Marine Bioprospecting: Enzymes and Stress Proteins from the Sea Anemones Anthopleura dowii and Lebrunia neglecta
Abstract
:1. Introduction
2. Results
2.1. Proteomic Analysis of Crude Venom Extracts (CVEs)
2.2. Differentially Expressed Cellular Proteins
2.3. Proteins with Many Biotechnology Applications
2.3.1. Peptidases
2.3.2. Chitinases
2.3.3. Antioxidant Enzymes
2.3.4. Heat Shock Proteins (HSPs)
2.3.5. Serine Protease Inhibitors (Serpins)
3. Discussion
4. Materials and Methods
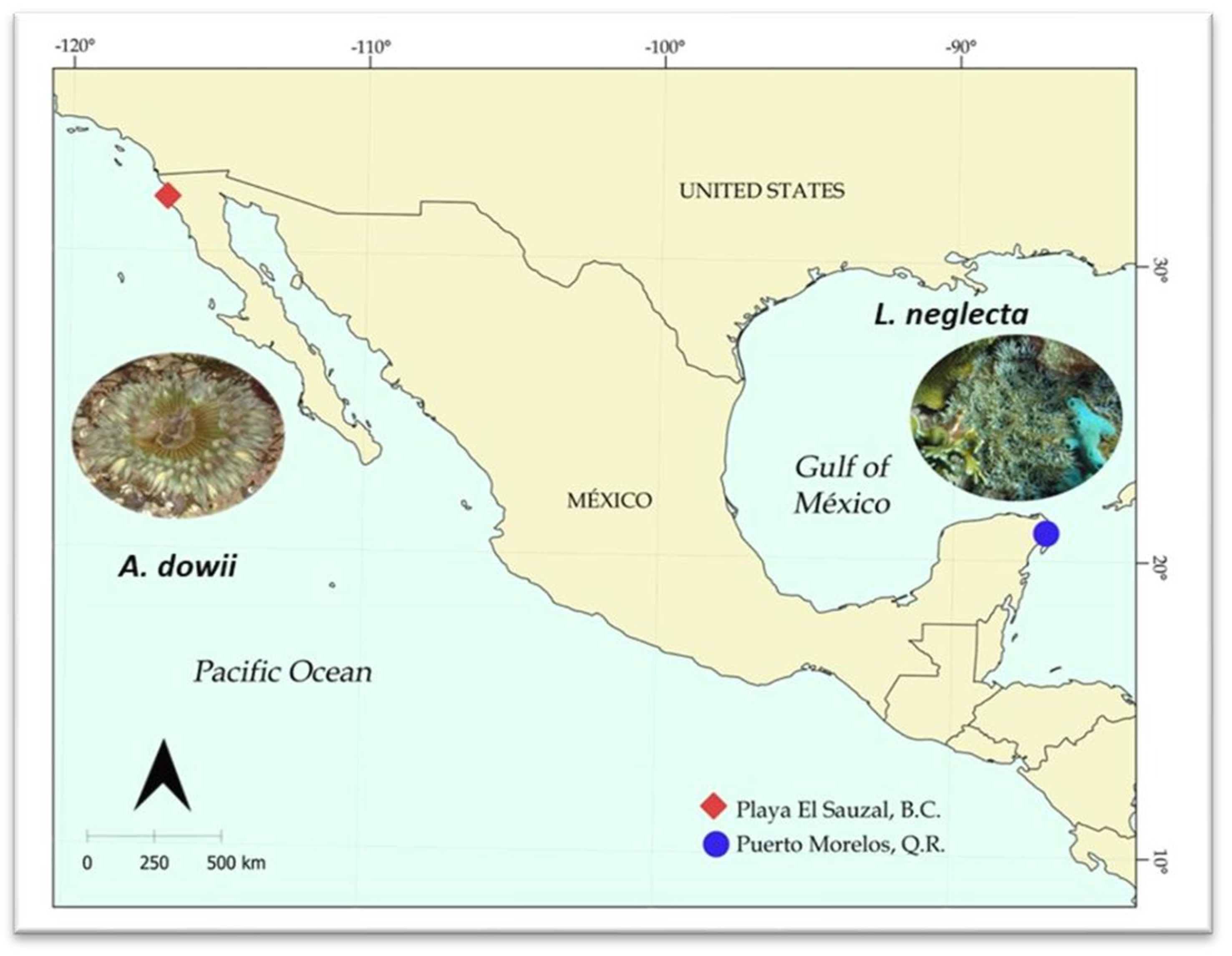
4.1. Sea Anemone Collection
4.2. Protein Extraction
4.3. Proteomic Samples
4.4. Protein Digestion
LC–MS/MS
4.5. Identification and Quantification of Proteins
4.6. Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Birolli, W.G.; Lima, R.N.; Porto, A.L.M. Applications of Marine-Derived Microorganisms and Their Enzymes in Biocatalysis and Biotransformation, the Underexplored Potentials. Front. Microbiol. 2019, 10, 1453. [Google Scholar] [CrossRef]
- Debashish, G.; Malay, S.; Barindra, S.; Joydeep, M. Marine Enzymes. In Marine Biotechnology I.; Ulber, R., Le Gal, Y., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2005; Volume 96, pp. 189–218. ISBN 978-3-540-25659-5. [Google Scholar]
- Bekiari, M. Marine Bioprospecting: Understanding the Activity and Some Challenges Related to Environmental Protection, Scientific Research, Ethics, and the Law. In Blue Planet Law: The Ecology of Our Economic and Technological World; da Gloria Garcia, M., Cortês, A., Eds.; Sustainable Development Goals Series; Springer International Publishing: Cham, Switzerland, 2023; pp. 237–252. ISBN 978-3-031-24888-7. [Google Scholar]
- Leal, M.C.; Madeira, C.; Brandão, C.A.; Puga, J.; Calado, R. Bioprospecting of Marine Invertebrates for New Natural Products-a Chemical and Zoogeographical Perspective. Molecules 2012, 17, 9842–9854. [Google Scholar] [CrossRef] [PubMed]
- Kvetkina, A.; Kostina, E.; Gladkikh, I.; Chausova, V.; Yurchenko, E.; Bakunina, I.; Pivkin, M.; Anastyuk, S.; Popov, R.; Monastyrnaya, M.; et al. Deep-Sea Anemones Are Prospective Source of New Antimicrobial and Cytotoxic Compounds. Mar. Drugs 2021, 19, 654. [Google Scholar] [CrossRef] [PubMed]
- Aertsen, A.; Meersman, F.; Hendrickx, M.E.G.; Vogel, R.F.; Michiels, C.W. Biotechnology under High Pressure: Applications and Implications. Trends Biotechnol. 2009, 27, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Avila, C.; Angulo-Preckler, C. A Minireview on Biodiscovery in Antarctic Marine Benthic Invertebrates. Front. Mar. Sci. 2021, 8, 686477. [Google Scholar] [CrossRef]
- Bellantuono, A.J.; Granados-Cifuentes, C.; Miller, D.J.; Hoegh-Guldberg, O.; Rodriguez-Lanetty, M. Coral Thermal Tolerance: Tuning Gene Expression to Resist Thermal Stress. PLoS ONE 2012, 7, e50685. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.-S.; Patel, A.K.; Singhania, R.R.; Chen, C.-W.; Dong, C.-D. Effects of Temperature and Salinity on Growth, Metabolism and Digestive Enzymes Synthesis of Goniopora columna. Biololgy 2022, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.I.; Majumdar, B.C.; Ehsan, R.; Hasan, M.; Baidya, A.; Bakky, M.A.H. Bioprospecting Potential of Marine Microbial Natural Bioactive Compounds. J. Appl. Biotechnol. Rep. 2021, 8, 96–108. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Zhang, P.; Gao, F.; Wang, J.; Lin, L.; Zhang, H. Characterization of a Novel Superoxide Dismutase from a Deep-Sea Sea Cucumber (Psychoropotes verruciaudatus). Antioxidants 2023, 12, 1227. [Google Scholar] [CrossRef]
- Fontanini, A.; Steckbauer, A.; Dupont, S.; Duarte, C.M. Variable metabolic responses of Skagerrak invertebrates to low O2 and high CO2 scenarios. Biogeosciences 2018, 15, 3717–3729. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Costa, P.M. The Hidden Biotechnological Potential of Marine Invertebrates: The Polychaeta Case Study. Environ. Res. 2019, 173, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Popkova, D.; Otstavnykh, N.; Sintsova, O.; Baldaev, S.; Kalina, R.; Gladkikh, I.; Isaeva, M.; Leychenko, E. Bioprospecting of Sea Anemones (Cnidaria, Anthozoa, Actiniaria) for β-Defensin-like α-Amylase Inhibitors. Biomedicines 2023, 11, 2682. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.G.; Turk, P.E.; Yang, W.T.; Hanlon, R.T. Biological Characteristics and Biomedical Applications of the Squid Sepioteuthis Lessoniana Cultured Through Multiple Generations. Biol. Bull. 1994, 186, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Linsmayer, L.B.; Deheyn, D.D.; Tomanek, L.; Tresguerres, M. Dynamic Regulation of Coral Energy Metabolism throughout the Diel Cycle. Sci. Rep. 2020, 10, 19881. [Google Scholar] [CrossRef] [PubMed]
- Dahlhoff, E.P.; Stillman, J.H.; Menge, B.A. Physiological Community Ecology: Variation in Metabolic Activity of Ecologically Important Rocky Intertidal Invertebrates Along Environmental Gradients. Integr. Comp. Biol. 2002, 42, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.S.; Smith, J.E.; Eynaud, Y. Comparative Metabolic Ecology of Tropical Herbivorous Echinoids on a Coral Reef. PLoS ONE 2018, 13, e0190470. [Google Scholar] [CrossRef] [PubMed]
- Sorte, C.J.B.; Jones, S.J.; Miller, L.P. Geographic Variation in Temperature Tolerance as an Indicator of Potential Population Responses to Climate Change. J. Exp. Mar. Biol. Ecol. 2011, 400, 209–217. [Google Scholar] [CrossRef]
- Kuan, P.-L.; You, J.-Y.; Wu, G.-C.; Tseng, Y.-C. Temperature Increases Induce Metabolic Adjustments in the Early Developmental Stages of Bigfin Reef Squid (Sepioteuthis Lessoniana). Sci. Total Environ. 2022, 844, 156962. [Google Scholar] [CrossRef]
- Sumi, M.S.; Thazeem, B.; Sunish, K.S. Bioprospective Studies of Pigmented Ink from Sepioteuthis lessoniana and Its Molecular Identification Using CO1 Gene. J. Basic Appl. Zool. 2023, 84, 5. [Google Scholar] [CrossRef]
- Doering, T.; Maire, J.; Chan, W.Y.; Perez-Gonzalez, A.; Meyers, L.; Sakamoto, R.; Buthgamuwa, I.; Blackall, L.L.; van Oppen, M.J.H. Comparing the Role of ROS and RNS in the Thermal Stress Response of Two Cnidarian Models, Exaiptasia diaphana and Galaxea fascicularis. Antioxidants 2023, 12, 1057. [Google Scholar] [CrossRef]
- Lee, Y.; Byeon, E.; Kim, D.-H.; Maszczyk, P.; Wang, M.; Wu, R.S.S.; Jeung, H.-D.; Hwang, U.-K.; Lee, J.-S. Hypoxia in Aquatic Invertebrates: Occurrence and Phenotypic and Molecular Responses. Aquat. Toxicol. 2023, 263, 106685. [Google Scholar] [CrossRef]
- Song, L.; Lv, J.; Wang, L.; Sun, D.; Gao, B.; Liu, P. Characterization of a Chitinase-1 Gene (PtCht-1) from a Marine Crab Portunus trituberculatus and Its Response to Immune Stress. Gene 2020, 741, 144523. [Google Scholar] [CrossRef]
- Shock, B.C.; Foran, C.M.; Stueckle, T.A. Effects of Salinity Stress on Survival, Metabolism, Limb Regeneration, and Ecdysis in Uca pugnax. J. Crustac. Biol. 2009, 29, 293–301. [Google Scholar] [CrossRef]
- Therapeutic Enzymes: Function and Clinical Implications; Labrou, N. (Ed.) Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; Volume 1148, ISBN 9789811377082. [Google Scholar]
- Bruhn, T.; Schaller, C.; Schulze, C.; Sanchez-Rodriguez, J.; Dannmeier, C.; Ravens, U.; Heubach, J.F.; Eckhardt, K.; Schmidtmayer, J.; Schmidt, H.; et al. Isolation and Characterisation of Five Neurotoxic and Cardiotoxic Polypeptides from the Sea Anemone Anthopleura elegantissima. Toxicon 2001, 39, 693–702. [Google Scholar] [CrossRef]
- Logashina, Y.A.; Mosharova, I.V.; Korolkova, Y.V.; Shelukhina, I.V.; Dyachenko, I.A.; Palikov, V.A.; Palikova, Y.A.; Murashev, A.N.; Kozlov, S.A.; Stensvåg, K.; et al. Peptide from Sea Anemone Metridium senile Affects Transient Receptor Potential Ankyrin-Repeat 1 (TRPA1) Function and Produces Analgesic Effect. J. Biol. Chem. 2017, 292, 2992–3004. [Google Scholar] [CrossRef]
- Madio, B.; King, G.F.; Undheim, E.A.B. Sea Anemone Toxins: A Structural Overview. Mar. Drugs 2019, 17, 325. [Google Scholar] [CrossRef]
- Xiao, M.; Brugler, M.R.; Broe, M.B.; Gusmão, L.C.; Daly, M.; Rodríguez, E. Mitogenomics suggests a sister relationship of Relicanthus daphneae (Cnidaria: Anthozoa: Hexacorallia: Incerti ordinis) with Actinaria. Sci. Rep. 2019, 9, 18182. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Vera-Estrella, R.; Portillo-Bobadilla, T.; Licea-Navarro, A.; Bernaldez-Sarabia, J.; Rudiño-Piñera, E.; Verleyen, J.J.; Rodríguez, E.; Rodríguez-Almazán, C. Transcriptomic and Proteomic Analysis of the Tentacles and Mucus of Anthopleura dowii Verrill, 1869. Mar. Drugs 2019, 17, E436. [Google Scholar] [CrossRef]
- Mitchell, M.L.; Tonkin-Hill, G.Q.; Morales, R.A.V.; Purcell, A.W.; Papenfuss, A.T.; Norton, R.S. Tentacle Transcriptomes of the Speckled Anemone (Actiniaria: Actiniidae: Oulactis Sp.): Venom-Related Components and Their Domain Structure. Mar. Biotechnol. 2020, 22, 207–219. [Google Scholar] [CrossRef]
- Monastyrnaya, M.M.; Kalina, R.S.; Kozlovskaya, E.P. The Sea Anemone Neurotoxins Modulating Sodium Channels: An Insight at Structure and Functional Activity after Four Decades of Investigation. Toxins 2022, 15, 8. [Google Scholar] [CrossRef]
- Prentis, P.J.; Pavasovic, A.; Norton, R.S. Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins 2018, 10, 36. Available online: https://www.mdpi.com/2072-6651/10/1/36 (accessed on 19 November 2023).
- Marino, A.; Valveri, V.; Muià, C.; Crupi, R.; Rizzo, G.; Musci, G.; La Spada, G. Cytotoxicity of the Nematocyst Venom from the Sea Anemone Aiptasia mutabilis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2004, 139, 295–301. [Google Scholar] [CrossRef]
- Sarian, F.D.; Janeček, Š.; Pijning, T.; Ihsanawati; Nurachman, Z.; Radjasa, O.K.; Dijkhuizen, L.; Natalia, D.; van der Maarel, M.J.E.C. A New Group of Glycoside hydrolase Family 13 α-Amylases with an Aberrant Catalytic Triad. Sci. Rep. 2017, 7, 44230. [Google Scholar] [CrossRef]
- Vandepas, L.E.; Tassia, M.G.; Halanych, K.M.; Amemiya, C.T. Unexpected Distribution of Chitin and Chitin Synthase across Soft-Bodied Cnidarians. Biomolecules 2023, 13, 777. [Google Scholar] [CrossRef]
- Macrander, J.; Broe, M.; Daly, M. Tissue-Specific Venom Composition and Differential Gene Expression in Sea Anemones. Genome Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef]
- Richier, S.; Sabourault, C.; Courtiade, J.; Zucchini, N.; Allemand, D.; Furla, P. Oxidative Stress and Apoptotic Events during Thermal Stress in the Symbiotic Sea Anemone, Anemonia viridis. FEBS J. 2006, 273, 4186–4198. [Google Scholar] [CrossRef]
- Tang, P.-C.; Watson, G.M. Proteomic Identification of Hair Cell Repair Proteins in the Model Sea Anemone Nematostella vectensis. Hear. Res. 2015, 327, 245–256. [Google Scholar] [CrossRef]
- Yang, Y.J.; Kim, C.S.; Choi, B.-H.; Cha, H.J. Mechanically Durable and Biologically Favorable Protein Hydrogel Based on Elastic Silklike Protein Derived from Sea Anemone. Biomacromolecules 2015, 16, 3819–3826. [Google Scholar] [CrossRef]
- Morlighem, J.R.L.; Huang, C.; Liao, Q.; Gomes, P.B.; Pérez, C.D.; Prieto-Da-Silva, R.D.B.; Lee, S.M.-Y.; Rádis-Baptista, G. The Holo-Transcriptome of the Zoantharian Protopalythoa variabilis (Cnidaria: Anthozoa): A Plentiful Source of Enzymes for Potential Application in Green Chemistry, Industrial and Pharmaceutical Biotechnology. Mar. Drugs 2018, 16, 207. [Google Scholar] [CrossRef]
- Merquiol, L.; Romano, G.; Ianora, A.; D’Ambra, I. Biotechnological Applications of Scyphomedusae. Mar. Drugs 2019, 17, 604. [Google Scholar] [CrossRef]
- Ayala-Sumuano, J.-T.; Licea-Navarro, A.; Rudiño-Piñera, E.; Rodríguez, E.; Rodríguez-Almazán, C. Sequencing and de Novo Transcriptome Assembly of Anthopleura dowii Verrill (1869), from Mexico. Genom. Data 2017, 11, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Pérez, D.; Campos, A.; Alexei Rodríguez, A.; Turkina, M.V.; Ribeiro, T.; Osorio, H.; Vasconcelos, V.; Antunes, A. Proteomic Analyses of the Unexplored Sea Anemone Bunodactis verrucosa. Mar. Drugs 2018, 16, 42. [Google Scholar] [CrossRef]
- Cassoli, J.S.; Verano-Braga, T.; Oliveira, J.S.; Montandon, G.G.; Cologna, C.T.; Peigneur, S.; de Castro Pimenta, A.M.; Kjeldsen, F.; Roepstorff, P.; Tytgat, J.; et al. The Proteomic Profile of Stichodactyla duerdeni Secretion Reveals the Presence of a Novel O-Linked Glycopeptide. J. Proteom. 2013, 87, 89–102. [Google Scholar] [CrossRef]
- Wu, C.C.; MacCoss, M.J.; Howell, K.E.; Yates, J.R. A Method for the Comprehensive Proteomic Analysis of Membrane Proteins. Nat. Biotechnol. 2003, 21, 532–538. [Google Scholar] [CrossRef]
- Zheng, Y.Z.; DeMarco, M.L. Manipulating Trypsin Digestion Conditions to Accelerate Proteolysis and Simplify Digestion Workflows in Development of Protein Mass Spectrometric Assays for the Clinical Laboratory. Clin. Mass Spectrom. 2017, 6, 1–12. [Google Scholar] [CrossRef]
- Liu, W.; Mo, F.; Jiang, G.; Liang, H.; Ma, C.; Li, T.; Zhang, L.; Xiong, L.; Mariottini, G.; Zhang, J.; et al. Stress-Induced Mucus Secretion and Its Composition by a Combination of Proteomics and Metabolomics of the Jellyfish Aurelia coerulea. Mar. Drugs 2018, 16, 341. [Google Scholar] [CrossRef]
- Sproles, A.E.; Oakley, C.A.; Matthews, J.L.; Peng, L.; Owen, J.G.; Grossman, A.R.; Weis, V.M.; Davy, S.K. Proteomics Quantifies Protein Expression Changes in a Model Cnidarian Colonised by a Thermally Tolerant but Suboptimal Symbiont—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/31118473/ (accessed on 19 November 2023).
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef]
- Filippova, I.Y.; Dvoryakova, E.A.; Sokolenko, N.I.; Simonyan, T.R.; Tereshchenkova, V.F.; Zhiganov, N.I.; Dunaevsky, Y.E.; Belozersky, M.A.; Oppert, B.; Elpidina, E.N. New Glutamine-Containing Substrates for the Assay of Cysteine Peptidases from the C1 Papain Family. Front. Mol. Biosci. 2020, 7, 578758. [Google Scholar] [CrossRef]
- Recombinant Cathepsin L of Tribolium Castaneum and Its Potential in the Hydrolysis of Immunogenic Gliadin Peptides—PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9266932/ (accessed on 19 November 2023).
- Dunaevsky, Y.E.; Tereshchenkova, V.F.; Belozersky, M.A.; Filippova, I.Y.; Oppert, B.; Elpidina, E.N. Effective Degradation of Gluten and Its Fragments by Gluten-Specific Peptidases: A Review on Application for the Treatment of Patients with Gluten Sensitivity. Pharmaceutics 2021, 13, 1603. [Google Scholar] [CrossRef] [PubMed]
- Kominami, E.; Ishido, K.; Muno, D.; Sato, N. The Primary Structure and Tissue Distribution of Cathepsin C. Biol. Chem. Hoppe Seyler 1992, 373, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Tewari, R.; Hoondal, G.S. Biotechnological Aspects of Chitinolytic Enzymes: A Review. Appl. Microbiol. Biotechnol. 2006, 71, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Stoykov, Y.M.; Pavlov, A.I.; Krastanov, A.I. Chitinase Biotechnology: Production, Purification, and Application. Eng. Life Sci. 2015, 15, 30–38. [Google Scholar] [CrossRef]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, Limitations, and Trends in Engineering for Suitable Applications. Biosci. Rep. 2018, 38, BSR2018032300. [Google Scholar] [CrossRef] [PubMed]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An Update. J. Pharm. Bioallied. Sci. 2013, 5, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Casadidio, C.; Peregrina, D.V.; Gigliobianco, M.R.; Deng, S.; Censi, R.; Di Martino, P. Chitin and Chitosans: Characteristics, Eco-Friendly Processes, and Applications in Cosmetic Science. Mar. Drugs 2019, 17, 369. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Woo, H.A.; Kil, I.S.; Bae, S.H. Peroxiredoxin Functions as a Peroxidase and a Regulator and Sensor of Local Peroxides. J. Biol. Chem. 2012, 287, 4403–4410. [Google Scholar] [CrossRef] [PubMed]
- Burnside, S.W.; Hardingham, G.E. Transcriptional Regulators of Redox Balance and Other Homeostatic Processes with the Potential to Alter Neurodegenerative Disease Trajectory. Biochem. Soc. Trans. 2017, 45, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Szeliga, M. Peroxiredoxins in Neurodegenerative Diseases. Antioxidants 2020, 9, 1203. [Google Scholar] [CrossRef] [PubMed]
- Mesika, R.; Reichmann, D. When Safeguarding Goes Wrong: Impact of Oxidative Stress on Protein Homeostasis in Health and Neurodegenerative Disorders. Adv. Protein Chem. Struct. Biol. 2019, 114, 221–264. [Google Scholar] [CrossRef]
- Rhee, S.G. Overview on Peroxiredoxin. Mol. Cells 2016, 39, 1–5. [Google Scholar] [CrossRef]
- Sharapov, M.G.; Novoselov, V.I.; Gudkov, S.V. Radioprotective Role of Peroxiredoxin 6. Antioxidants 2019, 8, 15. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, W.; Kim, E.E. Crystal Structures of Human Peroxiredoxin 6 in Different Oxidation States. Biochem. Biophys. Res. Commun. 2016, 477, 717–722. [Google Scholar] [CrossRef]
- Sharapov, M.G.; Gudkov, S.V.; Gordeeva, A.E.; Karp, O.E.; Ivanov, V.E.; Shelkovskaya, O.V.; Bruskov, V.I.; Novoselov, V.I.; Fesenko, E.E. Peroxiredoxin 6 Is a Natural Radioprotector. Dokl. Biochem. Biophys. 2016, 467, 110–112. [Google Scholar] [CrossRef]
- Sharapov, M.G.; Novoselov, V.I.; Fesenko, E.E.; Bruskov, V.I.; Gudkov, S.V. The Role of Peroxiredoxin 6 in Neutralization of X-Ray Mediated Oxidative Stress: Effects on Gene Expression, Preservation of Radiosensitive Tissues and Postradiation Survival of Animals. Free Radic. Res. 2017, 51, 148–166. [Google Scholar] [CrossRef]
- Mancini, A. Manganese Superoxide Dismutase Variants and Uses Thereof. U.S. Patent 9976127B2, 22 May 2018. [Google Scholar]
- Rosa, A.C.; Corsi, D.; Cavi, N.; Bruni, N.; Dosio, F. Superoxide Dismutase Administration: A Review of Proposed Human Uses. Molecules 2021, 26, 1844. [Google Scholar] [CrossRef]
- Guan, Y.; Hickey, M.J.; Borgstahl, G.E.; Hallewell, R.A.; Lepock, J.R.; O’Connor, D.; Hsieh, Y.; Nick, H.S.; Silverman, D.N.; Tainer, J.A. Crystal Structure of Y34F Mutant Human Mitochondrial Manganese Superoxide Dismutase and the Functional Role of Tyrosine 34. Biochemistry 1998, 37, 4722–4730. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-Shock Proteins, Molecular Chaperones, and the Stress Response: Evolutionary and Ecological Physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Schild, H.; Arnold-Schild, D.; Lammert, E.; Rammensee, H.G. Stress Proteins and Immunity Mediated by Cytotoxic T Lymphocytes. Curr. Opin. Immunol. 1999, 11, 109–113. [Google Scholar] [CrossRef]
- Kim, J.Y.; Barua, S.; Huang, M.Y.; Park, J.; Yenari, M.A.; Lee, J.E. Heat Shock Protein 70 (HSP70) Induction: Chaperonotherapy for Neuroprotection after Brain Injury. Cells 2020, 9, 2020. [Google Scholar] [CrossRef]
- Alberti, G.; Paladino, L.; Vitale, A.M.; Caruso Bavisotto, C.; Conway de Macario, E.; Campanella, C.; Macario, A.J.L.; Marino Gammazza, A. Functions and Therapeutic Potential of Extracellular Hsp60, Hsp70, and Hsp90 in Neuroinflammatory Disorders. Appl. Sci. 2021, 11, 736. [Google Scholar] [CrossRef]
- Weitzel, G.; Li, G.C. Thermal Response of Yeast Cells Overexpressing Hsp70 Genes. Int. J. Hyperth. 1993, 9, 783–797. [Google Scholar] [CrossRef]
- Jensen, T.K.; Ikonen, E.; Gungor, B. Heat Shock Proteins and Cholesterol Homeostasis. WO2017178029A1, 19 October 2017. [Google Scholar]
- Multhoff, G. Hsp70 Peptide Stimulating Natural Killer (NK) Cell Activity and Uses Thereof 2009. U.S. Patent 7517948B2, 1 April 2004. [Google Scholar]
- Miyakawa, T.; Oka, M.; Hazama, S.; Tamada, K.; Udaka, K. Hsp70-Derived Peptide, and Method of Manufacturing Pharmaceutical Composition, Immunity Inducer, and Antigen-Presenting Cell for Cancer Treatment or Prevention Using the Same. EP 15849707 A 20151007, 2016. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2016056596 (accessed on 19 November 2023).
- Le Poole, I.C.; Guevara-Patino, J.A.; Zloza, A. Mutant HSP70i to Prevent Autoimmune Disease 2013. U.S. Patent 2014 US 2018/0066030 A1, 20 November 2014. Available online: https://patentimages.storage.googleapis.com/e6/c1/ff/8aee26fe13c8bf/US20180066030A1.pdf (accessed on 19 November 2023).
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 Chaperone Network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional Enzymes in Life and Disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef]
- Wilkinson, D.J. Serpins in Cartilage and Osteoarthritis: What Do We Know? Biochem. Soc. Trans. 2021, 49, 1013–1026. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.; Wu, L.; Liu, S.; Zhang, Y.; Yang, X.; Zhu, P.; Zhang, H.; Zhang, K.; Lou, J.; et al. Identification of SERPINB1 as a Physiological Inhibitor of Human Granzyme H. J. Immunol. 2013, 190, 1319–1330. [Google Scholar] [CrossRef]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting Venom of the Sea Anemone Stichodactyla haddoni: Omics Techniques Reveal the Complete Toxin Arsenal of a Well-Studied Sea Anemone Genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Miranda-Zaragoza, B.; Rodríguez-Almazán, C. Actinoporins: From the Structure and Function to the Generation of Biotechnological and Therapeutic Tools. Biomolecules 2020, 10, E539. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Pérez-García, E.I.; Salazar-García, S.I.; Bernáldez-Sarabia, J.; Licea-Navarro, A.; Rudiño-Piñera, E.; Pérez-Martínez, L.; Pedraza-Alva, G.; Rodríguez-Almazán, C. Identification of a Pore-Forming Protein from Sea Anemone Anthopleura dowii Verrill (1869) Venom by Mass Spectrometry. J. Venom Anim. Toxins. Incl. Trop. Dis. 2019, 25, e147418. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical Statistical Model to Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Van Dongen, S. Graph Clustering Via a Discrete Uncoupling Process. SIAM J. Matrix Anal. Appl. 2008, 30, 121–141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Nowotny, J.; Cao, R.; Cheng, J. 3Drefine: An Interactive Web Server for Efficient Protein Structure Refinement. Nucleic Acids Res. 2016, 44, W406–W409. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Carreto, S.; Miranda-Zaragoza, B.; Simões, N.; González-Muñoz, R.; Rodríguez-Almazán, C. Marine Bioprospecting: Enzymes and Stress Proteins from the Sea Anemones Anthopleura dowii and Lebrunia neglecta. Mar. Drugs 2024, 22, 12. https://doi.org/10.3390/md22010012
Ramírez-Carreto S, Miranda-Zaragoza B, Simões N, González-Muñoz R, Rodríguez-Almazán C. Marine Bioprospecting: Enzymes and Stress Proteins from the Sea Anemones Anthopleura dowii and Lebrunia neglecta. Marine Drugs. 2024; 22(1):12. https://doi.org/10.3390/md22010012
Chicago/Turabian StyleRamírez-Carreto, Santos, Beatriz Miranda-Zaragoza, Nuno Simões, Ricardo González-Muñoz, and Claudia Rodríguez-Almazán. 2024. "Marine Bioprospecting: Enzymes and Stress Proteins from the Sea Anemones Anthopleura dowii and Lebrunia neglecta" Marine Drugs 22, no. 1: 12. https://doi.org/10.3390/md22010012
APA StyleRamírez-Carreto, S., Miranda-Zaragoza, B., Simões, N., González-Muñoz, R., & Rodríguez-Almazán, C. (2024). Marine Bioprospecting: Enzymes and Stress Proteins from the Sea Anemones Anthopleura dowii and Lebrunia neglecta. Marine Drugs, 22(1), 12. https://doi.org/10.3390/md22010012






