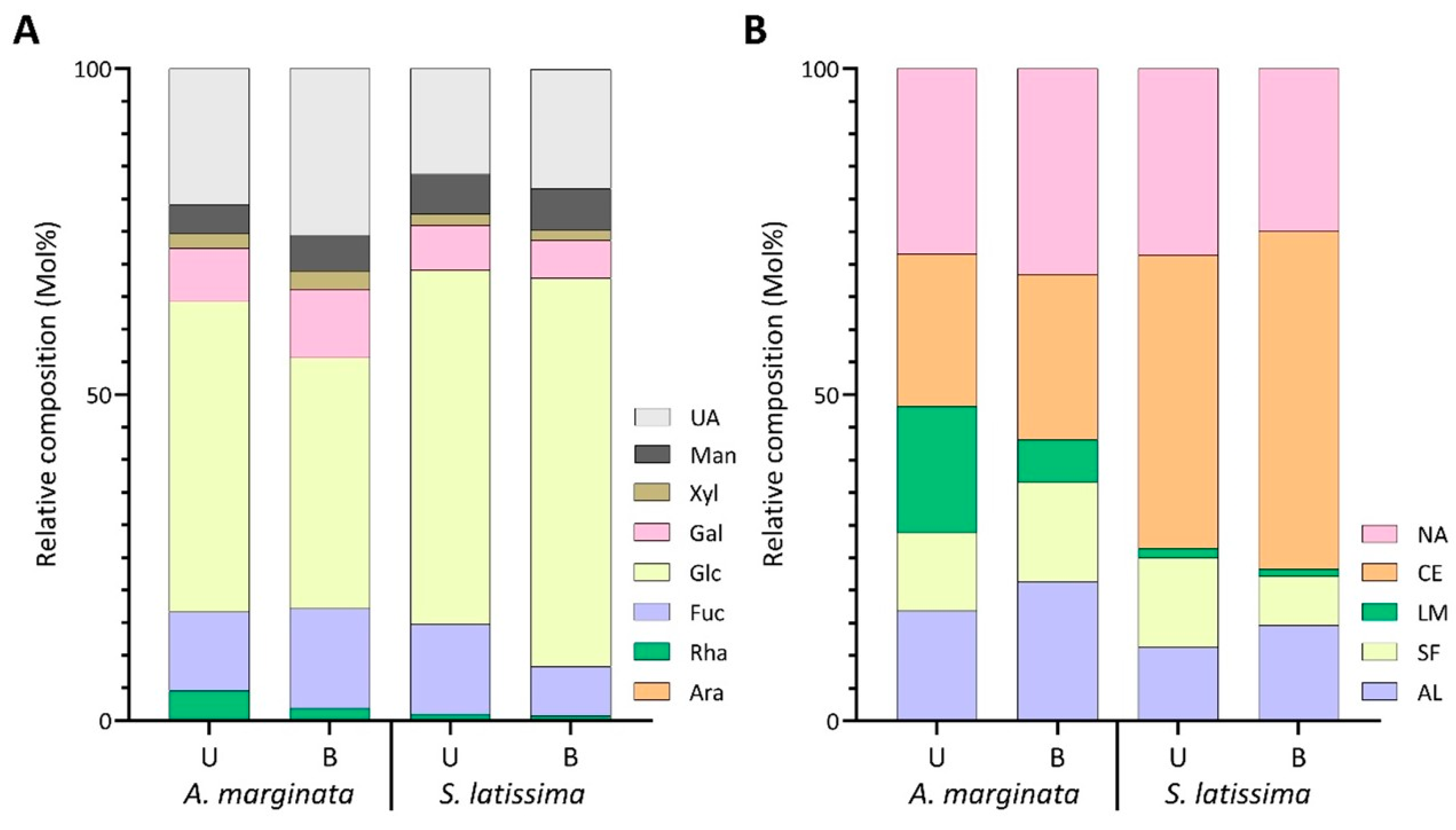
2.2.3. Effects of Blanching Treatments on Unfractionated Polysaccharide Linkage Compositions of Alaria marginata, Saccharina latissima, and Macrocystis tenuifolia
3. Discussions
3.1. Estimation of FCSPs in Various Brown Seaweed Species
3.2. Estimation of Cellulose in Various Brown Seaweed Species
3.3. Estimation of Alginate in Various Brown Seaweed Species
3.4. Further Discussions and Future Considerations
4. Materials and Methods
4.1. Brown Seaweed Materials
4.2. Preparation of Unfractionated Polysaccharides of Brown Seaweeds
4.3. Preparation of PMAA Derivatives from Dry AIR Powder of Brown Seaweeds
4.3.1. Permethylation
4.3.2. Acid Hydrolysis
4.3.3. Reduction
4.3.4. Peracetylation and Final Cleanup
4.4. GC-MS Analysis of PMAAs Prepared from Brown Seaweed AIRs
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bringloe, T.T.; Starko, S.; Wade, R.M.; Vieira, C.; Kawai, H.; De Clerck, O.; Cock, J.M.; Coelho, S.M.; Destombe, C.; Valero, M.; et al. Phylogeny and evolution of the brown algae. Crit. Rev. Plant Sci. 2020, 39, 281–321. [Google Scholar] [CrossRef]
- Mineur, F.; Arenas, F.; Assis, J.; Davies, A.J.; Engelen, A.H.; Fernandes, F.; Malta, E.-j.; Thibaut, T.; Van Nguyen, T.; Vaz-Pinto, F.; et al. European seaweeds under pressure: Consequences for communities and ecosystem functioning. J. Sea Res. 2015, 98, 91–108. [Google Scholar] [CrossRef]
- Sanjeewa, K.K.A.; You-Jin, J. Edible brown seaweeds: A review. J. Food Bioact. 2018, 2, 37–50. [Google Scholar] [CrossRef]
- Abbott, D.W.; Aasen, I.M.; Beauchemin, K.A.; Grondahl, F.; Gruninger, R.; Hayes, M.; Huws, S.; Kenny, D.A.; Krizsan, S.J.; Kirwan, S.F.; et al. Seaweed and seaweed bioactives for mitigation of enteric methane: Challenges and opportunities. Animals 2020, 10, 2432. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Zhang, Y.; Yang, Y.; Wang, P.; Imre, B.; Wong, A.C.Y.; Hsieh, Y.S.Y.; Wang, D. Brown algae carbohydrates: Structures, pharmaceutical properties, and research challenges. Mar. Drugs 2021, 19, 620. [Google Scholar] [CrossRef]
- Raimundo, S.C.; Pattathil, S.; Eberhard, S.; Hahn, M.G.; Popper, Z.A. β-1,3-Glucans are components of brown seaweed (Phaeophyceae) cell walls. Protoplasma 2017, 254, 997–1016. [Google Scholar] [CrossRef]
- Salmeán, A.A.; Duffieux, D.; Harholt, J.; Qin, F.; Michel, G.; Czjzek, M.; Willats, W.G.T.; Hervé, C. Insoluble (1 → 3), (1 → 4)-β-D-glucan is a component of cell walls in brown algae (Phaeophyceae) and is masked by alginates in tissues. Sci. Rep. 2017, 7, 2880. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.; Qin, Y.; Shen, P.; Peng, Q. Structures, properties and application of alginic acid: A review. Int. J. Biol. Macromol. 2020, 162, 618–628. [Google Scholar] [CrossRef]
- Gheorghita Puscaselu, R.; Lobiuc, A.; Dimian, M.; Covasa, M. Alginate: From food industry to biomedical applications and management of metabolic disorders. Polymers 2020, 12, 2417. [Google Scholar] [CrossRef]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef]
- Deniaud-Bouët, E.; Hardouin, K.; Potin, P.; Kloareg, B.; Hervé, C. A review about brown algal cell walls and fucose-containing sulfated polysaccharides: Cell wall context, biomedical properties and key research challenges. Carbohydr. Polym. 2017, 175, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Deniaud-Bouët, E.; Kervarec, N.; Michel, G.; Tonon, T.; Kloareg, B.; Hervé, C. Chemical and enzymatic fractionation of cell walls from Fucales: Insights into the structure of the extracellular matrix of brown algae. Ann. Bot. 2014, 114, 1203–1216. [Google Scholar] [CrossRef] [PubMed]
- Usov, A.I.; Bilan, M.I.; Ustyuzhanina, N.E.; Nifantiev, N.E. Fucoidans of Brown Algae: Comparison of Sulfated Polysaccharides from Fucus vesiculosus and Ascophyllum nodosum. Mar. Drugs 2022, 20, 638. [Google Scholar] [CrossRef] [PubMed]
- Benslima, A.; Sellimi, S.; Hamdi, M.; Nasri, R.; Jridi, M.; Cot, D.; Li, S.; Nasri, M.; Zouari, N. Brown seaweed Cystoseira schiffneri as a promising source of sulfated fucans: Seasonal variability of structural, chemical, and antioxidant properties. Food Sci. Nutr. 2021, 9, 1551–1563. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, K.; Manivasagan, P.; Venkatesan, J.; Kim, S.-K. Brown seaweed fucoidan: Biological activity and apoptosis, growth signaling mechanism in cancer. Int. J. Biol. Macromol. 2013, 60, 366–374. [Google Scholar] [CrossRef]
- Chizhov, A.O.; Dell, A.; Morris, H.R.; Reason, A.J.; Haslam, S.M.; McDowell, R.A.; Chizhov, O.S.; Usov, A.I. Structural analysis of laminarans by MALDI and FAB mass spectrometry. Carbohydr. Res. 1998, 310, 203–210. [Google Scholar] [CrossRef]
- Nelson, T.E.; Lewis, B.A. Separation and characterization of the soluble and insoluble components of insoluble laminaran. Carbohydr. Res. 1974, 33, 63–74. [Google Scholar] [CrossRef]
- Carpita, N.C.; Shea, E.M. Linkage structure of carbohydrates by gas chromatography-mass spectrometry (GC-MS) of partially methylated alditol acetates. In Analysis of Carbohydrates by GLC and MS; Biermann, C.J., McGinnis, G.D., Eds.; CRC Press, Inc.: Boca Raton, FL, USA, 1989; pp. 157–216. [Google Scholar]
- Jones, D.R.; Xing, X.; Tingley, J.P.; Klassen, L.; King, M.L.; Alexander, T.W.; Abbott, D.W. Analysis of Active Site Architecture and Reaction Product Linkage Chemistry Reveals a Conserved Cleavage Substrate for an Endo-alpha-mannanase within Diverse Yeast Mannans. J. Mol. Biol. 2020, 432, 1083–1097. [Google Scholar] [CrossRef]
- Pettolino, F.A.; Walsh, C.; Fincher, G.B.; Bacic, A. Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 2012, 7, 1590–1607. [Google Scholar] [CrossRef]
- Bajwa, B.; Xing, X.; Terry, S.A.; Gruninger, R.J.; Abbott, D.W. Methylation-GC-MS/FID-based glycosidic linkage analysis of unfractionated polysaccharides in red seaweeds. Mar. Drugs 2024, 22, 192. [Google Scholar] [CrossRef]
- Tingley, J.P.; Low, K.E.; Xing, X.; Abbott, D.W. Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues. Biotechnol. Biofuels 2021, 14, 16. [Google Scholar]
- Wood, J.A.; Tan, H.-T.; Collins, H.M.; Yap, K.; Khor, S.F.; Lim, W.L.; Xing, X.; Bulone, V.; Burton, R.A.; Fincher, G.B.; et al. Genetic and environmental factors contribute to variation in cell wall composition in mature desi chickpea (Cicer arietinum L.) cotyledons. Plant Cell Environ. 2018, 41, 2195–2208. [Google Scholar] [PubMed]
- Li, J.; Hsiung, S.-Y.; Kao, M.-R.; Xing, X.; Chang, S.-C.; Wang, D.; Hsieh, P.-Y.; Liang, P.-H.; Zhu, Z.; Cheng, T.-J.R.; et al. Structural compositions and biological activities of cell wall polysaccharides in the rhizome, stem, and leaf of Polygonatum odoratum (Mill.) Druce. Carbohydr. Res. 2022, 521, 108662. [Google Scholar] [CrossRef] [PubMed]
- Badhan, A.; Low, K.E.; Jones, D.R.; Xing, X.; Milani, M.R.M.; Polo, R.O.; Klassen, L.; Venketachalam, S.; Hahn, M.G.; Abbott, D.W.; et al. Mechanistic insights into the digestion of complex dietary fibre by the rumen microbiota using combinatorial high-resolution glycomics and transcriptomic analyses. Comput. Struct. Biotechnol. J. 2022, 20, 148–164. [Google Scholar] [CrossRef] [PubMed]
- Low, K.E.; Xing, X.; Moote, P.E.; Inglis, G.D.; Venketachalam, S.; Hahn, M.G.; King, M.L.; Tétard-Jones, C.Y.; Jones, D.R.; Willats, W.G.T.; et al. Combinatorial Glycomic Analyses to Direct CAZyme Discovery for the Tailored Degradation of Canola Meal Non-Starch Dietary Polysaccharides. Microorganisms 2020, 8, 1888. [Google Scholar] [CrossRef]
- Li, J.; Wang, D.; Xing, X.; Cheng, T.-J.R.; Liang, P.-H.; Bulone, V.; Park, J.H.; Hsieh, Y.S.Y. Structural analysis and biological activity of cell wall polysaccharides extracted from Panax ginseng marc. Int. J. Biol. Macromol. 2019, 135, 29–37. [Google Scholar] [CrossRef]
- Pham, T.A.T.; Kyriacou, B.A.; Schwerdt, J.G.; Shirley, N.J.; Xing, X.; Bulone, V.; Little, A. Composition and biosynthetic machinery of the Blumeria graminis f. sp. hordei conidia cell wall. Cell Surf. 2019, 5, 100029. [Google Scholar] [CrossRef]
- Pham, T.A.T.; Schwerdt, J.G.; Shirley, N.J.; Xing, X.; Hsieh, Y.S.Y.; Srivastava, V.; Bulone, V.; Little, A. Analysis of cell wall synthesis and metabolism during early germination of Blumeria graminis f. sp. hordei conidial cells induced in vitro. Cell Surf. 2019, 5, 100030. [Google Scholar] [CrossRef]
- Stevenson, T.T.; Furneaux, R.H. Chemical methods for the analysis of sulphated galactans from red algae. Carbohydr. Res. 1991, 210, 277–298. [Google Scholar] [CrossRef]
- Robb, C.S.; Hobbs, J.K.; Pluvinage, B.; Reintjes, G.; Klassen, L.; Monteith, S.; Giljan, G.; Amundsen, C.; Vickers, C.; Hettle, A.G.; et al. Metabolism of a hybrid algal galactan by members of the human gut microbiome. Nat. Chem. Biol. 2022, 18, 501–510. [Google Scholar] [CrossRef]
- Needs, P.W.; Selvendran, R.R. An improved methylation procedure for the analysis of complex polysaccharides including resistant starch and a critique of the factors which lead to undermethylation. Phytochem. Anal. 1993, 4, 210–216. [Google Scholar] [CrossRef]
- Kim, J.-B.; Carpita, N.C. Changes in esterification of the uronic acid groups of cell wall polysaccharides during elongation of maize coleoptiles 1. Plant Physiol. 1992, 98, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.H.; Cleary, M.T.; Dokoozlian, N.; Ford, C.M.; Fincher, G.B. Soluble cell wall carbohydrates and their relationship with sensory attributes in Cabernet Sauvignon wine. Food Chem. 2019, 298, 124745. [Google Scholar] [CrossRef] [PubMed]
- Muhidinov, Z.K.; Bobokalonov, J.T.; Ismoilov, I.B.; Strahan, G.D.; Chau, H.K.; Hotchkiss, A.T.; Liu, L. Characterization of two types of polysaccharides from Eremurus hissaricus roots growing in Tajikistan. Food Hydrocoll. 2020, 105, 105768. [Google Scholar] [CrossRef]
- Hosain, N.A.; Ghosh, R.; Bryant, D.L.; Arivett, B.A.; Farone, A.L.; Kline, P.C. Isolation, structure elucidation, and immunostimulatory activity of polysaccharide fractions from Boswellia carterii frankincense resin. Int. J. Biol. Macromol. 2019, 133, 76–85. [Google Scholar] [CrossRef]
- Schiano di Visconte, G.; Allen, M.J.; Spicer, A. Novel capsular polysaccharide from Lobochlamys segnis. Polysaccharides 2021, 2, 121–137. [Google Scholar] [CrossRef]
- Kelly, S.J.; Muszyński, A.; Kawaharada, Y.; Hubber, A.M.; Sullivan, J.T.; Sandal, N.; Carlson, R.W.; Stougaard, J.; Ronson, C.W. Conditional requirement for exopolysaccharide in the Mesorhizobium–Lotus symbiosis. Mol. Plant-Microbe Interact. 2013, 26, 319–329. [Google Scholar] [CrossRef]
- Muhidinov, Z.K.; Rahmonov, M.H.; Jonmurodov, A.; Bobokalonov, J.; Liu, L.S.; Strahan, G. Structural analyses of apricot pectin polysaccharides by NMR spectroscopy. Preprints 2023, 2023120151. [Google Scholar] [CrossRef]
- Graiff, A.; Ruth, W.; Kragl, U.; Karsten, U. Chemical characterization and quantification of the brown algal storage compound laminarin—A new methodological approach. J. Appl. Phycol. 2016, 28, 533–543. [Google Scholar] [CrossRef]
- Craigie, J.S.; Morris, E.R.; Rees, D.A.; Thom, D. Alginate block structure in phaeophyceae from Nova Scotia: Variation with species, environment and tissue-type. Carbohydr. Polym. 1984, 4, 237–252. [Google Scholar] [CrossRef]
- Rani, V. Influence of species, geographic location, seasonal variation and extraction method on the fucoidan yield of the brown seaweeds of Gulf of Mannar, India. Indian J. Pharm. Sci. 2017, 79, 65–71. [Google Scholar] [CrossRef]
- Bruhn, A.; Janicek, T.; Manns, D.; Nielsen, M.M.; Balsby, T.J.S.; Meyer, A.S.; Rasmussen, M.B.; Hou, X.; Saake, B.; Göke, C.; et al. Crude fucoidan content in two North Atlantic kelp species, Saccharina latissima and Laminaria digitata—Seasonal variation and impact of environmental factors. J. Appl. Phycol. 2017, 29, 3121–3137. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sathuvan, M.; Zhang, X.; Zhang, W.; Tang, S.; Liu, Y.; Cheong, K.-L. Characterization of polysaccharides from different species of brown seaweed using saccharide mapping and chromatographic analysis. BMC Chem. 2021, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, H.R.; Biller, P.; Ross, A.B.; Adams, J.M.M. The seasonal variation of fucoidan within three species of brown macroalgae. Algal Res. 2017, 22, 79–86. [Google Scholar] [CrossRef]
- Chandía, N.P.; Matsuhiro, B.; Mejías, E.; Moenne, A. Alginic acids in Lessonia vadosa: Partial hydrolysis and elicitor properties of the polymannuronic acid fraction. J. Appl. Phycol. 2004, 16, 127–133. [Google Scholar] [CrossRef]
- Kelly, B.J.; Brown, M.T. Variations in the alginate content and composition of Durvillaea antarctica and D. willana from southern New Zealand. J. Appl. Phycol. 2000, 12, 317–324. [Google Scholar] [CrossRef]
- Skriptsova, A.V. Seasonal variations in the fucoidan content of brown algae from Peter the Great Bay, Sea of Japan. Russ. J. Mar. Biol. 2016, 42, 351–356. [Google Scholar] [CrossRef]
- Qari, R.; Siyal, F.; Razak, M. A comparative study on the seasonal variation in alginic acid extracted from two brown seaweed species P. pavonica Linnaeus and P. tetrastromatica Hauck. Acta Sci. Microbiol. 2019, 2, 90–99. [Google Scholar] [CrossRef]
- Jönsson, M.; Allahgholi, L.; Sardari, R.R.R.; Hreggviðsson, G.O.; Nordberg Karlsson, E. Extraction and modification of macroalgal polysaccharides for current and next-generation applications. Molecules 2020, 25, 930. [Google Scholar] [CrossRef]
- Xiao, H.-W.; Pan, Z.; Deng, L.-Z.; El-Mashad, H.M.; Yang, X.-H.; Mujumdar, A.S.; Gao, Z.-J.; Zhang, Q. Recent developments and trends in thermal blanching—A comprehensive review. Inf. Process. Agric. 2017, 4, 101–127. [Google Scholar] [CrossRef]
- Zhu, X.; Healy, L.E.; Sevindik, O.; Sun, D.-W.; Selli, S.; Kelebek, H.; Tiwari, B.K. Impacts of novel blanching treatments combined with commercial drying methods on the physicochemical properties of Irish brown seaweed Alaria esculenta. Food Chem. 2022, 369, 130949. [Google Scholar] [CrossRef] [PubMed]
- Susanto, E.; Fahmi, A.S.; Agustini, T.W.; Rosyadi, S.; Wardani, A.D. Effects of different heat processing on fucoxanthin, antioxidant activity and colour of Indonesian brown seaweeds. IOP Conf. Ser. Earth Environ. Sci. 2017, 55, 012063. [Google Scholar] [CrossRef]
- Cox, S.; Gupta, S.; Abu-Ghannam, N. Effect of different rehydration temperatures on the moisture, content of phenolic compounds, antioxidant capacity and textural properties of edible Irish brown seaweed. LWT 2012, 47, 300–307. [Google Scholar] [CrossRef]
- Akomea-Frempong, S.; Perry, J.J.; Skonberg, D.I. Effects of pre-freezing blanching procedures on the physicochemical properties and microbial quality of frozen sugar kelp. J. Appl. Phycol. 2022, 34, 609–624. [Google Scholar] [CrossRef]
- Liu, P.; Hu, J.; Wang, Q.; Tan, J.; Wei, J.; Yang, H.; Tang, S.; Huang, H.; Zou, Y.; Huang, Z. Physicochemical characterization and cosmetic application of kelp blanching water polysaccharides. Int. J. Biol. Macromol. 2023, 248, 125981. [Google Scholar] [CrossRef]
- Nielsen, C.W.; Holdt, S.L.; Sloth, J.J.; Marinho, G.S.; Sæther, M.; Funderud, J.; Rustad, T. Reducing the High Iodine Content of Saccharina latissima and Improving the Profile of Other Valuable Compounds by Water Blanching. Foods 2020, 9, 569. [Google Scholar] [CrossRef]
- Stévant, P.; Marfaing, H.; Duinker, A.; Fleurence, J.; Rustad, T.; Sandbakken, I.; Chapman, A. Biomass soaking treatments to reduce potentially undesirable compounds in the edible seaweeds sugar kelp (Saccharina latissima) and winged kelp (Alaria esculenta) and health risk estimation for human consumption. J. Appl. Phycol. 2018, 30, 2047–2060. [Google Scholar] [CrossRef]
- Ragaza, A.R.; Hurtado, A. Sargassum Studies in Currimao, Ilocos Norte, Northern Philippines II. Seasonal Variations in Alginate Yield and Viscosity of Sargassum carpophyllum J. Agardh, Sargassum ilicifolium (Turner) C. Agardh and Sargassum siliquosum J. Agardh (Phaeophyta, Sargassaceae). Bot. Mar.-BOT MAR 1999, 42, 327–331. [Google Scholar]
- Rodríguez-Montesinos, Y.E.; Arvizu-Higuera, D.L.; Hernández-Carmona, G. Seasonal variation on size and chemical constituents of Sargassum sinicola Setchell et Gardner from Bahía de La Paz, Baja California Sur, Mexico. Phycol. Res. 2008, 56, 33–38. [Google Scholar] [CrossRef]
- Rioux, L.E.; Turgeon, S.L.; Beaulieu, M. Characterization of polysaccharides extracted from brown seaweeds. Carbohydr. Polym. 2007, 69, 530–537. [Google Scholar] [CrossRef]
- García-Ríos, V.; Ríos-Leal, E.; Robledo, D.; Freile-Pelegrin, Y. Polysaccharides composition from tropical brown seaweeds. Phycol. Res. 2012, 60, 305–315. [Google Scholar] [CrossRef]
- Karmakar, P.; Ghosh, T.; Sinha, S.; Saha, S.; Mandal, P.; Ghosal, P.K.; Ray, B. Polysaccharides from the brown seaweed Padina tetrastromatica: Characterization of a sulfated fucan. Carbohydr. Polym. 2009, 78, 416–421. [Google Scholar] [CrossRef]
- Pomin, V.H.; Valente, A.P.; Pereira, M.S.; Mourão, P.A.S. Mild acid hydrolysis of sulfated fucans: A selective 2-desulfation reaction and an alternative approach for preparing tailored sulfated oligosaccharides. Glycobiology 2005, 15, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Takano, R. Desulfation of sulfated polysaccharides. Trends Glycosci. Glycotechnol. 2002, 14, 343–351. [Google Scholar] [CrossRef]
- Lechner, J.; Wieland, F.; Sumper, M. Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J. Biol. Chem. 1985, 260, 860–866. [Google Scholar] [CrossRef]
- Li, B.; Wei, X.J.; Sun, J.L.; Xu, S.Y. Structural investigation of a fucoidan containing a fucose-free core from the brown seaweed, Hizikia fusiforme. Carbohydr. Res. 2006, 341, 1135–1146. [Google Scholar] [CrossRef]
- Duarte, M.E.R.; Cardoso, M.A.; Noseda, M.D.; Cerezo, A.S. Structural studies on fucoidans from the brown seaweed Sargassum stenophyllum. Carbohydr. Res. 2001, 333, 281–293. [Google Scholar] [CrossRef]
- Chizhov, A.O.; Dell, A.; Morris, H.R.; Haslam, S.M.; McDowell, R.A.; Shashkov, A.S.; Nifant’ev, N.E.; Khatuntseva, E.A.; Usov, A.I. A study of fucoidan from the brown seaweed Chorda filum. Carbohydr. Res. 1999, 320, 108–119. [Google Scholar] [CrossRef]
- Mateos-Aparicio, I.; Martera, G.; Goñi, I.; Villanueva-Suárez, M.-J.; Redondo-Cuenca, A. Chemical structure and molecular weight influence the in vitro fermentability of polysaccharide extracts from the edible seaweeds Himathalia elongata and Gigartina pistillata. Food Hydrocoll. 2018, 83, 348–354. [Google Scholar] [CrossRef]
- Bilan, M.I.; Grachev, A.A.; Shashkov, A.S.; Nifantiev, N.E.; Usov, A.I. Structure of a fucoidan from the brown seaweed Fucus serratus L. Carbohydr. Res. 2006, 341, 238–245. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Y.; Lai, Z.; Hu, X.; Wang, L.; Wang, X.; Li, Z.; Gao, M.; Yang, Y.; Wang, Q.; et al. Effect of monosaccharide composition and proportion on the bioactivity of polysaccharides: A review. Int. J. Biol. Macromol. 2024, 254, 127955. [Google Scholar] [CrossRef] [PubMed]
- Conchie, J.; Percival, E.G.V. 167. Fucoidin. Part II. The hydrolysis of a methylated fucoidin prepared from Fucus vesiculosus. J. Chem. Soc. (Resumed) 1950, 167, 827–832. [Google Scholar] [CrossRef]
- Chevolot, L.; Mulloy, B.; Ratiskol, J.; Foucault, A.; Colliec-Jouault, S. A disaccharide repeat unit is the major structure in fucoidans from two species of brown algae. Carbohydr. Res. 2001, 330, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.U.; Nagahawatta, D.P.; Fernando, I.P.S.; Kim, Y.T.; Kim, J.S.; Kim, W.S.; Lee, J.S.; Jeon, Y.J. A Review on Fucoidan Structure, Extraction Techniques, and Its Role as an Immunomodulatory Agent. Mar. Drugs 2022, 20, 755. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.N.G.; Rosenau, T.; Böhmdorfer, S. The structure of fucoidan by linkage analysis tailored for fucose in four algae species: Fucus serratus, Fucus evanescens, Fucus vesiculosus and Laminaria hyperborea. Carbohydr. Polym. Technol. Appl. 2024, 7, 100455. [Google Scholar] [CrossRef]
- Jia, R.-B.; Yang, G.; Lai, H.; Zheng, Q.; Xia, W.; Zhao, M. Structural characterization and human gut microbiota fermentation in vitro of a polysaccharide from Fucus vesiculosus. Int. J. Biol. Macromol. 2024, 275, 133369. [Google Scholar] [CrossRef]
- Ptak, S.H.; Hjuler, A.L.; Ditlevsen, S.I.; Fretté, X.; Errico, M.; Christensen, K.V. The effect of seasonality and geographic location on sulphated polysaccharides from brown algae. Aquac. Res. 2021, 52, 6235–6243. [Google Scholar] [CrossRef]
- Usoltseva, R.V.; Anastyuk, S.D.; Shevchenko, N.M.; Zvyagintseva, T.N.; Ermakova, S.P. The comparison of structure and anticancer activity in vitro of polysaccharides from brown algae Alaria marginata and A. angusta. Carbohydr. Polym. 2016, 153, 258–265. [Google Scholar] [CrossRef]
- Bilan, M.I.; Klochkova, N.G.; Shashkov, A.S.; Usov, A.I. Polysaccharides of Algae 71*. Polysaccharides of the Pacific brown alga Alaria marginata. Russ. Chem. Bull. 2018, 67, 137–143. [Google Scholar] [CrossRef]
- Ponce, N.M.A.; Pujol, C.A.; Damonte, E.B.; Flores, M.A.L.; Stortz, C.A. Fucoidans from the brown seaweed Adenocystis utricularis: Extraction methods, antiviral activity and structural studies. Carbohydr. Res. 2003, 338, 153–165. [Google Scholar] [CrossRef]
- Lim, S.J.; Wan Aida, W.M.; Schiehser, S.; Rosenau, T.; Böhmdorfer, S. Structural elucidation of fucoidan from Cladosiphon okamuranus (Okinawa mozuku). Food Chem. 2019, 272, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, F.; Jia, G.; Zhang, W.; Wu, F. Extraction optimization and analysis of monosaccharide composition of fucoidan from Saccharina japonica by capillary zone electrophoresis. J. Appl. Phycol. 2013, 25, 1903–1908. [Google Scholar] [CrossRef]
- Ermakova, S.; Sokolova, R.; Kim, S.-M.; Um, B.-H.; Isakov, V.; Zvyagintseva, T. Fucoidans from Brown Seaweeds Sargassum hornery, Eclonia cava, Costaria costata: Structural Characteristics and Anticancer Activity. Appl. Biochem. Biotechnol. 2011, 164, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Dulong, V.; Rihouey, C.; Gaignard, C.; Bridiau, N.; Gourvil, P.; Laroche, C.; Pierre, G.; Varacavoudin, T.; Probert, I.; Maugard, T.; et al. Exopolysaccharide from marine microalgae belonging to the Glossomastix genus: Fragile gel behavior and suspension stability. Bioengineered 2024, 15, 2296257. [Google Scholar] [CrossRef]
- Moreira, A.S.P.; Gaspar, D.; Ferreira, S.S.; Correia, A.; Vilanova, M.; Perrineau, M.M.; Kerrison, P.D.; Gachon, C.M.M.; Domingues, M.R.; Coimbra, M.A.; et al. Water-Soluble Saccharina latissima Polysaccharides and Relation of Their Structural Characteristics with In Vitro Immunostimulatory and Hypocholesterolemic Activities. Mar. Drugs 2023, 21, 183. [Google Scholar] [CrossRef]
- Croci, D.O.; Cumashi, A.; Ushakova, N.A.; Preobrazhenskaya, M.E.; Piccoli, A.; Totani, L.; Ustyuzhanina, N.E.; Bilan, M.I.; Usov, A.I.; Grachev, A.A.; et al. Fucans, but not fucomannoglucuronans, determine the biological activities of sulfated polysaccharides from Laminaria saccharina brown seaweed. PLoS ONE 2011, 6, e17283. [Google Scholar] [CrossRef]
- Bilan, M.I.; Grachev, A.A.; Shashkov, A.S.; Kelly, M.; Sanderson, C.J.; Nifantiev, N.E.; Usov, A.I. Further studies on the composition and structure of a fucoidan preparation from the brown alga Saccharina latissima. Carbohydr. Res. 2010, 345, 2038–2047. [Google Scholar] [CrossRef]
- Ehrig, K.; Alban, S. Sulfated Galactofucan from the Brown Alga Saccharina latissima—Variability of Yield, Structural Composition and Bioactivity. Mar. Drugs 2015, 13, 76–101. [Google Scholar] [CrossRef]
- Gonzalez, S.T.; Alberto, F.; Molano, G. Whole-genome sequencing distinguishes the two most common giant kelp ecomorphs. Evolution 2023, 77, 1354–1369. [Google Scholar] [CrossRef]
- Lindstrom, S.C. A reinstated species name for north-eastern Pacific Macrocystis (Laminariaceae, Phaeophyceae). Not. Algarum 2023, 290, 1–2. [Google Scholar]
- Silva, M.M.C.L.; dos Santos Lisboa, L.; Paiva, W.S.; Batista, L.A.N.C.; Luchiari, A.C.; Rocha, H.A.O.; Camara, R.B.G. Comparison of in vitro and in vivo antioxidant activities of commercial fucoidans from Macrocystis pyrifera, Undaria pinnatifida, and Fucus vesiculosus. Int. J. Biol. Macromol. 2022, 216, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Oda, T.; Yu, Q.; Jin, J.-O. Fucoidan from Macrocystis pyrifera Has Powerful Immune-Modulatory Effects Compared to Three Other Fucoidans. Mar. Drugs 2015, 13, 1084–1104. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Yang, X.; Yuan, Y.; Jing, C.; Cao, J.; Wang, Y.; Zhang, L.; Zhang, C.; Li, Y. Purification and characterization of a fucoidan from the brown algae Macrocystis pyrifera and the activity of enhancing salt-stress tolerance of wheat seedlings. Int. J. Biol. Macromol. 2021, 180, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Zueva, A.O.; Usoltseva, R.V.; Malyarenko, O.S.; Surits, V.V.; Silchenko, A.S.; Anastyuk, S.D.; Rasin, A.B.; Khanh, H.H.N.; Thinh, P.D.; Ermakova, S.P. Structure and chemopreventive activity of fucoidans from the brown alga Alaria angusta. Int. J. Biol. Macromol. 2023, 225, 648–657. [Google Scholar] [CrossRef]
- Schiener, P.; Black, K.D.; Stanley, M.S.; Green, D.H. The seasonal variation in the chemical composition of the kelp species Laminaria digitata, Laminaria hyperborea, Saccharina latissima and Alaria esculenta. J. Appl. Phycol. 2015, 27, 363–373. [Google Scholar] [CrossRef]
- Azuma, K.; Osaki, T.; Ifuku, S.; Maeda, H.; Morimoto, M.; Takashima, O.; Tsuka, T.; Imagawa, T.; Okamoto, Y.; Saimoto, H.; et al. Suppressive effects of cellulose nanofibers—Made from adlay and seaweed—On colon inflammation in an inflammatory bowel-disease model. Bioact. Carbohydr. Diet. Fibre 2013, 2, 65–72. [Google Scholar] [CrossRef]
- Quellmalz, A.; Mihranyan, A. Citric Acid Cross-Linked Nanocellulose-Based Paper for Size-Exclusion Nanofiltration. ACS Biomater. Sci. Eng. 2015, 1, 271–276. [Google Scholar] [CrossRef]
- Doh, H.; Lee, M.H.; Whiteside, W.S. Physicochemical characteristics of cellulose nanocrystals isolated from seaweed biomass. Food Hydrocoll. 2020, 102, 105542. [Google Scholar] [CrossRef]
- Black, I.; Heiss, C.; Azadi, P. Comprehensive Monosaccharide Composition Analysis of Insoluble Polysaccharides by Permethylation to Produce Methyl Alditol Derivatives for Gas Chromatography/Mass Spectrometry. Anal. Chem. 2019, 91, 13787–13793. [Google Scholar] [CrossRef]
- Black, W.A.P. The seasonal variation in the cellulose content of the common Scottish Laminariaceae and Fucaceae. J. Mar. Biol. Assoc. UK 1950, 29, 379–387. [Google Scholar] [CrossRef]
- Ching, S.H.; Bansal, N.; Bhandari, B. Alginate gel particles–A review of production techniques and physical properties. Crit. Rev. Food Sci. Nutr. 2017, 57, 1133–1152. [Google Scholar] [CrossRef] [PubMed]
- Rinaudo, M. Seaweed Polysaccharides. Compr. Glycosci. 2007, 2, 691–735. [Google Scholar]
- Grant, G.T.; Morris, E.R.; Rees, D.A.; Smith, P.J.C.; Thom, D. Biological interactions between polysaccharides and divalent cations: The egg-box model. FEBS Lett. 1973, 32, 195–198. [Google Scholar] [CrossRef]
- Ramos, P.E.; Silva, P.; Alario, M.M.; Pastrana, L.M.; Teixeira, J.A.; Cerqueira, M.A.; Vicente, A.A. Effect of alginate molecular weight and M/G ratio in beads properties foreseeing the protection of probiotics. Food Hydrocoll. 2018, 77, 8–16. [Google Scholar] [CrossRef]
- Davis, T.A.; Llanes, F.; Volesky, B.; Diaz-Pulido, G.; McCook, L.; Mucci, A. 1H-NMR study of Na alginates extracted from Sargassum spp. in relation to metal biosorption. Appl. Biochem. Biotechnol. 2003, 110, 75–90. [Google Scholar] [CrossRef]
- Manns, D.; Nielsen, M.M.; Bruhn, A.; Saake, B.; Meyer, A.S. Compositional variations of brown seaweeds Laminaria digitata and Saccharina latissima in Danish waters. J. Appl. Phycol. 2017, 29, 1493–1506. [Google Scholar] [CrossRef]
- Manns, D.; Deutschle, A.L.; Saake, B.; Meyer, A.S. Methodology for quantitative determination of the carbohydrate composition of brown seaweeds (Laminariaceae). RSC Adv. 2014, 4, 25736–25746. [Google Scholar] [CrossRef]
- Zvyagintseva, T.N.; Shevchenko, N.M.; Nazarenko, E.L.; Gorbach, V.I.; Urvantseva, A.M.; Kiseleva, M.I.; Isakov, V.V. Water-soluble polysaccharides of some brown algae of the Russian Far-East. Structure and biological action of low-molecular mass polyuronans. J. Exp. Mar. Biol. Ecol. 2005, 320, 123–131. [Google Scholar] [CrossRef]
- Tanaka, Y.; Waldron-Edward, D.; Skoryna, S.C. Studies on inhibition of intestinal absorption of radioactive strontium. VII. Relationship of biological activity to chemical composition of alginates obtained from North American seaweeds. Can. Med. Assoc. J. 1968, 99, 169–175. [Google Scholar]
- Flórez-Fernández, N.; Domínguez, H.; Torres, M.D. Functional Features of Alginates Recovered from Himanthalia elongata Using Subcritical Water Extraction. Molecules 2021, 26, 4726. [Google Scholar] [CrossRef]
- Fawzy, M.A.; Gomaa, M.; Hifney, A.F.; Abdel-Gawad, K.M. Optimization of alginate alkaline extraction technology from Sargassum latifolium and its potential antioxidant and emulsifying properties. Carbohydr. Polym. 2017, 157, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolli, C.; Espindola, A.P.D.M.; Kleinübing, S.J.; Tasic, L.; Silva, M.G.C. Sargassum filipendula alginate from Brazil: Seasonal influence and characteristics. Carbohydr. Polym. 2014, 111, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Khajouei, R.A.; Keramat, J.; Hamdami, N.; Ursu, A.-V.; Delattre, C.; Laroche, C.; Gardarin, C.; Lecerf, D.; Desbrières, J.; Djelveh, G.; et al. Extraction and characterization of an alginate from the Iranian brown seaweed Nizimuddinia zanardini. Int. J. Biol. Macromol. 2018, 118, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Nyvall, P.; Corre, E.; Boisset, C.; Barbeyron, T.; Rousvoal, S.; Scornet, D.; Kloareg, B.; Boyen, C. Characterization of mannuronan C-5-epimerase genes from the brown alga Laminaria digitata. Plant Physiol. 2003, 133, 726–735. [Google Scholar] [CrossRef]
- Wirenfeldt, C.B.; Sørensen, J.S.; Kreissig, K.J.; Hyldig, G.; Holdt, S.L.; Hansen, L.T. Post-harvest quality changes and shelf-life determination of washed and blanched sugar kelp (Saccharina latissima). Front. Food Sci. Technol. 2022, 2, 1030229. [Google Scholar] [CrossRef]
- Sowinski, E.E.; Gilbert, S.; Lam, E.; Carpita, N.C. Linkage structure of cell-wall polysaccharides from three duckweed species. Carbohydr. Polym. 2019, 223, 115119. [Google Scholar] [CrossRef]
- Nayak, B.K.; Hazra, A. How to choose the right statistical test? Indian J. Ophthalmol. 2011, 59, 85–86. [Google Scholar] [CrossRef]
- Li, B.; Lu, F.; Wei, X.; Zhao, R. Fucoidan: Structure and bioactivity. Molecules 2008, 13, 1671–1695. [Google Scholar] [CrossRef]
- Usov, A.I. Polysaccharides of the red algae. Adv. Carbohydr. Chem. Biochem. 2011, 65, 115–217. [Google Scholar]
- Rioux, L.-E.; Turgeon, S.L.; Beaulieu, M. Structural characterization of laminaran and galactofucan extracted from the brown seaweed Saccharina longicruris. Phytochemistry 2010, 71, 1586–1595. [Google Scholar] [CrossRef]
- Krizsan, S.J.; Hayes, M.; Gröndahl, F.; Ramin, M.; O’Hara, P.; Kenny, O. Characterization and in vitro assessment of seaweed bioactives with potential to reduce methane production. Front. Anim. Sci. 2022, 3, 1062324. [Google Scholar] [CrossRef]
- Klassen, L.; Reintjes, G.; Li, M.; Jin, L.; Amundsen, C.; Xing, X.; Dridi, L.; Castagner, B.; Alexander, T.W.; Abbott, D.W. Fluorescence activated cell sorting and fermentation analysis to study rumen microbiome responses to administered live microbials and yeast cell wall derived prebiotics. Front. Microbiol. 2023, 13, 1020250. [Google Scholar] [CrossRef] [PubMed]
- Voiges, K.; Adden, R.; Rinken, M.; Mischnick, P. Critical re-investigation of the alditol acetate method for analysis of substituent distribution in methyl cellulose. Cellulose 2012, 19, 993–1004. [Google Scholar] [CrossRef]
- Yu, L.; Yakubov, G.E.; Zeng, W.; Xing, X.; Stenson, J.; Bulone, V.; Stokes, J.R. Multi-layer mucilage of Plantago ovata seeds: Rheological differences arise from variations in arabinoxylan side chains. Carbohydr. Polym. 2017, 165, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Gaphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Wickham, H. Reshaping data with the reshape package. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]



| Linkage | HE | FV | AM | SL | MT | ||
|---|---|---|---|---|---|---|---|
| Receptacle | Blade | Stipe | |||||
| t-Fucp | 0.8 ± 0.1 | 5.2 ± 0.6 | 1.7 ± 0.1 | 1.8 ± 0.7 | 1.2 ± 0.1 | 1.6 ± 0.2 | 1.2 ± 0.2 |
| 2-Fucp | 0.8 ± 0.1 | 3.5 ± 0.2 | 1.0 ± 0.1 | 0.8 ± 0.0 | 0.7 ± 0.0 | 1.1 ± 0.1 | 0.8 ± 0.2 |
| 3-Fucp | 2.1 ± 0.0 | 1.8 ± 0.1 | 2.8 ± 0.1 | 2.7 ± 0.6 | 2.7 ± 0.2 | 3.0 ± 0.5 | 2.6 ± 0.7 |
| 4-Fucp | 2.7 ± 0.0 | 3.7 ± 0.2 | 1.0 ± 0.2 | 1.6 ± 0.1 | 1.0 ± 0.1 | 1.4 ± 0.2 | 0.9 ± 0.0 |
| 2,3-Fucp | 2.9 ± 0.9 | 12.1 ± 1.5 | 1.0 ± 0.1 | 0.8 ± 0.6 | 0.7 ± 0.0 | 1.5 ± 0.0 | 0.7 ± 0.1 |
| 2,4-Fucp | 2.7 ± 0.0 | 6.6 ± 0.9 | 0.6 ± 0.1 | Trace | Trace | 0.7 ± 0.0 | Trace |
| 3,4-Fucp | 12.0 ± 0.4 | 2.2 ± 0.0 | 2.3 ± 0.2 | 2.6 ± 1.9 | 2.2 ± 0.6 | 2.5 ± 0.6 | 2.5 ± 0.2 |
| 2,3,4-Fucp | 6.9 ± 0.6 | 18.8 ± 0.8 | 1.7 ± 0.2 | 3.1 ± 0.1 | 5.8 ± 0.1 | 7.7 ± 0.1 | 4.9 ± 0.8 |
| 2-Galp | Trace | Trace | 1.0 ± 0.0 | 0.7 ± 0.1 | 0.6 ± 0.0 | 0.6 ± 0.0 | Trace |
| 3,4-Galp | Trace | Trace | 3.3 ± 0.4 | 1.9 ± 0.2 | 1.9 ± 0.1 | 2.6 ± 0.2 | 1.5 ± 0.2 |
| 3,6-Galp | 1.0 ± 0.1 | Trace | 0.6 ± 0.0 | 1.2 ± 0.5 | 0.9 ± 0.0 | 1.4 ± 0.2 | 0.6 ± 0.1 |
| 3,4,6-Galp | 0.6 ± 0.0 | Trace | 1.3 ± 0.1 | 1.8 ± 0.7 | 1.3 ± 0.2 | 1.8 ± 0.2 | 1.1 ± 0.1 |
| 2,4-Glcp + 2,4-Galp | 1.3 ± 0.3 | 0.6 ± 0.0 | 0.8 ± 0.0 | 1.8 ± 0.7 | 1.7 ± 0.2 | 1.5 ± 0.1 | 1.8 ± 0.3 |
| t-Glcp | 1.2 ± 0.1 | Trace | 1.4 ± 0.1 | Trace | Trace | Trace | Trace |
| 3-Glcp | 6.6 ± 0.8 | 3.9 ± 0.6 | 18.7 ± 0.2 | 1.2 ± 0.1 | 0.9 ± 0.2 | 1.1 ± 0.0 | 1.2 ± 0.2 |
| 4-Glcp | 13.8 ± 0.4 | 7.3 ± 1.5 | 23.3 ± 0.3 | 45.0 ± 3.4 | 41.9 ± 5.0 | 46.5 ± 1.1 | 37.6 ± 2.0 |
| 3,4-Glcp | 1.5 ± 0.0 | Trace | 0.7 ± 0.1 | 1.5 ± 0.1 | 1.4 ± 0.0 | 1.3 ± 0.1 | 1.7 ± 0.4 |
| 3,6-Glcp | 1.2 ± 0.2 | 0.6 ± 0.0 | 0.6 ± 0.1 | Trace | Trace | Trace | Trace |
| 4,6-Glcp | 1.6 ± 0.0 | Trace | Trace | 1.4 ± 0.1 | 1.3 ± 0.0 | 1.4 ± 0.0 | 1.6 ± 0.6 |
| 3,4,6-Glcp | 1.3 ± 0.3 | 0.8 ± 0.4 | 0.7 ± 0.3 | 1.2 ± 0.6 | 0.6 ± 0.0 | 1.5 ± 0.1 | 0.7 ± 0.1 |
| 2,3,4,6-Glcp | 0.7 ± 0.0 | Trace | Trace | 1.0 ± 0.9 | 0.6 ± 0.3 | Trace | 0.8 ± 0.3 |
| 2-Manp | Trace | Trace | 1.0 ± 0.1 | 1.2 ± 0.1 | 1.0 ± 0.0 | 0.9 ± 0.1 | 1.3 ± 0.5 |
| 4-Manp | Trace | 0.6 ± 0.0 | Trace | Trace | Trace | 0.7 ± 0.0 | Trace |
| 2,3-Manp | 0.8 ± 0.1 | 0.9 ± 0.1 | Trace | Trace | Trace | 0.7 ± 0.1 | Trace |
| 2,4-Manp | Trace | Trace | 0.8 ± 0.0 | 0.6 ± 0.7 | Trace | Trace | Trace |
| 2,6-Manp | Trace | Trace | Trace | Trace | Trace | 0.7 ± 0.1 | Trace |
| 2,3,6-Manp | Trace | 0.6 ± 0.0 | 0.6 ± 0.1 | 1.0 ± 0.6 | 0.6 ± 0.0 | 0.9 ± 0.0 | 0.7 ± 0.2 |
| 2,4,6-Manp | Trace | Trace | Trace | Trace | Trace | 0.9 ± 0.1 | Trace |
| 3,4,6-Manp | 1.9 ± 1.7 | Trace | Trace | Trace | Trace | Trace | Trace |
| 2,3,4,6-Manp | 1.4 ± 0.8 | 0.6 ± 0.0 | Trace | Trace | 0.8 ± 0.4 | Trace | 0.6 ± 0.4 |
| 2,4-Rhap | 1.5 ± 0.2 | 0.9 ± 0.1 | 4.3 ± 0.1 | Trace | Trace | Trace | Trace |
| t-Xylp | 1.4 ± 0.1 | 2.2 ± 0.1 | 1.4 ± 0.2 | 1.0 ± 0.1 | 0.8 ± 0.1 | 1.5 ± 0.2 | 0.9 ± 0.1 |
| 2-Xylp | 3.0 ± 0.3 | 1.6 ± 0.0 | Trace | Trace | Trace | Trace | Trace |
| 3-Xylp | 0.8 ± 0.2 | Trace | Trace | Trace | Trace | Trace | Trace |
| 4-Xylp | 1.8 ± 0.1 | 0.7 ± 0.1 | Trace | Trace | Trace | Trace | Trace |
| 3-GlcpA | Trace | 0.8 ± 0.0 | 1.8 ± 0.3 | 2.1 ± 0.1 | 0.8 ± 0.1 | 0.6 ± 0.2 | 0.8 ± 0.0 |
| 4-GlcpA | 1.4 ± 0.0 | 2.3 ± 0.4 | 2.0 ± 0.5 | 2.5 ± 0.3 | 2.3 ± 0.2 | 1.6 ± 0.1 | 2.4 ± 0.1 |
| 4-GulpA | 8.5 ± 0.1 | 10.2 ± 3.1 | 12.4 ± 0.7 | 8.1 ± 0.3 | 10.8 ± 2.9 | 5.1 ± 0.7 | 12.1 ± 2.2 |
| 4-ManpA | 8.3 ± 0.6 | 2.4 ± 0.5 | 4.1 ± 0.2 | 2.8 ± 0.1 | 8.9 ± 2.6 | 1.4 ± 0.2 | 11.9 ± 2.8 |
| AM | SL | HE | FV | MP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2021 | 2022 | 2021 | Blade | Stipe | Receptacle | ||||||||
| U | B | U | B | U | B | U | U | U | B | U | B | U | B |
| 3.0 ± 0.3 | 2.7 ± 0.7 | 4.3 ± 1.0 | 3.6 ± 0.6 | 2.8 ± 0.1 | 2.9 ± 1.2 | 1.0 ± 0.1 | 4.1 ± 0.4 | 3.6 ± 0.1 | 1.8 ± 0.4 | 1.0 ± 0.1 | 1.8 ± 1.1 | 1.2 ± 0.0 | 1.0 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajwa, B.; Xing, X.; Serin, S.C.; Hayes, M.; Terry, S.A.; Gruninger, R.J.; Abbott, D.W. Characterization of Unfractionated Polysaccharides in Brown Seaweed by Methylation-GC-MS-Based Linkage Analysis. Mar. Drugs 2024, 22, 464. https://doi.org/10.3390/md22100464
Bajwa B, Xing X, Serin SC, Hayes M, Terry SA, Gruninger RJ, Abbott DW. Characterization of Unfractionated Polysaccharides in Brown Seaweed by Methylation-GC-MS-Based Linkage Analysis. Marine Drugs. 2024; 22(10):464. https://doi.org/10.3390/md22100464
Chicago/Turabian StyleBajwa, Barinder, Xiaohui Xing, Spencer C. Serin, Maria Hayes, Stephanie A. Terry, Robert J. Gruninger, and D. Wade Abbott. 2024. "Characterization of Unfractionated Polysaccharides in Brown Seaweed by Methylation-GC-MS-Based Linkage Analysis" Marine Drugs 22, no. 10: 464. https://doi.org/10.3390/md22100464
APA StyleBajwa, B., Xing, X., Serin, S. C., Hayes, M., Terry, S. A., Gruninger, R. J., & Abbott, D. W. (2024). Characterization of Unfractionated Polysaccharides in Brown Seaweed by Methylation-GC-MS-Based Linkage Analysis. Marine Drugs, 22(10), 464. https://doi.org/10.3390/md22100464







