Venomics Reveals the Venom Complexity of Sea Anemone Heteractis magnifica
Abstract
1. Introduction
2. Results
2.1. Summary of the Transcriptome Sequencing and Assembly
2.2. Statistics of Transcriptome Annotation
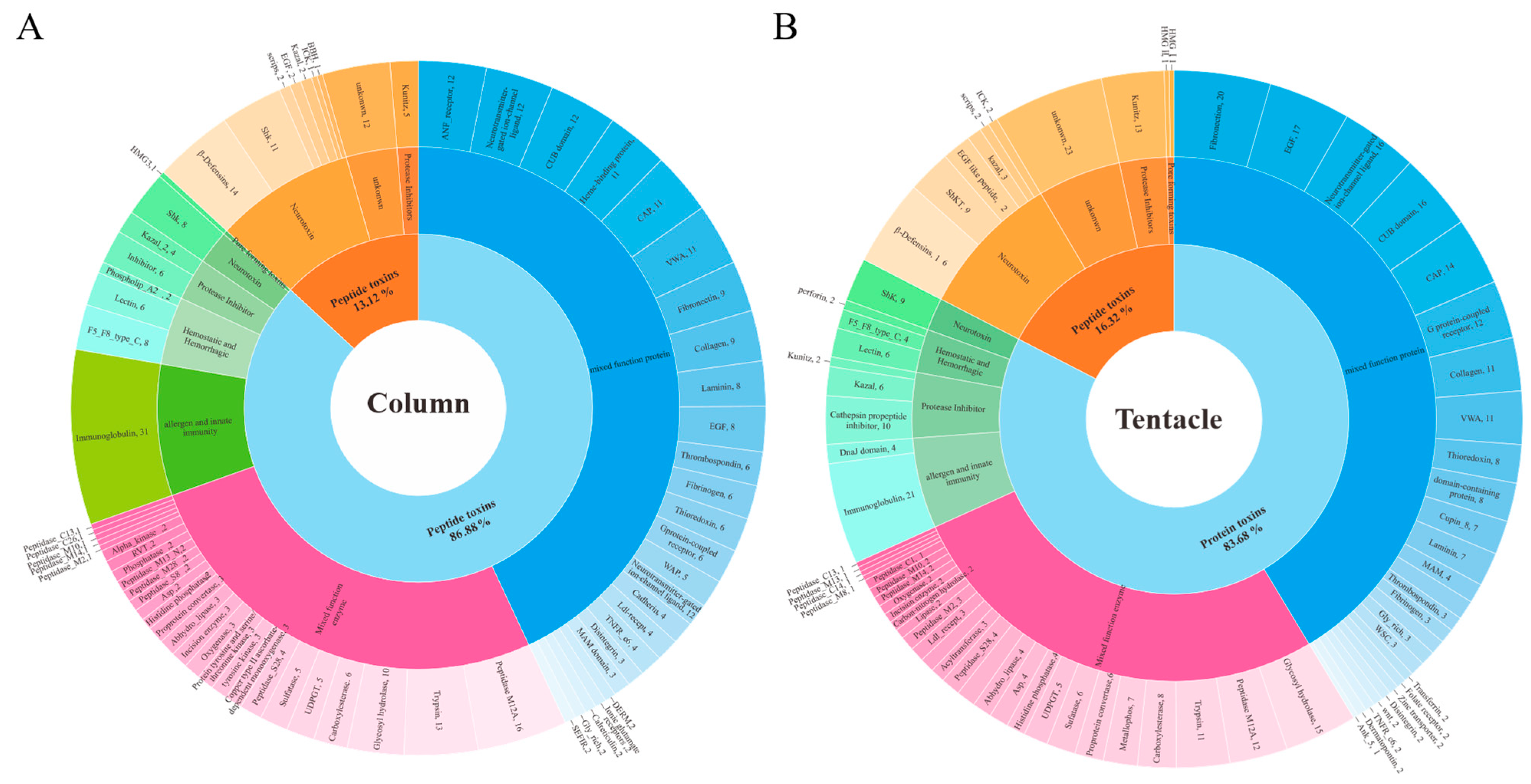
2.3. Toxins Identified in the Transcriptomes of H. magnifica
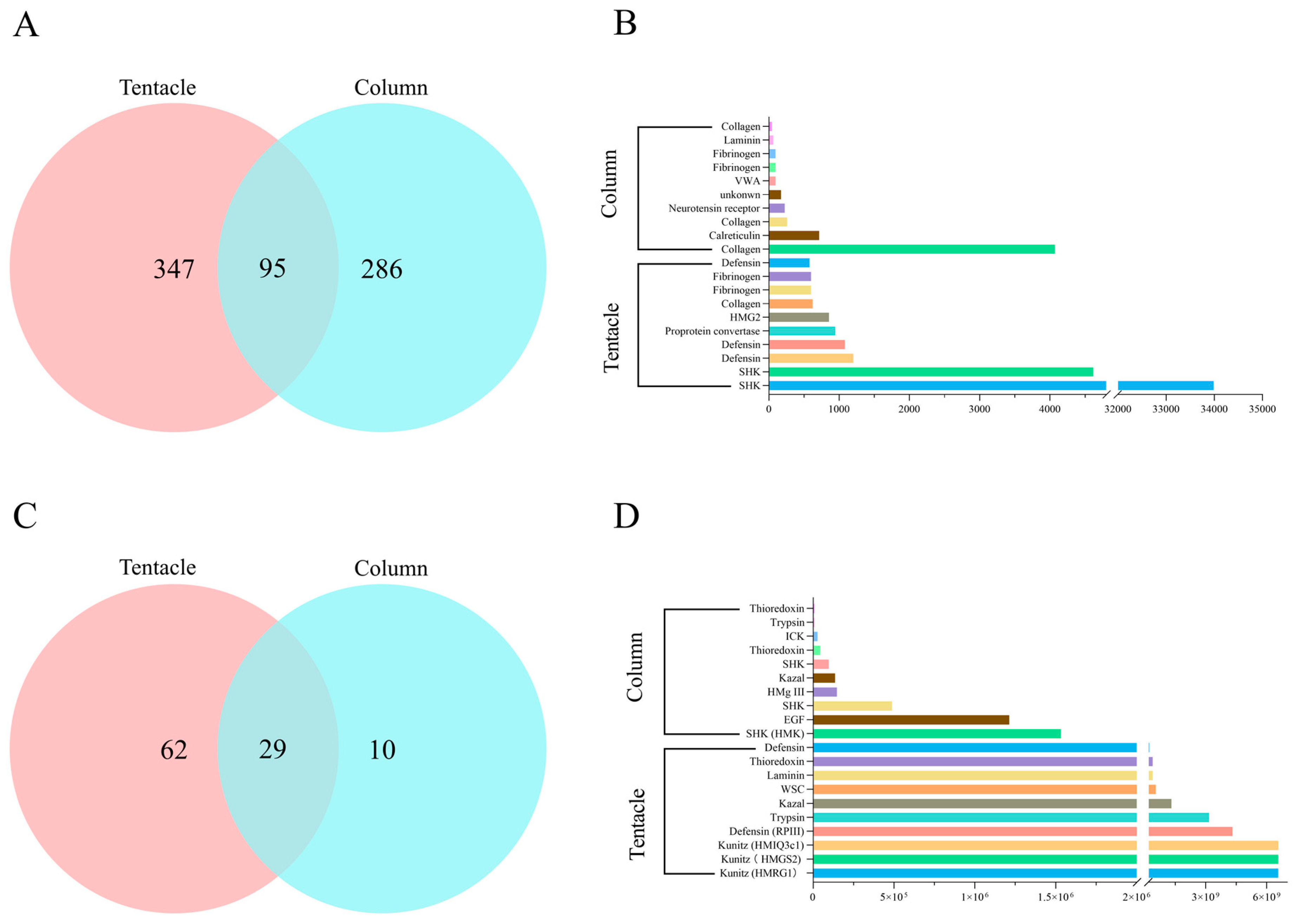
2.4. Proteomic Analysis of H. magnifica Venom Proteins and Peptides
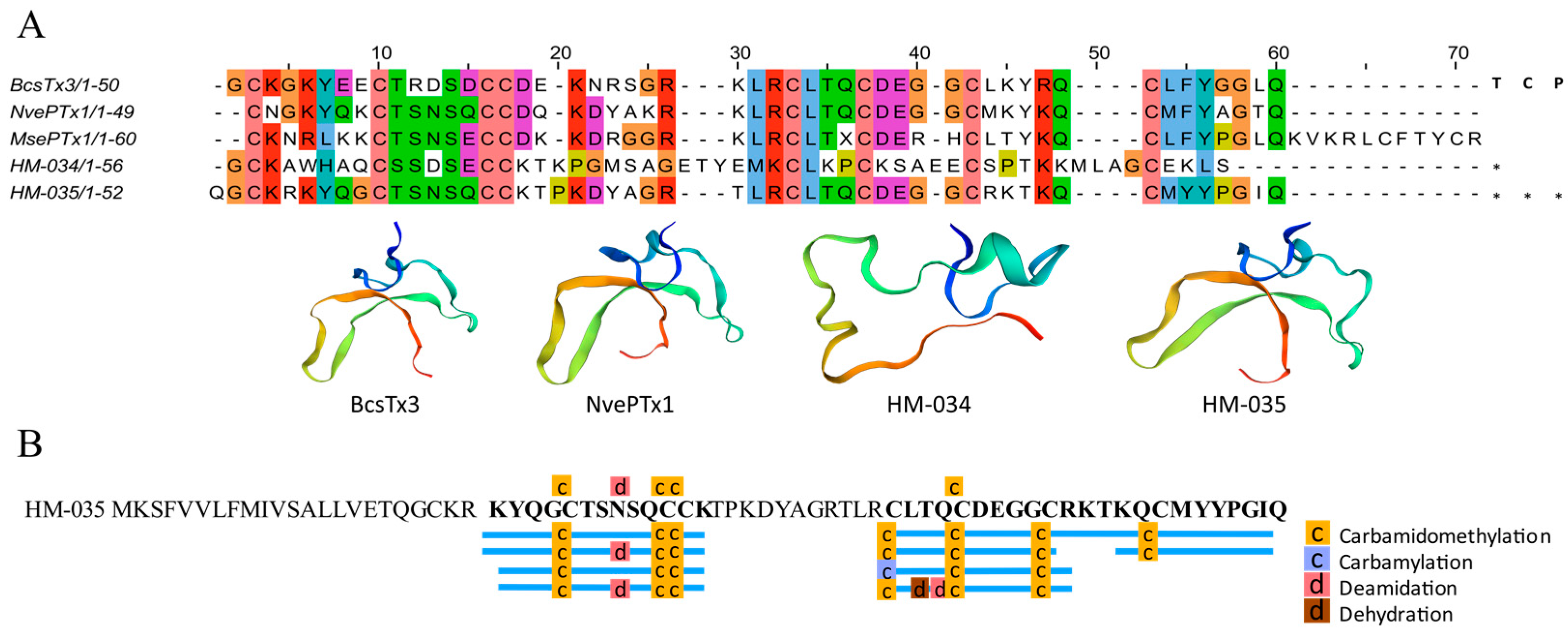
2.4.1. ShK-like Peptides
2.4.2. β-Defensin Peptides
2.4.3. Inhibitor Cystine Knot Fold Peptides
2.4.4. EGF-like Peptides
2.4.5. Kunitz-Type Peptides
2.4.6. Kazal Domain Peptides
2.4.7. Pore-Forming Toxins
3. Discussion
4. Materials and Methods
4.1. Sea Anemone Collection
4.2. Transcriptome Construction and Quality Checking
4.3. Functional Annotation of the Assembled Transcriptome
4.4. Candidate Toxin Gene Identification
4.5. Identification of Protein and Peptide Toxins
4.6. Venom Sample Preparation for the Proteomic Analysis
4.7. Tandem Mass Spectrometry (MS/MS)
4.8. Spectral Searches and Bioinformatics Analysis
4.9. Alignment and Homology Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jouiaei, M.; Yanagihara, A.A.; Madio, B.; Nevalainen, T.J.; Alewood, P.F.; Fry, B.G. Ancient venom systems: A review on Cnidaria toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef] [PubMed]
- Richet, C. De l’action de la congestine (virus des Actinies) sur les lapins et de ses effects anaphylactiques. CR Soc. Biol. Paris 1905, 58, 109–112. [Google Scholar]
- Martin, E.J. Toxicity of dialyzed extracts of some California anemones (Coelenterata). Pac. Sci. 1963, 17, 302–304. [Google Scholar]
- Martinez, G.; Kopeyan, C.; Schweitz, H.; Lazdunski, M. Toxin III from Anemonia sulcata: Primary structure. FEBS Lett. 1977, 84, 247–252. [Google Scholar] [CrossRef]
- Béress, L.; Béress, R. Reinigung zweier krabbenlähmender Toxine aus der Seeanemone Anemonia sulcata. Kiel. Meeresforsch 1971, 27, 117–127. [Google Scholar]
- Schweitz, H.; Bidard, J.N.; Frelin, C.; Pauron, D.; Vijverberg, H.P.M.; Mahasneh, D.M.; Lazdunski, M.; Vilbois, F.; Tsugita, A. Purification, sequence, and pharmacological properties of sea anemone toxins from Radianthus paumotensis. A new class of sea anemone toxins acting on the sodium channel. Biochemistry 1985, 24, 3554–3561. [Google Scholar] [CrossRef]
- Wemmer, D.E.; Kumar, N.V.; Metrione, R.M.; Lazdunski, M.; Drobny, G.; Kallenbach, N.R. NMR analysis and sequence of toxin II from the sea anemone Radianthus paumotensis. Biochemistry 1986, 25, 6842–6849. [Google Scholar] [CrossRef] [PubMed]
- Metrione, R.M.; Schweitz, H.; Walsh, K.A. The amino acid sequence of toxin RpIII from the sea anemone, Radianthus paumotensis. FEBS Lett. 1987, 218, 59–62. [Google Scholar] [CrossRef]
- Pease, J.H.; Kumar, N.V.; Schweitz, H.; Kallenbach, N.R.; Wemmer, D.E. NMR studies of toxin III from the sea anemone Radianthus paumotensis and comparison of its secondary structure with related toxins. Biochemistry 1989, 28, 2199–2204. [Google Scholar] [CrossRef]
- Kalina, R.S.; Kasheverov, I.E.; Koshelev, S.G.; Sintsova, O.; Peigneur, S.; Pinheiro-Junior, E.L.; Popov, R.S.; Chausova, V.E.; Monastyrnaya, M.M.; Dmitrenok, P.S.; et al. Nicotinic acetylcholine receptors are Novel targets of APETx-like toxins from the sea anemone Heteractis magnifica. Toxins 2022, 14, 697. [Google Scholar] [CrossRef]
- Gendeh, G.S.; Young, L.C.; de Medeiros, C.L.C.; Jeyaseelan, K.; Harvey, A.L.; Chung, M.C.M. A new potassium channel toxin from the sea anemone Heteractis magnifica: Isolation, cDNA cloning, and functional expression. Biochemistry 1997, 36, 11461–11471. [Google Scholar] [CrossRef]
- Sintsova, O.; Gladkikh, I. Peptide fingerprinting of the sea anemone Heteractis magnifica mucus revealed neurotoxins, Kunitz-type proteinase inhibitors and a new β-defensin α-amylase inhibitor. J. Proteom. 2018, 173, 12–21. [Google Scholar] [CrossRef]
- Khoo, K.S.; Kam, W.K.; Khoo, H.E.; Gopalakrishnakone, P.; Chung, M.C. Purification and partial characterization of two cytolysins from a tropical sea anemone, Heteractis magnifica. Toxicon 1993, 31, 1567–1579. [Google Scholar] [CrossRef]
- Mebs, D.; Claus, I.; Schroter, A.; Takeya, H.; Iwanga, S. Haemolytic proteins from sea anemones. Recent Adv. Toxinol. Res. 1992, 2, 392–395. [Google Scholar] [CrossRef]
- Wang, Y.; Chua, K.L.; Khoo, H.E. A new cytolysin from the sea anemone, Heteractis magnifica: Isolation, cDNA cloning and functional expression. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1478, 9–18. [Google Scholar] [CrossRef]
- Samejima, Y.; Yanagisawa, M.; Aoki-Tomomatsu, Y.; Iwasaki, E.; Ando, J.; Mebs, D. Amino acid sequence studies on cytolytic toxins from sea anemone Heteractis magnifica, Entacmaea quadricolor and Stichodactyla mertensii (Anthozoa). Toxicon 2000, 38, 259–264. [Google Scholar] [CrossRef]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.X.; He, Y.B.; Peng, C.; Tang, T.L.; Jin, A.H.; Liao, Y.L.; Shi, Q.; Gao, B.M. Transcriptome sequencing of the pale anemones (Exaiptasia diaphana) revealed functional peptide gene resources of sea anemone. Front. Mar. Sci. 2022, 9, 15. [Google Scholar] [CrossRef]
- Macrander, J.; Broe, M.; Daly, M. Tissue-specific venom composition and differential gene expression in sea anemones. Genome Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef]
- Delgado, A.; Benedict, C.; Macrander, J.; Daly, M. Never, nver make an enemy horizontal ellipsis Out of an Anemone: Transcriptomic comparison of Clownfish hosting sea anemone venoms. Mar. Drugs 2022, 20, 730. [Google Scholar] [CrossRef]
- Klompen, A.M.; Macrander, J.; Reitzel, A.M.; Stampar, S.N. Transcriptomic analysis of four cerianthid (Cnidaria, Ceriantharia) venoms. Mar. Drugs 2020, 18, 413. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Pérez, D.; Campos, A.; Alexei Rodríguez, A.; Turkina, M.V.; Ribeiro, T.; Osorio, H.; Vasconcelos, V.; Antunes, A. Proteomic analyses of the unexplored sea anemone Bunodactis verrucosa. Mar. Drugs 2018, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Levitan, S.; Sher, N.; Brekhman, V.; Ziv, T.; Lubzens, E.; Lotan, T. The making of an embryo in a basal metazoan: Proteomic analysis in the sea anemone Nematostella vectensis. Proteomics 2015, 15, 4096–4104. [Google Scholar] [CrossRef]
- Mazzi Esquinca, M.E.; Correa, C.N.; Marques de Barros, G.; Montenegro, H.; Mantovani de Castro, L. Multiomic approach for bioprospection: Investigation of toxins and peptides of Brazilian sea anemone Bunodosoma caissarum. Mar. Drugs 2023, 21, 197. [Google Scholar] [CrossRef]
- Madio, B.; Undheim, E.A.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Vera-Estrella, R.; Portillo-Bobadilla, T.; Licea-Navarro, A.; Bernaldez-Sarabia, J.; Rudiño-Piñera, E.; Verleyen, J.J.; Rodríguez, E.; Rodríguez-Almazán, C. Transcriptomic and proteomic analysis of the tentacles and mucus of Anthopleura dowii Verrill, 1869. Mar. Drugs 2019, 17, 436. [Google Scholar] [CrossRef]
- King, G.F.; Hardy, M.C. Spider-venom peptides: Structure, pharmacology, and potential for control of insect pests. Annu. Rev. Entomol. 2013, 58, 475–496. [Google Scholar] [CrossRef]
- Guiguet, A.; Tooker, J.F.; Deans, A.R.; Mikó, I.; Ning, G.; Schwéger, S.; Hines, H.M. Comparative anatomy of venom glands suggests a role of maternal secretions in gall induction by cynipid wasps (Hymenoptera: Cynipidae). Insect Syst. Divers. 2023, 7, 3. [Google Scholar] [CrossRef]
- Ashwood, L.M.; Undheim, E.A.; Madio, B.; Hamilton, B.R.; Daly, M.; Hurwood, D.A.; King, G.F.; Prentis, P.J. Venoms for all occasions: The functional toxin profiles of different anatomical regions in sea anemones are related to their ecological function. Mol. Ecol. 2022, 31, 866–883. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, L.M.; Mitchell, M.L.; Madio, B.; Hurwood, D.A.; King, G.F.; Undheim, E.A.; Norton, R.S.; Prentis, P.J. Tentacle morphological variation coincides with differential expression of toxins in sea anemones. Toxins 2021, 13, 452. [Google Scholar] [CrossRef] [PubMed]
- Fautin, D.G. Structural diversity, systematics, and evolution of cnidae. Toxicon 2009, 54, 1054–1064. [Google Scholar] [CrossRef]
- Madio, B.; Peigneur, S.; Chin, Y.K.; Hamilton, B.R.; Henriques, S.T.; Smith, J.J.; Cristofori-Armstrong, B.; Dekan, Z.; Boughton, B.A.; Alewood, P.F. PHAB toxins: A unique family of predatory sea anemone toxins evolving via intra-gene concerted evolution defines a new peptide fold. Cell Mol. Life Sci. 2018, 75, 4511–4524. [Google Scholar] [CrossRef]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: An overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef]
- Columbus-Shenkar, Y.Y.; Sachkova, M.Y.; Macrander, J.; Fridrich, A.; Modepalli, V.; Reitzel, A.M.; Sunagar, K.; Moran, Y. Dynamics of venom composition across a complex life cycle. eLife 2018, 7, e35014. [Google Scholar] [CrossRef]
- Surm, J.M.; Smith, H.L.; Madio, B.; Undheim, E.A.; King, G.F.; Hamilton, B.R.; van der Burg, C.A.; Pavasovic, A.; Prentis, P.J. A process of convergent amplification and tissue-specific expression dominates the evolution of toxin and toxin-like genes in sea anemones. Mol. Ecol. 2019, 28, 2272–2289. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.G.; Surm, J.M.; Macrander, J.; Simhi, A.; Amir, G.; Sachkova, M.Y.; Lewandowska, M.; Reitzel, A.M.; Moran, Y. Micro and macroevolution of sea anemone venom phenotype. Nat. Commun. 2023, 14, 249. [Google Scholar] [CrossRef]
- Shao, P.-J.; Chiu, Y.-L.; Tsai, P.-H.; Shikina, S. Possible germline progenitor cells in extra-gonadal tissues of the sea anemone, Exaiptasia diaphana. Front. Mar. Sci. 2023, 10, 1278022. [Google Scholar] [CrossRef]
- Menezes, C.; Thakur, N.L. Sea anemone venom: Ecological interactions and bioactive potential. Toxicon 2022, 208, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Gendeh, G.S.; Chung, M.C.; Jeyaseelan, K. Genomic structure of a potassium channel toxin from Heteractis magnifica. FEBS Lett. 1997, 418, 183–188. [Google Scholar] [CrossRef]
- Santana, F.L.; Estrada, K.; Ortiz, E.; Corzo, G. Reptilian β-defensins: Expanding the repertoire of known crocodylian peptides. Peptides 2021, 136, 170473. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.M.A.; Quimbar, P.; van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Kim, C.H.; Lee, Y.J.; Go, H.J.; Oh, H.Y.; Lee, T.K.; Park, J.B.; Park, N.G. Defensin-neurotoxin dyad in a basally branching metazoan sea anemone. FEBS J. 2017, 284, 3320–3338. [Google Scholar] [CrossRef]
- Kalina, R.; Gladkikh, I.; Dmitrenok, P.; Chernikov, O.; Koshelev, S.; Kvetkina, A.; Kozlov, S.; Kozlovskaya, E.; Monastyrnaya, M. New APETx-like peptides from sea anemone Heteractis crispa modulate ASIC1a channels. Peptides 2018, 104, 41–49. [Google Scholar] [CrossRef]
- Orts, D.J.; Moran, Y.; Cologna, C.T.; Peigneur, S.; Madio, B.; Praher, D.; Quinton, L.; De Pauw, E.; Bicudo, J.E.; Tytgat, J. Bcs Tx3 is a founder of a novel sea anemone toxin family of potassium channel blocker. FEBS J. 2013, 280, 4839–4852. [Google Scholar] [CrossRef]
- Muskavitch, M.A.; Hoffmann, F.M. Homologs of vertebrate growth factors in Drosophila melanogaster and other invertebrates. Curr. Top. Dev. Biol. 1990, 24, 289–328. [Google Scholar] [CrossRef]
- Honma, T.; Nagai, H.; Nagashima, Y.; Shiomi, K. Molecular cloning of an epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Biochim Biophys. Acta 2003, 1652, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Tolle, D.P.; Barrett, A.J. Evolutionary families of peptidase inhibitors. Biochem. J. 2004, 378, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Kvetkina, A.; Leychenko, E.; Yurchenko, E.; Pislyagin, E.; Peigneur, S.; Tytgat, Y.; Isaeva, M.; Aminin, D.; Kozlovskaya, E. A new Iq-peptide of the Kunitz type from the Heteractis magnifica sea anemone exhibits neuroprotective activity in a model of Alzheimer’s disease. Russ. J. Bioorg. Chem. 2018, 44, 416–423. [Google Scholar] [CrossRef]
- Fritz, H.; Brey, B.; Béress, L. Polyvalent isoinhibitors for trypsin, chymotrypsin, plasmin and kallikreins of sea anemones (Anemonia sulcata), isolation, inhibitory behavior and amino acid composition. Hoppe-Seyler’s Z. Fur Physiol. Chem. 1972, 353, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Wunderer, G.; Kummer, K.; Fritz, H.; Béress, L.; Machleidt, W. Broad specificity inhibitors from sea anemones. In Proceedings of the Proteinase Inhibitors, Berlin, Germany, 16–20 October 1973; pp. 277–281. [Google Scholar]
- Schweitz, H.; Bruhn, T.; Guillemare, E.; Moinier, D.; Lancelin, J.-M.; Béress, L.; Lazdunski, M. Kalicludines and kaliseptine: Two different classes of sea anemone toxins for voltage-sensitive K+ channels. J. Biol. Chem. 1995, 270, 25121–25126. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, R.; Peigneur, S.; Pons, T.; Alvarez, C.; González, L.; Chávez, M.A.; Tytgat, J. The Kunitz-type protein ShPI-1 inhibits serine proteases and voltage-gated potassium channels. Toxins 2016, 8, 110. [Google Scholar] [CrossRef]
- Mishra, M. Evolutionary aspects of the structural convergence and functional diversification of Kunitz-domain inhibitors. J. Mol. Evol. 2020, 88, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Rimphanitchayakit, V.; Tassanakajon, A. Structure and function of invertebrate Kazal-type serine proteinase inhibitors. Dev. Comp. Immunol. 2010, 34, 377–386. [Google Scholar] [CrossRef]
- Tschesche, H.; Kolkenbrock, H.; Bode, W. The covalent structure of the elastase inhibitor from Anemonia sulcata-a “non-classical” Kazal-type protein. Biol. Chem. Hoppe-Seyler 1987, 368, 1297–1304. [Google Scholar] [CrossRef]
- Podobnik, M.; Anderluh, G. Pore-forming toxins in Cnidaria. Semin. Cell Dev. Biol. 2017, 72, 133–141. [Google Scholar] [CrossRef]
- Castañeda, O.; Sotolongo, V.; Amor, A.M.; Stöcklin, R.; Anderson, A.J.; Harvey, A.L.; Engström, Å.; Wernstedt, C.; Karlsson, E. Characterization of a potassium channel toxin from the Caribbean sea anemone Stichodactyla helianthus. Toxicon 1995, 33, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Chi, V.; Pennington, M.W.; Norton, R.S.; Tarcha, E.J.; Londono, L.M.; Sims-Fahey, B.; Upadhyay, S.K.; Lakey, J.T.; Iadonato, S.; Wulff, H.; et al. Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon 2012, 59, 529–546. [Google Scholar] [CrossRef]
- Wang, Z.L.; Zhang, S.Y.; Hao, S.L.; Yang, W.X. Neurotoxins and pore forming toxins in sea anemones: Potential candidates for new drug development. Histol. Histopathol. 2023, 38, 9–28. [Google Scholar] [CrossRef]
- Wang, X.L.; Li, G.Y.; Guo, J.K.; Zhang, Z.P.; Zhang, S.Z.; Zhu, Y.D.; Cheng, J.W.; Yu, L.; Ji, Y.H.; Tao, J. Kv1.3 channel as a key therapeutic target for neuroinflammatory diseases: State of the art and beyond. Front. Neurosci. 2020, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Chandy, K.G.; Sanches, K.; Norton, R.S. Structure of the voltage-gated potassium channel Kv1.3: Insights into the inactivated conformation and binding to therapeutic leads. Channels 2023, 17, 2253104. [Google Scholar] [CrossRef]
- Varanita, T.; Angi, B.; Scattolini, V.; Szabo, I. Kv1.3 K+ channel physiology assessed by genetic and pharmacological modulation. Physiology 2023, 38, 25–41. [Google Scholar] [CrossRef]
- Manolios, N.; Papaemmanouil, J.; Adams, D. The role of ion channels in T cell function and disease. Front. Immunol. 2023, 14, 14. [Google Scholar] [CrossRef]
- Basulto, A.; Pérez, V.M.; Noa, Y.; Varela, C.; Otero, A.J.; Pico, M.C. Immunohistochemical targeting of sea anemone cytolysins on tentacles, mesenteric filaments and isolated nematocysts of Stichodactyla helianthus. J. Exp. Zool. Part A Comp. Exp. Biol. 2006, 305, 253–258. [Google Scholar] [CrossRef]
- Suarez-Carmona, M.; Hubert, P.; Delvenne, P.; Herfs, M. Defensins:“Simple” antimicrobial peptides or broad-spectrum molecules? Cytokine Growth Factor Rev. 2015, 26, 361–370. [Google Scholar] [CrossRef]
- Shafee, T.M.; Lay, F.T.; Hulett, M.D.; Anderson, M.A. The defensins consist of two independent, convergent protein superfamilies. Mol. Biol. Evol. 2016, 33, 2345–2356. [Google Scholar] [CrossRef]
- Chagot, B.; Diochot, S.; Pimentel, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx1 from the sea anemone Anthopleura elegantissima: A new fold for an HERG toxin. Proteins Struct. Funct. Bioinform. 2005, 59, 380–386. [Google Scholar] [CrossRef]
- Chagot, B.; Escoubas, P.; Diochot, S.; Bernard, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx2, a specific peptide inhibitor of ASIC3 proton-gated channels. Protein Sci. 2005, 14, 2003–2010. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Baron, A.; Rash, L.D.; Deval, E.; Escoubas, P.; Scarzello, S.; Salinas, M.; Lazdunski, M. A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 2004, 23, 1516–1525. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Blumenthal, K.M. Site-3 sea anemone toxins: Molecular probes of gating mechanisms in voltage-dependent sodium channels. Toxicon 2007, 49, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Župunski, V.; Kordiš, D.; Gubenšek, F. Adaptive evolution in the snake venom Kunitz/BPTI protein family. FEBS Lett. 2003, 547, 131–136. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Pinheiro-Junior, E.L.; Béress, L.; Gladkikh, I.; Leychenko, E.; Undheim, E.A.B.; Peigneur, S.; Tytgat, J. AsKC11, a aunitz peptide from Anemonia sulcata, Is a novel activator of G Protein-coupled inward-rectifier potassium channels. Mar. Drugs 2022, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- Gladkikh, I.; Monastyrnaya, M.; Leychenko, E.; Zelepuga, E.; Chausova, V.; Isaeva, M.; Anastyuk, S.; Andreev, Y.; Peigneur, S.; Tytgat, J.; et al. Atypical Reactive Center Kunitz-Type Inhibitor from the Sea Anemone Heteractis crispa. Marine Drugs 2012, 10, 1545–1565. [Google Scholar] [CrossRef] [PubMed]
- Kalinovskii, A.P.; Sintsova, O.V.; Gladkikh, I.N.; Leychenko, E.V. Natural inhibitors of mammalian α-Amylases as promising drugs for the treatment of metabolic diseases. Int. J. Mol. Sci. 2023, 24, 16514. [Google Scholar] [CrossRef] [PubMed]
- Mouchbahani-Constance, S.; Sharif-Naeini, R. Proteomic and transcriptomic techniques to decipher the molecular evolution of venoms. Toxins 2021, 13, 154. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Morgenstern, D.; Reitzel, A.M.; Moran, Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016, 135, 62–72. [Google Scholar] [CrossRef] [PubMed]
- von Reumont, B.M.; Anderluh, G.; Antunes, A.; Ayvazyan, N.; Beis, D.; Caliskan, F.; Crnković, A.; Damm, M.; Dutertre, S.; Ellgaard, L. Modern venomics—Current insights, novel methods, and future perspectives in biological and applied animal venom research. GigaScience 2022, 11, 27. [Google Scholar] [CrossRef]
- Macrander, J.; Brugler, M.R.; Daly, M. A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genom. 2015, 16, 1–19. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Law, S.T.S.; Yu, Y.; Nong, W.; So, W.L.; Li, Y.; Swale, T.; Ferrier, D.E.; Qiu, J.; Qian, P.; Hui, J.H.L. The genome of the deep-sea anemone Actinernus sp. contains a mega-array of ANTP-class homeobox genes. Proc. R. Soc. B-Biol. Sci. 2023, 290, 20231563. [Google Scholar] [CrossRef]
- Liao, Q.; Gong, G.; Poon, T.C.; Ang, I.L.; Lei, K.M.; Siu, S.W.I.; Wong, C.T.T.; Rádis-Baptista, G.; Lee, S.M.-Y. Combined transcriptomic and proteomic analysis reveals a diversity of venom-related and toxin-like peptides expressed in the mat anemone Zoanthus natalensis (Cnidaria, Hexacorallia). Arch. Toxicol. 2019, 93, 1745–1767. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liao, X.; Han, T.; Su, A.; Guo, Z.; Lu, N.; He, C.; Lu, Z. Full-length transcriptome sequencing of the scleractinian Coral Montipora foliosa reveals the gene expression profile of Coral–Zooxanthellae Holobiont. Biology 2021, 10, 1274. [Google Scholar] [CrossRef]
- Jungo, F.; Bairoch, A. Tox-Prot, the toxin protein annotation program of the Swiss-Prot protein knowledgebase. Toxicon 2005, 45, 293–301. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, C.; Bell, E.W.; Zhang, Y. I-TASSER gateway: A protein structure and function prediction server powered by XSEDE. Future Gener. Comput. Syst. 2019, 99, 73–85. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Mao, K.; Huang, M.; Liao, Y.; Fu, J.; Pan, K.; Shi, Q.; Gao, B. Venomics Reveals the Venom Complexity of Sea Anemone Heteractis magnifica. Mar. Drugs 2024, 22, 71. https://doi.org/10.3390/md22020071
Li M, Mao K, Huang M, Liao Y, Fu J, Pan K, Shi Q, Gao B. Venomics Reveals the Venom Complexity of Sea Anemone Heteractis magnifica. Marine Drugs. 2024; 22(2):71. https://doi.org/10.3390/md22020071
Chicago/Turabian StyleLi, Ming, Kailin Mao, Meiling Huang, Yanling Liao, Jinxing Fu, Kun Pan, Qiong Shi, and Bingmiao Gao. 2024. "Venomics Reveals the Venom Complexity of Sea Anemone Heteractis magnifica" Marine Drugs 22, no. 2: 71. https://doi.org/10.3390/md22020071
APA StyleLi, M., Mao, K., Huang, M., Liao, Y., Fu, J., Pan, K., Shi, Q., & Gao, B. (2024). Venomics Reveals the Venom Complexity of Sea Anemone Heteractis magnifica. Marine Drugs, 22(2), 71. https://doi.org/10.3390/md22020071





