Pinctada martensii Hydrolysate Modulates the Brain Neuropeptidome and Proteome in Diabetic (db/db) Mice via the Gut–Brain Axis
Abstract
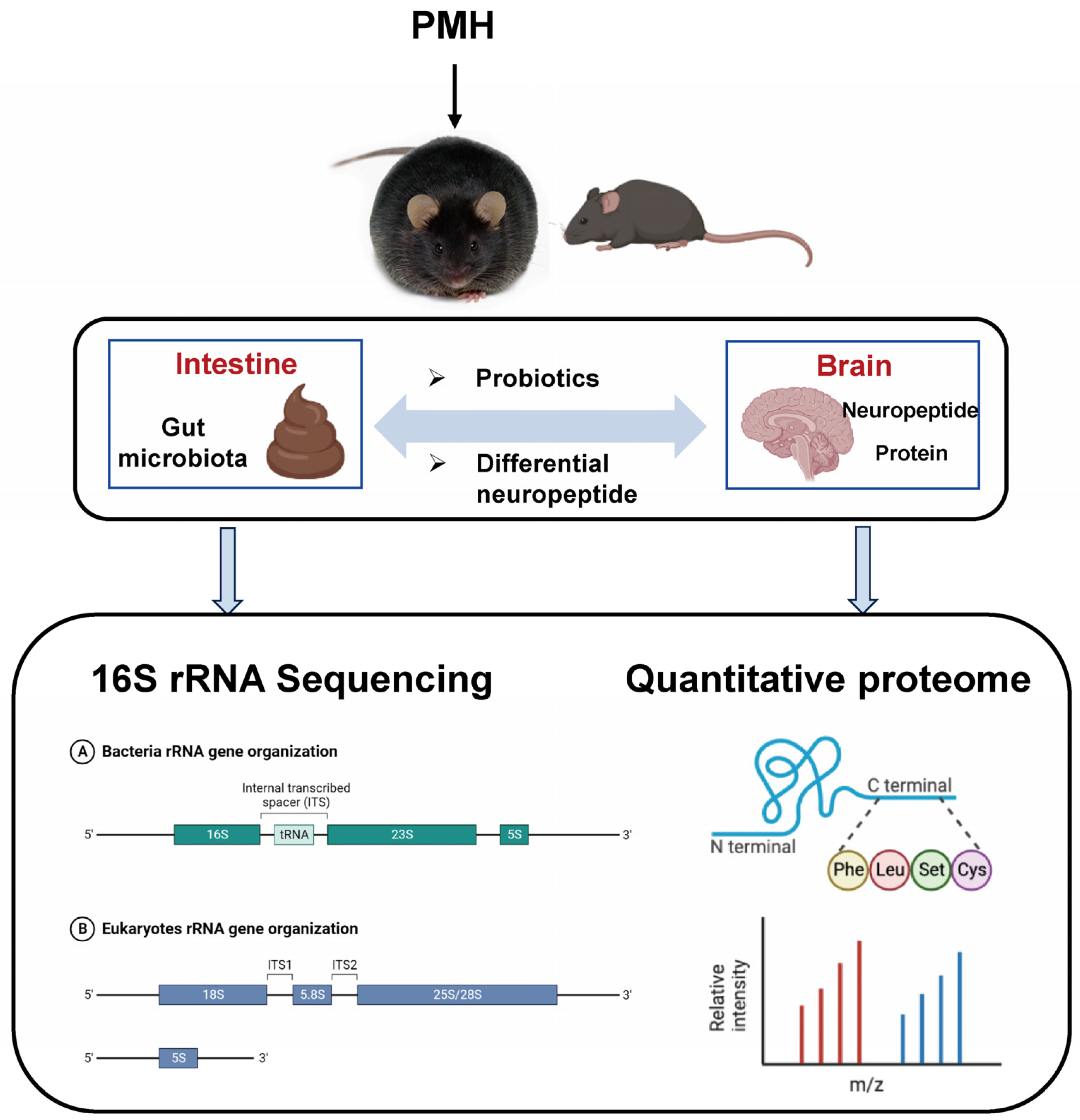
:1. Introduction
2. Results and Discussion
2.1. Identification and Characterization of PMH
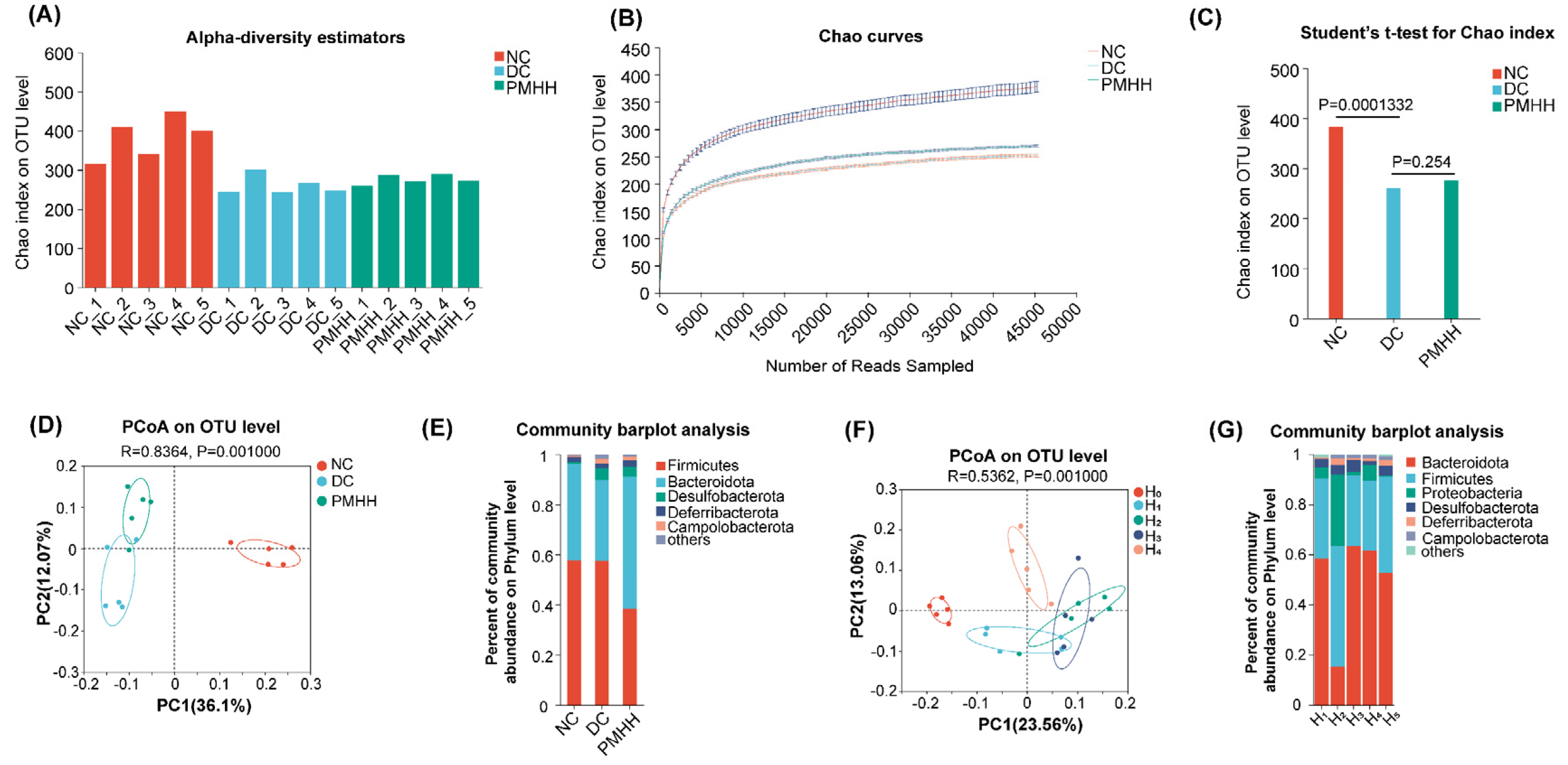
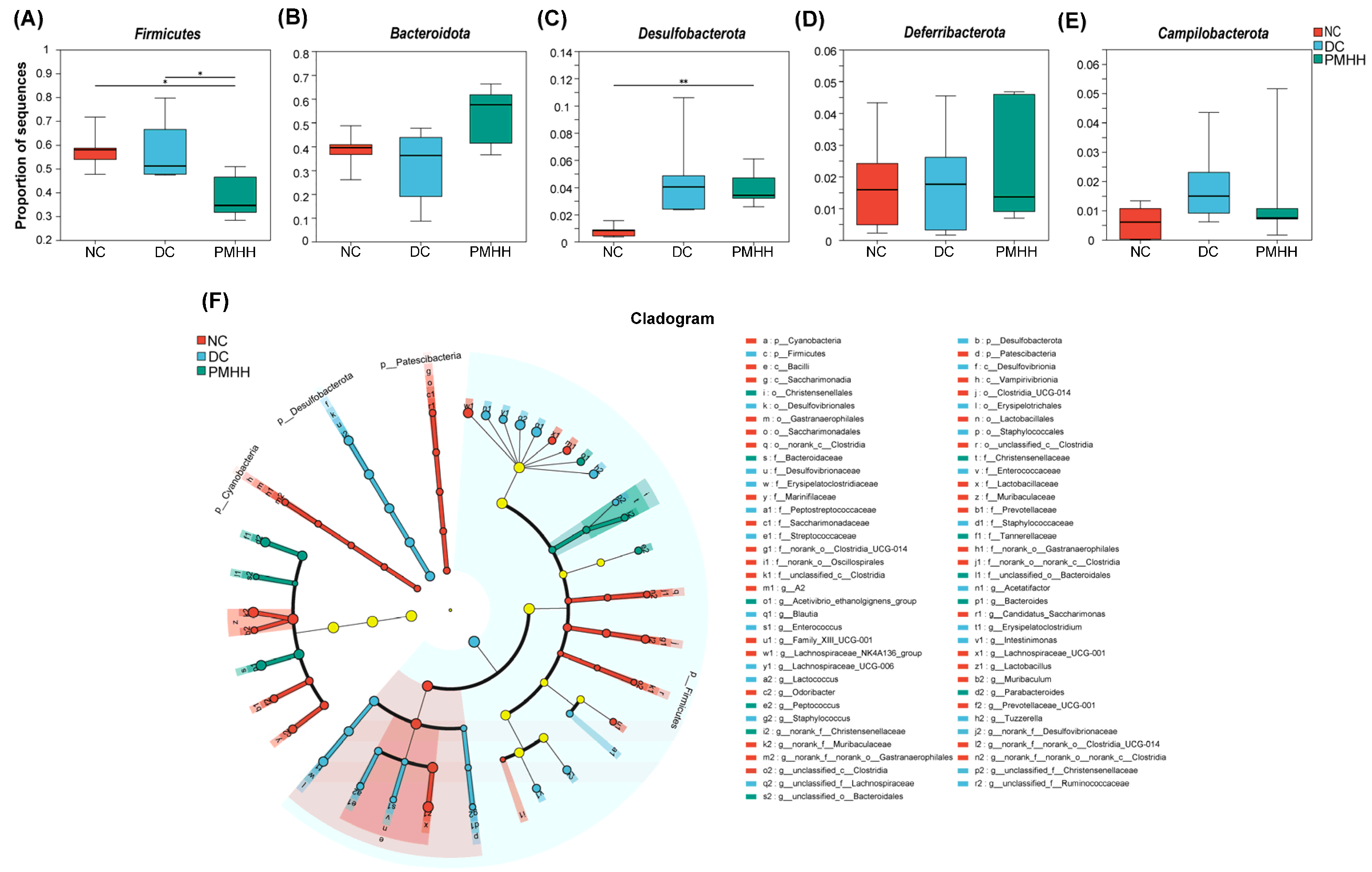
2.2. Gut Microbial Diversity Analysis
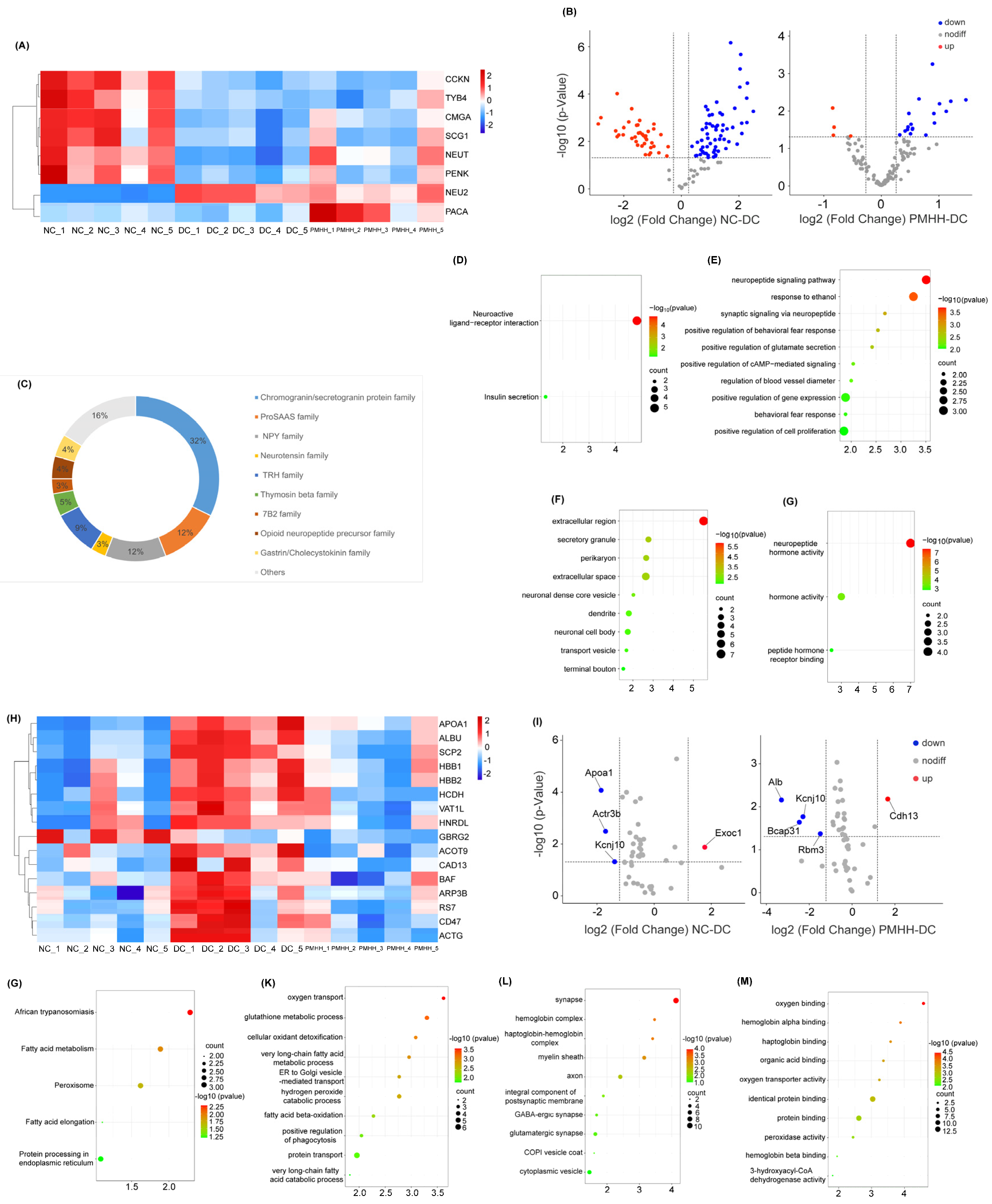
2.3. Label-Free Quantification of Endogenous Peptides in the Hypothalamus
2.4. Proteomic Analysis
2.5. KEGG Pathway Enrichment Analysis
2.6. GO Enrichment Analysis
3. Materials and Methods
3.1. Materials and Sample Preparation
3.2. Animal and Experimental Procedures
3.3. Sample Collection
3.4. DNA Extraction and Polymerase Chain Reaction Amplification
3.5. 16S rRNA Sequencing and Data Analysis
3.6. Neuropeptidomic and Proteomic Analysis of Hypothalamus
3.7. Potential Target Pathway Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Shen, J.; Zeng, M.; Huang, P.; Chen, B.; Xia, Z.; Cao, Y.; Miao, J. Purification and activity evaluation of novel anti-inflammatory peptides from pearl oyster (Pinctada martensii) hydrolysates. Food Funct. 2023, 14, 4242–4253. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Guo, Y.; Chen, M.; Li, N.; Sun, Y.; Ren, S.; Xiao, J.; Wang, D.; Liu, X.; Pan, Y. Hypoglycemic activities of flowers of Xanthoceras sorbifolia and identification of anti-oxidant components by off-line UPLC-QTOF-MS/MS-free radical scavenging detection. Chin. Herbal Med. 2023, 16, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wei, Y.; Huang, S.; Yan, S.; Zhao, B.; Wang, X.; Sun, J.; Chen, T.; Lai, Y.; Liu, R. Hyperglycemia effect of Pinctada martensii hydrolysate in diabetic db/db mice. J. Ethnopharmacol. 2023, 319 Pt 1, 117104. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yan, H.; Li, W. Recent advances in neuropeptide-related omics and gene editing: Spotlight on NPY and somatostatin and their roles in growth and food intake of fish. Front. Endocrinol. 2022, 13, 1023842. [Google Scholar] [CrossRef] [PubMed]
- de Clercq, N.C.; Frissen, M.N.; Groen, A.K.; Nieuwdorp, M. Gut Microbiota and the Gut-Brain Axis: New Insights in the Pathophysiology of Metabolic Syndrome. Psychosom. Med. 2017, 79, 874–879. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, H.; Zhu, Q. The Impact of Microbiota-Gut-Brain Axis on Diabetic Cognition Impairment. Front. Aging Neurosci. 2017, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Richards, P.; Thornberry, N.A.; Pinto, S. The gut-brain axis: Identifying new therapeutic approaches for type 2 diabetes, obesity, and related disorders. Mol. Metab. 2021, 46, 101175. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Yang, Y.; Zhao, X.; Cheng, J.; Li, Y.; Wu, S.; Wu, C. Potential efficacy and mechanism of eight mild-natured and bitter-flavored TCMs based on gut microbiota: A review. Chin. Herb. Med. 2023, 16, 42–55. [Google Scholar] [CrossRef]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ (Clin. Res. Ed.) 2018, 361, k2179. [Google Scholar] [CrossRef]
- Kim, W.J.; Kil, B.J.; Lee, C.; Kim, T.Y.; Han, G.; Choi, Y.; Kim, K.; Shin, C.H.; Park, S.Y.; Kim, H.; et al. B. longum CKD1 enhances the efficacy of anti-diabetic medicines through upregulation of IL- 22 response in type 2 diabetic mice. Gut Microbes 2024, 16, 2319889. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.L.; Li, S.Z.; Xiao, P.T.; Cai, Y.Y.; Chu, C.; Chen, B.Z.; Li, P.; Li, J.; Liu, E.H. Citrus polymeth-oxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci. Adv. 2020, 6, eaax6208. [Google Scholar] [CrossRef]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014, 5, e00889. [Google Scholar] [CrossRef]
- Liu, K.; Zou, J.; Fan, H.; Hu, H.; You, Z. Causal effects of gut microbiota on diabetic retinopathy: A Mendelian randomization study. Front. Immunol. 2022, 13, 930318. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Gálvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Ren, D.; Li, T.; Niu, P.; Zhang, X.; Yang, X.; Xiao, J. Fu Brick Tea Manages HFD/STZ-Induced Type 2 Diabetes by Regulating the Gut Microbiota and Activating the IRS1/PI3K/Akt Signaling Pathway. J. Agric. Food Chem. 2022, 70, 8274–8287. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. Intestinal microbiota and their metabolic contribution to type 2 diabetes and obesity. J. Diabetes Metab. Disord. 2021, 20, 1855–1870. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, H.; Kolodziejczyk, A.A.; Halstuch, D.; Elinav, E. Bile acids in glucose metabolism in health and disease. J. Exp. Med. 2018, 215, 383–396. [Google Scholar] [CrossRef]
- Mayer, E.A.; Nance, K.; Chen, S. The Gut-Brain Axis. Annu. Rev. Med. 2022, 73, 439–453. [Google Scholar] [CrossRef]
- Ahn, W.; Latremouille, J.; Harris, R.B.S. Leptin receptor-expressing cells in the ventromedial nucleus of the hypothalamus contribute to enhanced CCK-induced satiety following central leptin injection. Am. J. Physiol. Endocrinol. Metab. 2022, 323, E267–E280. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Bugescu, R.; Mayer, T.A.; Gata-Garcia, A.; Kurt, G.; Woodworth, H.L.; Leinninger, G.M. Loss of Action via Neurotensin-Leptin Receptor Neurons Disrupts Leptin and Ghrelin-Mediated Control of Energy Balance. Endocrinology 2017, 158, 1271–1288. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Bello, N.T.; Pang, Z.P. Presynaptic Regulation of Leptin in a Defined Lateral Hypothalamus-Ventral Tegmental Area Neurocircuitry Depends on Energy State. J. Neurosci. 2017, 37, 11854–11866. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Virella, J.; Leinninger, G.M. The Role of Central Neurotensin in Regulating Feeding and Body Weight. Endocrinology 2021, 162, bqab038. [Google Scholar] [CrossRef] [PubMed]
- Barchetta, I.; Baroni, M.G.; Melander, O.; Cavallo, M.G. New Insights in the Control of Fat Homeostasis: The Role of Neurotensin. Int. J. Mol. Sci. 2022, 23, 2209. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.M.; Welch, C.C.; Grace, M.K.; Billington, C.J.; Levine, A.S. Chronic food restriction and acute food deprivation decrease mRNA levels of opioid peptides in arcuate nucleus. Am. J. Physiol. 1996, 270 Pt 2, R1019–R1024. [Google Scholar] [CrossRef] [PubMed]
- Resch, J.M.; Maunze, B.; Phillips, K.A.; Choi, S. Inhibition of food intake by PACAP in the hypothalamic ventromedial nuclei is mediated by NMDA receptors. Physiol. Behav. 2014, 133, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Kullmann, S.; Kleinridders, A.; Small, D.M.; Fritsche, A.; Häring, H.-U.; Preissl, H.; Heni, M. Central nervous pathways of insulin action in the control of metabolism and food intake. Lancet Diabetes Endocrinol. 2020, 8, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Matozaki, T.; Murata, Y.; Okazawa, H.; Ohnishi, H. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 2009, 19, 72–80. [Google Scholar] [CrossRef]
- Li, D.; Gwag, T.; Wang, S. Absence of CD47 maintains brown fat thermogenic capacity and protects mice from aging-related obesity and metabolic disorder. Biochem. Biophys. Res. Commun. 2021, 575, 14–19. [Google Scholar] [CrossRef]
- Fiorentino, T.V.; Monroy, A.; Kamath, S.; Sotero, R.; Cas, M.D.; Daniele, G.; Chavez, A.O.; Abdul-Ghani, M.; Hribal, M.L.; Sesti, G.; et al. Pioglitazone corrects dysregulation of skeletal muscle mitochondrial proteins involved in ATP synthesis in type 2 diabetes. Metabolism 2021, 114, 154416. [Google Scholar] [CrossRef]
- Curley, S.; Gall, J.; Byrne, R.; Yvan-Charvet, L.; McGillicuddy, F.C. Metabolic Inflammation in Obesity-At the Crossroads between Fatty Acid and Cholesterol Metabolism. Mol. Nutr. Food Res. 2021, 65, e1900482. [Google Scholar] [CrossRef]
- Machate, D.J.; Figueiredo, P.S.; Marcelino, G.; Guimarães, R.C.A.; Hiane, P.A.; Bogo, D.; Pinheiro, V.A.Z.; Oliveira, L.C.S.; Pott, A. Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis. Int. J. Mol. Sci. 2020, 21, 4093. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El Dine, R.S.; Mohamed, G.A.; Ibrahim SR, M.; Shehata, I.A.; El-Halawany, A.M. Natural Peroxisome Proliferator-Activated Receptor γ (PPARγ) Activators for Diabetes. Altern. Ther. Health Med. 2020, 26, 28–44. [Google Scholar]
- Phu, T.A.; Ng, M.; Vu, N.K.; Bouchareychas, L.; Raffai, R.L. IL-4 polarized human macrophage exosomes control cardiometabolic inflammation and diabetes in obesity. Mol. Ther. 2022, 30, 2274–2297. [Google Scholar] [CrossRef]
- Herrema, H.; Guan, D.; Choi, J.W.; Feng, X.; Salazar Hernandez, M.A.; Faruk, F.; Auen, T.; Boudett, E.; Tao, R.; Chun, H.; et al. FKBP11 rewires UPR signaling to promote glucose homeostasis in type 2 diabetes and obesity. Cell Metab. 2022, 34, 1004–1022.e1008. [Google Scholar] [CrossRef] [PubMed]
- Teskey, G.; Abrahem, R.; Cao, R.; Gyurjian, K.; Islamoglu, H.; Lucero, M.; Martinez, A.; Paredes, E.; Salaiz, O.; Robinson, B.; et al. Glutathione as a Marker for Human Disease. Adv. Clin. Chem. 2018, 87, 141–159. [Google Scholar]
- Lappalainen, Z.; Lappalainen, J.; Oksala, N.K.; Laaksonen, D.E.; Khanna, S.; Sen, C.K.; Atalay, M. Diabetes impairs exercise training-associated thioredoxin response and glutathione status in rat brain. J. Appl. Physiol. (1985) 2009, 106, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hirokawa, J.; Tagami, S.; Kawakami, Y.; Urata, Y.; Kondo, T. Weakened cellular scavenging activity against oxidative stress in diabetes mellitus: Regulation of glutathione synthesis and efflux. Diabetologia 1995, 38, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Bi, Q.; Huang, D.; Li, Y.; Yao, C.; Zhang, J.; Wei, W.; Li, J.; Li, Z.; Zhang, J.; et al. Characterization of natural peptides in Pheretima by integrating proteogenomics and label-free peptidomics. J. Pharm. Anal. 2023, 13, 1070–1079. [Google Scholar] [CrossRef]
- Qu, C.; Xu, Q.Q.; Yang, W.; Zhong, M.; Yuan, Q.; Xian, Y.F.; Lin, Z.X. Gut dysbiosis aggravates cognitive deficits, amyloid pathology and lipid metabolism dysregulation in a transgenic mouse model of Alzheimer’s disease. J. Pharm. Anal. 2023, 13, 1526–1547. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.V.; Natu, A.; Ramelow, C.C.; Rayaprolu, S.; Xiao, H.; Kumar, V.; Kumar, P.; Seyfried, N.T.; Rangaraju, S. Identification of State-Specific Proteomic and Transcriptomic Signatures of Microglia-Derived Extracellular Vesicles. Mol. Cell. Proteom. 2023, 22, 100678. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Yang, A.; Lu, Q.; Zhang, X.; Liu, G.; Liu, G. Study on the mechanism of Baihe Dihuang decoction in treating menopausal syndrome based on network pharmacology. Medicine 2023, 102, e33189. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Lv, Y.; Wei, Y.; Wang, X.; Yan, S.; Zhao, B.; Sun, J.; Liu, R.; Lai, Y. Pinctada martensii Hydrolysate Modulates the Brain Neuropeptidome and Proteome in Diabetic (db/db) Mice via the Gut–Brain Axis. Mar. Drugs 2024, 22, 249. https://doi.org/10.3390/md22060249
Li J, Lv Y, Wei Y, Wang X, Yan S, Zhao B, Sun J, Liu R, Lai Y. Pinctada martensii Hydrolysate Modulates the Brain Neuropeptidome and Proteome in Diabetic (db/db) Mice via the Gut–Brain Axis. Marine Drugs. 2024; 22(6):249. https://doi.org/10.3390/md22060249
Chicago/Turabian StyleLi, Jiayun, Yijun Lv, Yuanqing Wei, Xinzhi Wang, Shenghan Yan, Binyuan Zhao, Jipeng Sun, Rui Liu, and Yueyang Lai. 2024. "Pinctada martensii Hydrolysate Modulates the Brain Neuropeptidome and Proteome in Diabetic (db/db) Mice via the Gut–Brain Axis" Marine Drugs 22, no. 6: 249. https://doi.org/10.3390/md22060249
APA StyleLi, J., Lv, Y., Wei, Y., Wang, X., Yan, S., Zhao, B., Sun, J., Liu, R., & Lai, Y. (2024). Pinctada martensii Hydrolysate Modulates the Brain Neuropeptidome and Proteome in Diabetic (db/db) Mice via the Gut–Brain Axis. Marine Drugs, 22(6), 249. https://doi.org/10.3390/md22060249





