Purification and Structural Analyses of Sulfated Polysaccharides from Low-Value Sea Cucumber Stichopus naso and Anticoagulant Activities of Its Oligosaccharides
Abstract
:1. Introduction
2. Results and Discussion
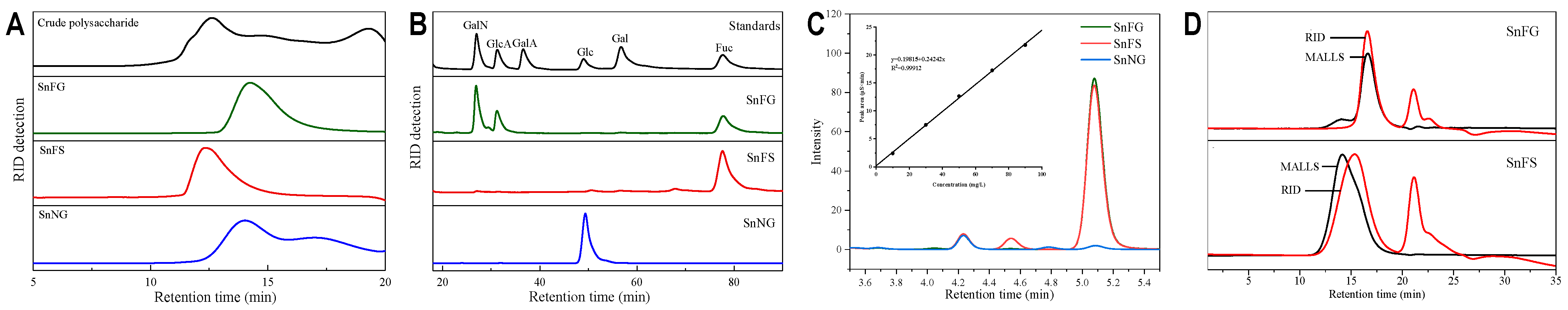
2.1. Extraction and Purification of Polysaccharides
2.2. Analysis of Physicochemical Properties
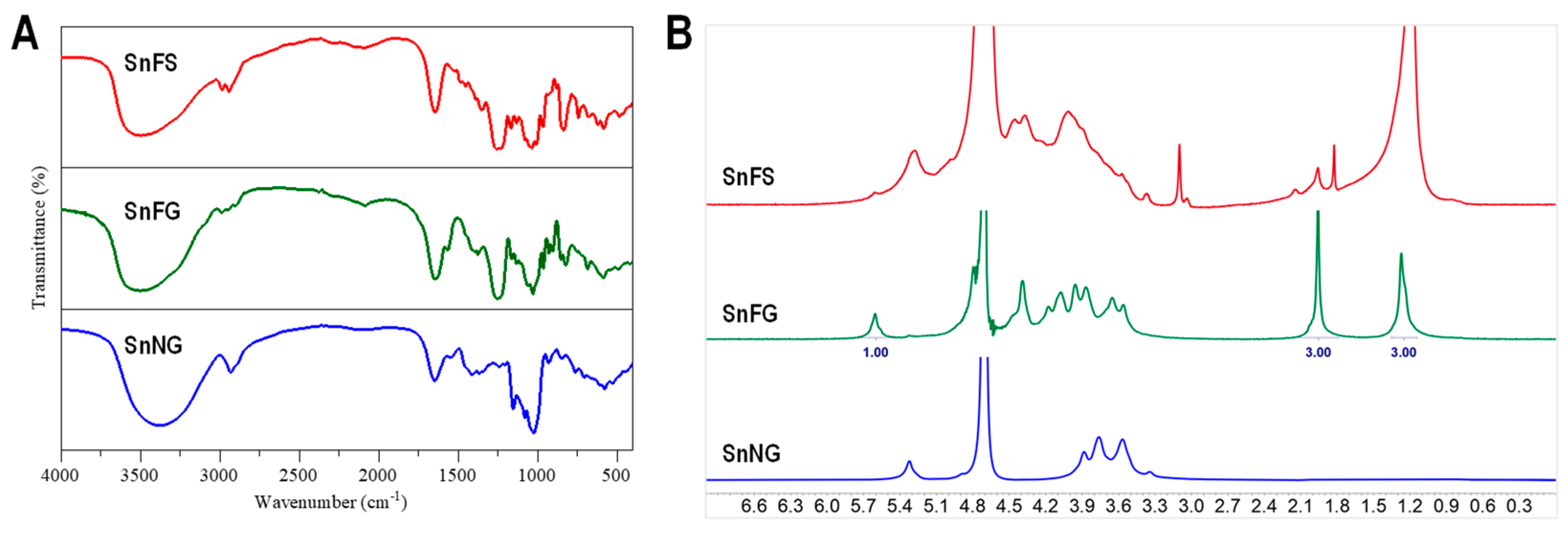
2.3. Spectral Analysis of the Natural Polysaccharides
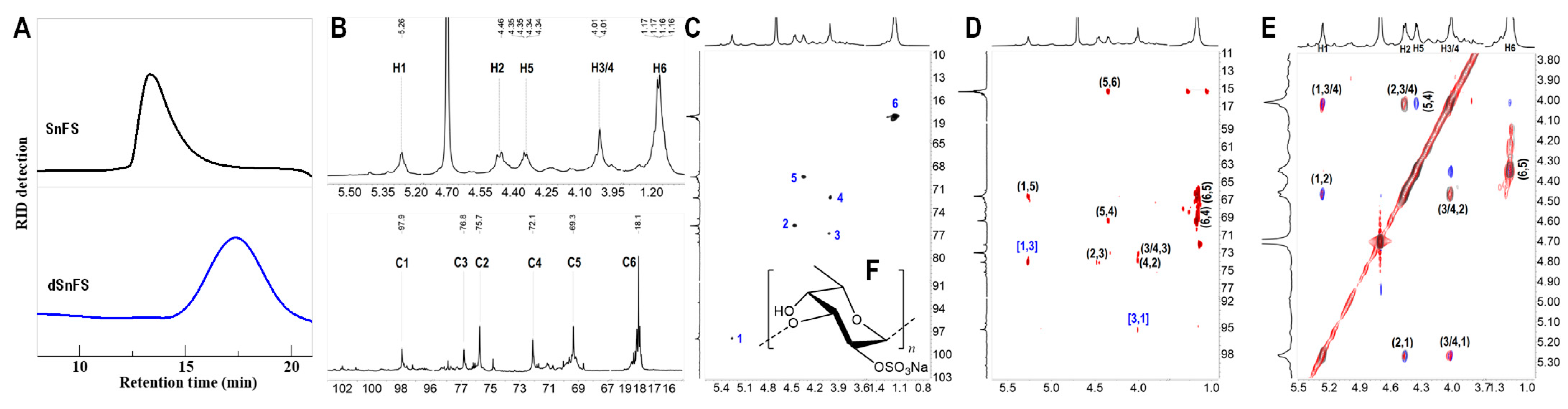
2.4. Hydrogen Peroxide Depolymerization of SnFS and Its Structural Elucidation
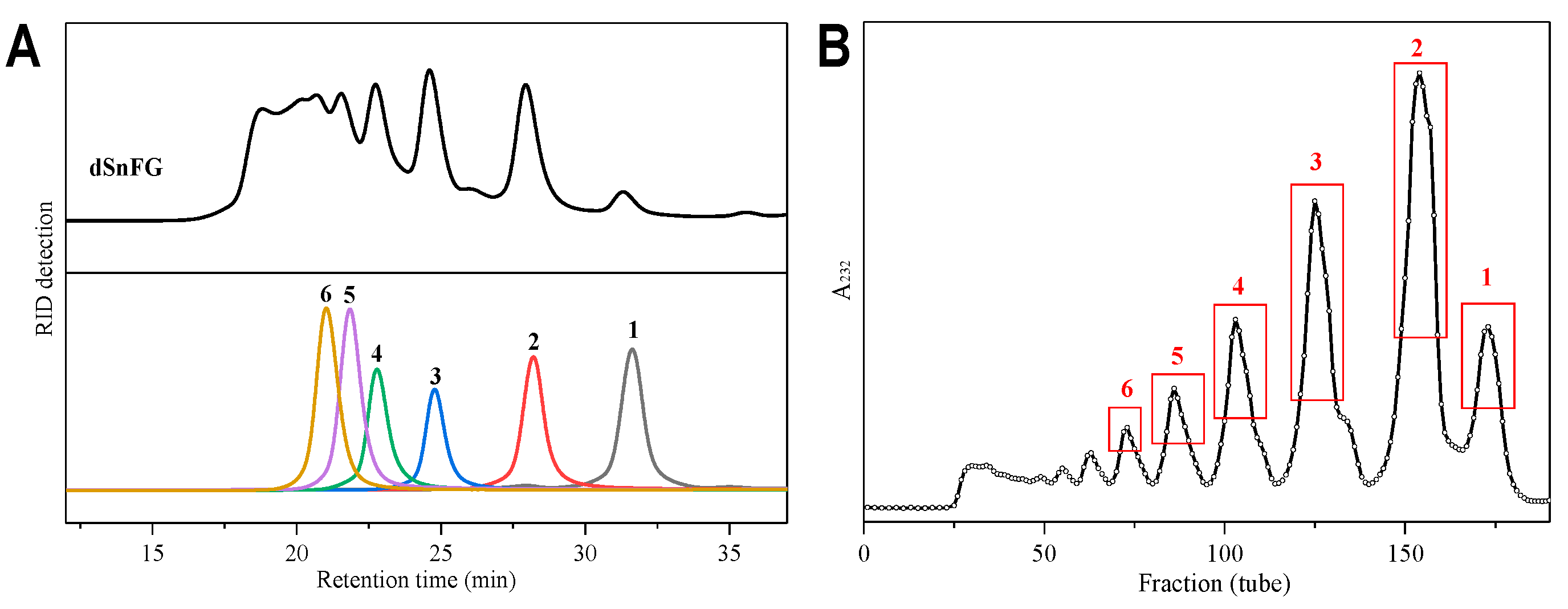
2.5. β-Eliminative Depolymerization of SnFG and the Purification of Oligosaccharides
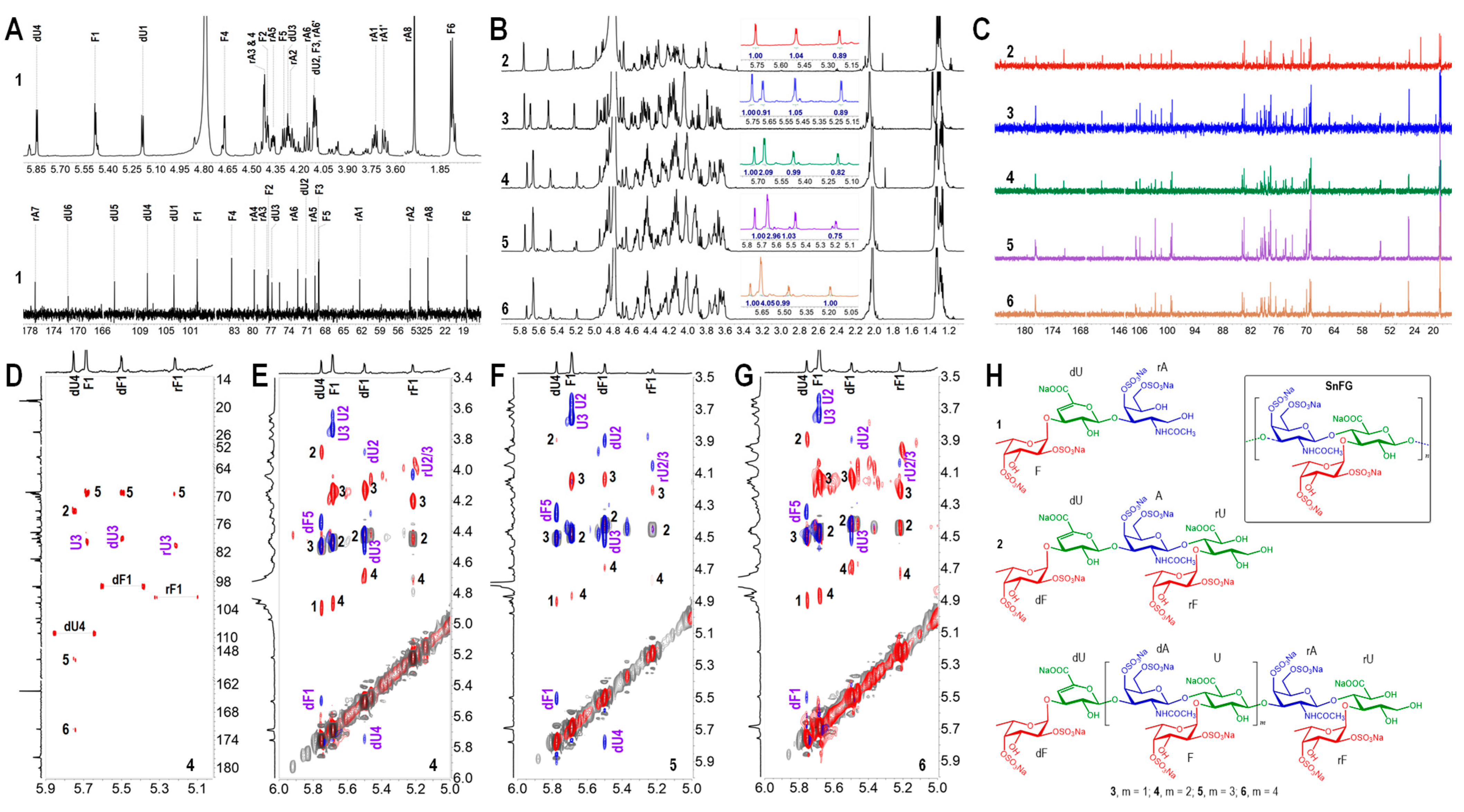
2.6. Structural Analyses of Oligosaccharides 1–6 and SnFG
2.7. Anticoagulation and Inhibition of iXase of Oligosaccharides 3–6 and SnFG
3. Materials and Methods
3.1. Materials and Reagents
3.2. Purification of the Polysaccharides
3.3. Analysis of Physicochemical Properties
3.4. IR, NMR and MS Spectral Analyses
3.5. Chemical Depolymerization of Polysaccharides
3.5.1. Hydrogen Peroxide Depolymerization of SnFS
3.5.2. β-Eliminative Depolymerization of SnFG
3.6. Purification of Oligosaccharides from dSnFG
3.7. Anticoagulant Activity and Inhibition of iXase of SnFG and Its Derived Oligosaccharides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hossain, A.; Dave, D.; Shahidi, F. Sulfated polysaccharides in sea cucumbers and their biological properties: A review. Int. J. Biol. Macromol. 2023, 253, 127329. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H. Holothurian fucosylated chondroitin sulfate. Mar. Drugs 2014, 12, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wu, M.; Xiao, C.; Yang, L.; Zhou, L.; Gao, N.; Li, Z.; Chen, J.; Chen, J.; Liu, J.; et al. Discovery of an intrinsic tenase complex inhibitor: Pure nonasaccharide from fucosylated glycosaminoglycan. Proc. Natl. Acad. Sci. USA 2015, 112, 8284–8289. [Google Scholar] [CrossRef] [PubMed]
- Ustyuzhanina, N.E.; Bilan, M.I.; Dmitrenok, A.S.; Borodina, E.Y.; Stonik, V.A.; Nifantiev, N.E.; Usov, A.I. A highly regular fucosylated chondroitin sulfate from the sea cucumber Massinium magnum: Structure and effects on coagulation. Carbohydr. Polym. 2017, 167, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Peng, Y.; Zhou, L.; Zheng, W.; Liu, X.; Wang, P.; Yuan, Q.; Gao, N.; Zhao, L.; Zhao, J. Precise structure and anticoagulant activity of fucosylated glycosaminoglycan from Apostichopus japonicus: Analysis of its depolymerized fragments. Mar. Drugs 2019, 17, 195. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Zhou, L.; Gao, N.; Lin, L.; Sun, H.; Chen, D.; Cai, Y.; Zuo, Z.; Hu, K.; Huang, S.; et al. Unveiling the disaccharide-branched glycosaminoglycan and anticoagulant potential of its derivatives. Biomacromolecules 2021, 22, 1244–1255. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Cai, Y.; Li, S.; Sun, H.; Lin, L.; Pan, Y.; Yang, W.; He, Z.; Chen, R.; Zhou, L.; et al. A new fucosylated glycosaminoglycan containing disaccharide branches from Acaudina molpadioides: Unusual structure and anti-intrinsic tenase activity. Carbohydr. Polym. 2020, 245, 116503. [Google Scholar] [CrossRef]
- Li, S.; Zhong, W.; Pan, Y.; Lin, L.; Cai, Y.; Mao, H.; Zhang, T.; Li, S.; Chen, R.; Zhou, L.; et al. Structural characterization and anticoagulant analysis of the novel branched fucosylated glycosaminoglycan from sea cucumber Holothuria nobilis. Carbohydr. Polym. 2021, 269, 118290. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Pan, Y.; Cai, Y.; Yang, F.; Gao, N.; Ruzemaimaiti, D.; Zhao, J. Re-understanding of structure and anticoagulation: Fucosylated chondroitin sulfate from sea cucumber Ludwigothurea grisea. Carbohydr. Polym. 2022, 294, 119826. [Google Scholar] [CrossRef]
- Lan, D.; Zhang, J.; Shang, X.; Yu, L.; Xu, C.; Wang, P.; Cui, L.; Cheng, N.; Sun, H.; Ran, J.; et al. Branch distribution pattern and anticoagulant activity of a fucosylated chondroitin sulfate from Phyllophorella kohkutiensis. Carbohydr. Polym. 2023, 321, 121304. [Google Scholar] [CrossRef]
- Ustyuzhanina, N.E.; Bilan, M.I.; Dmitrenok, A.S.; Shashkov, A.S.; Nifantiev, N.E.; Usov, A.I. The structure of a fucosylated chondroitin sulfate from the sea cucumber Cucumaria frondosa. Carbohydr. Polym. 2017, 165, 7–12. [Google Scholar] [CrossRef]
- Fonseca, R.J.; Oliveira, S.N.; Pomin, V.H.; Mecawi, A.S.; Araujo, I.G.; Mourão, P.A. Effects of oversulfated and fucosylated chondroitin sulfates on coagulation. Challenges for the study of anticoagulant polysaccharides. Thromb. Haemost. 2010, 103, 994–1004. [Google Scholar] [CrossRef]
- Mourao, P.A.; Pereira, M.S.; Pavao, M.S.; Mulloy, B.; Tollefsen, D.M.; Mowinckel, M.C.; Abildgaard, U. Structure and anticoagulant activity of a fucosylated chondroitin sulfate from echinoderm. Sulfated fucose branches on the polysaccharide account for its high anticoagulant action. J. Biol. Chem. 1996, 271, 23973–23984. [Google Scholar] [CrossRef]
- Wu, M.; Wen, D.; Gao, N.; Xiao, C.; Yang, L.; Xu, L.; Lian, W.; Peng, W.; Jiang, J.; Zhao, J. Anticoagulant and antithrombotic evaluation of native fucosylated chondroitin sulfates and their derivatives as selective inhibitors of intrinsic factor Xase. Eur. J. Med. Chem. 2015, 92, 257–269. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Hao, J.; Zhao, X.; Lang, Y.; Fan, F.; Cai, C.; Li, G.; Zhang, L.; Yu, G. In vivo anti-cancer mechanism of low-molecular-weight fucosylated chondroitin sulfate (LFCS) from sea cucumber Cucumaria frondosa. Molecules 2016, 21, 625. [Google Scholar] [CrossRef]
- Nadezhda, E.U.; Maria, I.B.; Andrey, S.D.; Alexander, S.S.; Nora, M.A.P.; Carlos, A.S.; Nikolay, E.N.; Anatolii, I.U. Fucosylated chondroitin sulfate from the sea cucumber Hemioedema spectabilis: Structure and influence on cell adhesion and tubulogenesis. Carbohydr. Polym. 2020, 234, 115895. [Google Scholar] [CrossRef]
- Huang, N.; Wu, M.Y.; Zheng, C.B.; Zhu, L.; Zhao, J.H.; Zheng, Y.T. The depolymerized fucosylated chondroitin sulfate from sea cucumber potently inhibits HIV replication via interfering with virus entry. Carbohydr. Res. 2013, 380, 64–69. [Google Scholar] [CrossRef]
- Lian, W.; Wu, M.; Huang, N.; Gao, N.; Xiao, C.; Li, Z.; Zhang, Z.; Zheng, Y.; Peng, W.; Zhao, J. Anti-HIV-1 activity and structure-activity-relationship study of a fucosylated glycosaminoglycan from an echinoderm by targeting the conserved CD4 induced epitope. Biochim. Biophys. Acta 2013, 1830, 4681–4691. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Mao, G.; Wu, T.; Lin, D.; Hu, Y.; Ye, X.; Tian, D.; Chai, W.; Linhardt, R.J.; et al. Fucosylated chondroitin sulfate from Isostichopus badionotus alleviates metabolic syndromes and gut microbiota dysbiosis induced by high-fat and high-fructose diet. Int. J. Biol. Macromol. 2019, 124, 377–388. [Google Scholar] [CrossRef]
- Mou, J.; Li, Q.; Shi, W.; Qi, X.; Song, W.; Yang, J. Chain conformation, physicochemical properties of fucosylated chondroitin sulfate from sea cucumber Stichopus chloronotus and its in vitro fermentation by human gut microbiota. Carbohydr. Polym. 2020, 228, 115359. [Google Scholar] [CrossRef]
- Nagase, H.; Enjyoji, K.; Minamiguchi, K.; Kitazato, K.T.; Kitazato, K.; Saito, H.; Kato, H. Depolymerized holothurian glycosaminoglycan with novel anticoagulant actions: Antithrombin III- and heparin cofactor II-independent inhibition of factor X activation by factor IXa-factor VIIIa complex and heparin cofactor II-dependent inhibition of thrombin. Blood 1995, 85, 1527–1534. [Google Scholar]
- Sheehan, J.P.; Walke, E.N. Depolymerized holothurian glycosaminoglycan and heparin inhibit the intrinsic tenase complex by a common antithrombin-independent mechanism. Blood 2006, 107, 3876–3882. [Google Scholar] [CrossRef]
- Xiao, C.; Zhao, L.; Gao, N.; Wu, M.; Zhao, J. Nonasaccharide inhibits intrinsic factor Xase complex by binding to factor IXa and disrupting factor IXa-factor VIIIa interactions. Thromb. Haemost. 2019, 119, 705–715. [Google Scholar] [CrossRef]
- Li, J.; Li, S.; Yan, L.; Ding, T.; Linhardt, R.J.; Yu, Y.; Liu, X.; Liu, D.; Ye, X.; Chen, S. Fucosylated chondroitin sulfate oligosaccharides exert anticoagulant activity by targeting at intrinsic tenase complex with low FXII activation: Importance of sulfation pattern and molecular size. Eur. J. Med. Chem. 2017, 139, 191–200. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Y.; Jiang, T.; Lv, L.; Zhang, B.; Lv, Z. Depolymerized glycosaminoglycan and its anticoagulant activities from sea cucumber Apostichopus japonicus. Int. J. Biol. Macromol. 2015, 72, 699–705. [Google Scholar] [CrossRef]
- Li, X.; Luo, L.; Cai, Y.; Yang, W.; Lin, L.; Li, Z.; Gao, N.; Purcell, S.W.; Wu, M.; Zhao, J. Structural elucidation and biological activity of a highly regular fucosylated glycosaminoglycan from the edible sea cucumber Stichopus herrmanni. J. Agric. Food Chem. 2017, 65, 9315–9323. [Google Scholar] [CrossRef]
- Chen, S.; Li, G.; Wu, N.; Guo, X.; Liao, N.; Ye, X.; Liu, D.; Xue, C.; Chai, W. Sulfation pattern of the fucose branch is important for the anticoagulant and antithrombotic activities of fucosylated chondroitin sulfates. Biochim. Biophys. Acta 2013, 1830, 3054–3066. [Google Scholar] [CrossRef]
- Yin, R.; Zhou, L.; Gao, N.; Li, Z.; Zhao, L.; Shang, F.; Wu, M.; Zhao, J. Oligosaccharides from depolymerized fucosylated glycosaminoglycan: Structures and minimum size for intrinsic factor Xase complex inhibition. J. Biol. Chem. 2018, 293, 14089–14099. [Google Scholar] [CrossRef]
- Cai, Y.; Yang, W.; Li, X.; Zhou, L.; Wang, Z.; Lin, L.; Chen, D.; Zhao, L.; Li, Z.; Liu, S.; et al. Precise structures and anti-intrinsic tenase complex activity of three fucosylated glycosaminoglycans and their fragments. Carbohydr. Polym. 2019, 224, 115146. [Google Scholar] [CrossRef]
- Sun, H.; Gao, N.; Ren, L.; Liu, S.; Lin, L.; Zheng, W.; Zhou, L.; Yin, R.; Zhao, J. The components and activities analysis of a novel anticoagulant candidate dHG-5. Eur. J. Med. Chem. 2020, 207, 112796. [Google Scholar] [CrossRef]
- Ustyuzhanina, N.E.; Bilan, M.I.; Dmitrenok, A.S.; Shashkov, A.S.; Nifantiev, N.E.; Usov, A.I. Two structurally similar fucosylated chondroitin sulfates from the holothurian species Stichopus chloronotus and Stichopus horrens. Carbohydr. Polym. 2018, 189, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Mou, R.; Zhang, Z.; Gao, N.; Lin, L.; Li, Z.; Wu, M.; Zhao, J. Structural analysis and anticoagulant activities of three highly regular fucan sulfates as novel intrinsic factor Xase inhibitors. Carbohydr. Polym. 2018, 195, 257–266. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, S.; Shi, W.; Qi, X.; Song, W.; Mou, J.; Yang, J. Structure characterization, antioxidant and immunoregulatory properties of a novel fucoidan from the sea cucumber Stichopus chloronotus. Carbohydr. Polym. 2020, 231, 115767. [Google Scholar] [CrossRef]
- Li, X.; Li, S.; Liu, J.; Lin, L.; Sun, H.; Yang, W.; Cai, Y.; Gao, N.; Zhou, L.; Qin, H.; et al. A regular fucan sulfate from Stichopus herrmanni and its peroxide depolymerization: Structure and anticoagulant activity. Carbohydr. Polym. 2021, 256, 117513. [Google Scholar] [CrossRef]
- Ruzemaimaiti, D.; Sun, H.; Zhang, J.; Xu, C.; Chen, L.; Yin, R.; Zhao, J. Oligomer-guided recognition of two fucan sulfate from Bohadschia argus and inhibition of P-selectin binding to its ligand. Carbohydr. Polym. 2023, 317, 121080. [Google Scholar] [CrossRef]
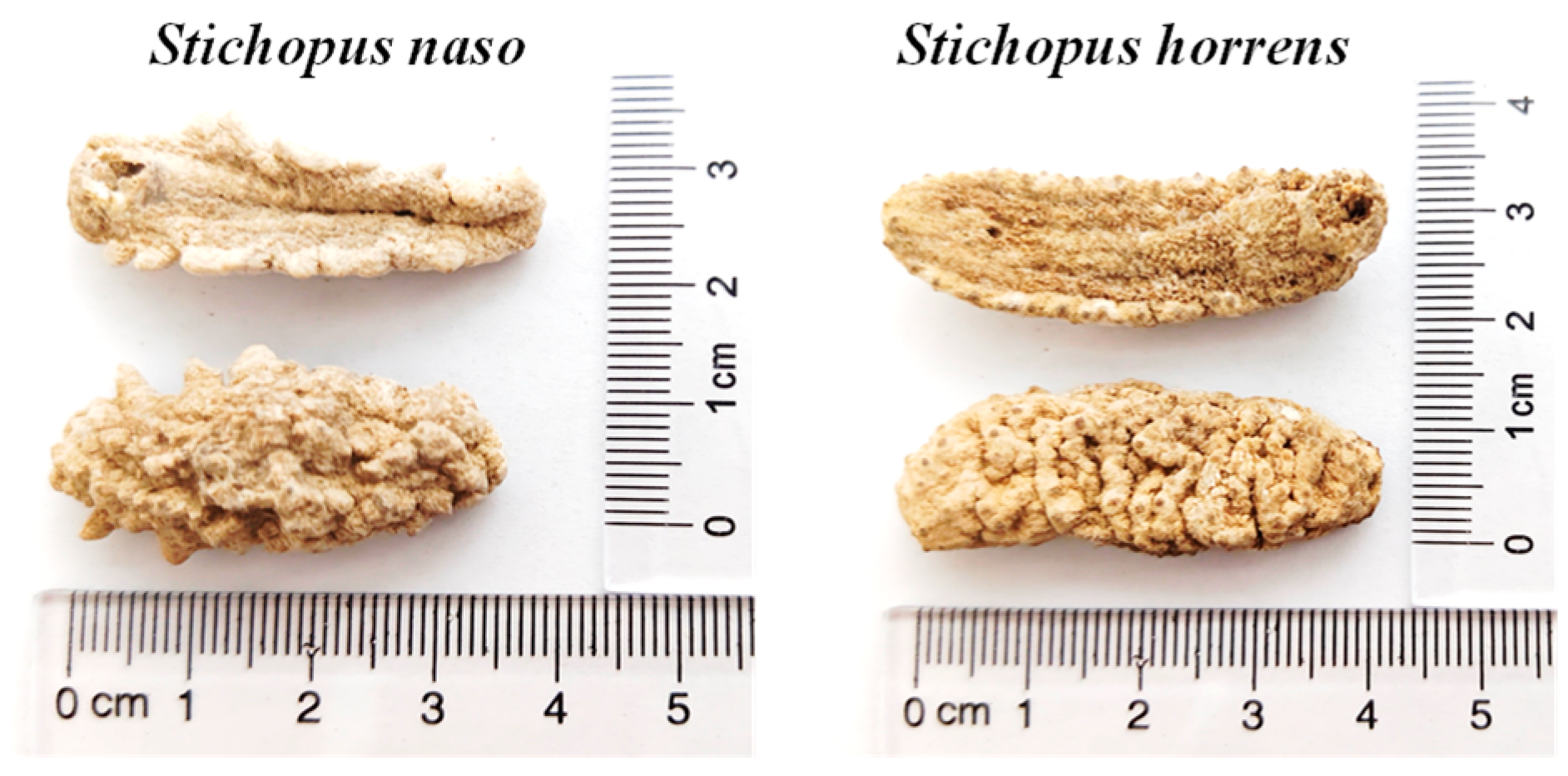
- Purcell, S.W.; Samyn, Y.; Conand, C. Commercially Important Sea Cucumbers of the World; FAO: Rome, Italy, 2012; pp. 102–103. [Google Scholar]
- Zheng, W.; Zhou, L.; Lin, L.; Cai, Y.; Sun, H.; Zhao, L.; Gao, N.; Yin, R.; Zhao, J. Physicochemical characteristics and anticoagulant activities of the polysaccharides from sea cucumber Pattalus mollis. Mar. Drugs 2019, 17, 198. [Google Scholar] [CrossRef]
- Shang, F.; Gao, N.; Yin, R.; Lin, L.; Xiao, C.; Zhou, L.; Li, Z.; Purcell, S.W.; Wu, M.; Zhao, J. Precise structures of fucosylated glycosaminoglycan and its oligosaccharides as novel intrinsic factor Xase inhibitors. Eur. J. Med. Chem. 2018, 148, 423–435. [Google Scholar] [CrossRef]
- Luo, L.; Wu, M.; Xu, L.; Lian, W.; Xiang, J.; Lu, F.; Gao, N.; Xiao, C.; Wang, S.; Zhao, J. Comparison of physicochemical characteristics and anticoagulant activities of polysaccharides from three sea cucumbers. Mar. Drugs 2013, 11, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Lu, F.; Xiao, C.; Yang, L.; Chen, J.; Zhou, K.; Wen, D.; Li, Z.; Wu, M.; Jiang, J.; et al. β-Eliminative depolymerization of the fucosylated chondroitin sulfate and anticoagulant activities of resulting fragments. Carbohydr. Polym. 2015, 127, 427–437. [Google Scholar] [CrossRef]
- Ning, Z.; Wang, P.; Zuo, Z.; Tao, X.; Gao, L.; Xu, C.; Wang, Z.; Wu, B.; Gao, N.; Zhao, J. A fucan sulfate with pentasaccharide repeating units from the sea cucumber Holothuria floridana and its anticoagulant activity. Mar. Drugs 2022, 20, 377. [Google Scholar] [CrossRef]
- Gao, N.; Chen, R.; Mou, R.; Xiang, J.; Zhou, K.; Li, Z.; Zhao, J. Purification, structural characterization and anticoagulant activities of four sulfated polysaccharides from sea cucumber Holothuria fuscopunctata. Int. J. Biol. Macromol. 2020, 164, 3421–3428. [Google Scholar] [CrossRef] [PubMed]

| Monosaccharide Composition | [α] | Mw | Mn | OSO3− (%) | ||||
|---|---|---|---|---|---|---|---|---|
| GlcA | GalN | Fuc | Glc | (kDa) | ||||
| SnFS | ˗ | ˗ | + | ˗ | −163 | 411.3 | 304.9 | 31.4 |
| SnFG | + | + | + | ˗ | −73 | 61.1 | 58.6 | 34.5 |
| SnNG | ˗ | ˗ | ˗ | + | +139 | / | / | 0.6 |
| Comp. | dF | rF | dU | rU | A | F | U | dA | rA | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | H1 | 5.47 | 5.20 | 4.88 | 3.77 | 4.76 | ||||
| C1 | 99.1 | 101.3 | 105.8 | 65.0 | 103.4 | |||||
| H2 | 4.38 a | 4.40 | 3.85 | 4.01 | 4.07 | |||||
| C2 | 77.7 | 78.0 | 73.0 | 73.3 | 54.3 | |||||
| H3 | 4.10 | 4.18 | 4.46 b | 4.01 | 4.19 | |||||
| C3 | 69.0 | 69.1 | 79.0 | 80.7 | 78.5 | |||||
| H4 | 4.66 | 4.70 | 5.73 | 4.13 | 4.96 | |||||
| C4 | 83.4 | 83.7 | 109.1 | 82.9 | 79.1 | |||||
| H5 | 4.32 | 4.55 | 4.28 | 4.08 | ||||||
| C5 | 69.0 | 69.4 | 149.6 | 74.9 | 75.0 | |||||
| H6/6’ | 1.27 | 1.29 | 180.0 | 4.31/4.17 | ||||||
| C6 | 18.4 | 18.7 | 171.6 | 70.5 | ||||||
| C7 | 177.6 | |||||||||
| H8 | 2.02 | |||||||||
| C8 | 25.2 | |||||||||
| 3 | H1 | 5.49 | 5.22 | 4.89 | 3.79 | 5.69 | 4.45 | 4.60 | 4.76 | |
| C1 | 99.0 | 101.3 | 106.6 | 65.0 | 99.2 | 105.9 | 102.5 | 103.4 | ||
| H2 | 4.41 | 4.43 | 3.89 | 4.04 | 4.49 | 3.65 | 4.13 | 4.04 | ||
| C2 | 77.7 | 78.0 | 73.0 | 73.2 | 77.7 | 76.5 | 54.0 | 54.2 | ||
| H3 | 4.14 | 4.21 | 4.51 | 4.04 | 4.14 | 3.74 | 4.15 | 4.04 | ||
| C3 | 69.2 | 69.1 | 79.1 | 80.6 | 69.2 | 79.9 | 78.7 | 78.8 | ||
| H4 | 4.69 | 4.72 | 5.75 | 4.18 | 4.86 | 3.95 | 4.97 | 4.82 | ||
| C4 | 83.4 | 83.7 | 109.0 | 83.0 | 83.9 | 78.2 | 79.0 | 79.0 | ||
| H5 | 4.35 | 4.56 | 4.31 | 4.93 | 3.68 | 4.05 | 4.05 | |||
| C5 | 69.0 | 69.4 | 149.7 | 74.8 | 69.0 | 79.7 | 74.9 | 74.5 | ||
| H6/6’ | 1.30 | 1.32 | 1.38 | 4.28/4.20 | 4.30/4.20 | |||||
| C6 | 18.4 | 18.6 | 171.6 | 180.0 | 18.5 | 177.7 | 70.0 | 70.5 | ||
| C7 | 177.7 | 177.7 | ||||||||
| H8 | 2.05 | 2.05 | ||||||||
| C8 | 25.2 | 25.2 |
| Comp. | Molecular Ion | m/z | Molecular Formula | Mw | |
|---|---|---|---|---|---|
| Observed | Calculated | ||||
| 2 | [M-2Na]2− | 729.4473 | 729.4465 | C32H43NNa6O43S62− | 1505.9382 |
| [M-4Na + 2H]2− | 707.4650 | 707.4645 | C32H45NNa4O43S62− | ||
| [M-6Na + 4H]2− | 685.4821 | 685.4826 | C32H47NNa2O43S62− | ||
| [M-5Na + 2H]3− | 463.9813 | 463.9800 | C32H45NNa3O43S63− | ||
| 3 | [M-5Na + 2H]3− | 782.2808 | 782.2821 | C52H71N2Na8O70S103− | 2461.5350 |
| [M-6Na + 3H]3− | 774.9552 | 774.9548 | C52H72N2Na7O70S103− | ||
| [M-7Na + 4H]3− | 767.6271 | 767.6274 | C52H73N2Na6O70S103− | ||
| [M-8Na + 4H]4− | 569.9748 | 569.9733 | C52H73N2Na5O70S104− | ||
| 4 | [M-9Na + 5H]4− | 803.7056 | 803.2044 | C72H100N3Na9O97S144− | 3414.6842 |
| [M-8Na-SO3Na + 5H]4− | 783.2174 | 783.2152 | C72H100N3Na9O94S134− | ||
| [M-9Na + 4H]5− | 642.7641 | 642.3620 | C72H99N3Na9O97S145− | ||
| [M-11Na + 6H]5− | 633.5697 | 633.5693 | C72H101N3Na7O97S145− | ||
| 5 | [M-6Na]6− | 705.8212 | 705.2759 | C92H121N4Na17O124S186− | 4369.5905 |
| [M-7Na]7− | 601.0156 | 601.2380 | C92H121N4Na16O124S187− | ||
| 6 | [M-11Na + 4H]7− | 725.8486 | 725.2354 | C112H150N5Na21O152S226− | 5325.5002 |
| [M-7Na + H + H2O]6− | 863.1834 | 863.9322 | C112H150N5Na21O152S226− | ||
| [M-4Na-3SO3Na + H + 2H2O]6− | 826.7962 | 826.9556 | C112H152N5Na21O144S196− | ||
| Sample | Resource | Branch Type | dp | Mw a (Da) | 2APTT c (μg/mL) | Anti-iXase d (ng/mL) |
|---|---|---|---|---|---|---|
| 3 | Stichopus naso | Fuc2S4S | 8 | 2462 | 20.94 ± 0.27 | 335.0 ± 24.73 |
| 4 | 11 | 3417 | 16.68 ± 0.29 | 194.1 ± 18.81 | ||
| 5 | 14 | 4373 | 10.19 ± 0.05 | 61.14 ± 5.37 | ||
| 6 | 17 | 5328 | 6.41 ± 0.16 | 21.99 ± 3.04 | ||
| SnFG | 61,100 b | 2.07 ± 0.02 | 8.80 ± 0.28 | |||
| oHG-8 | Holothuria fuscopunctata | Fuc3S4S | 8 | 2462 | 32.35 ± 0.30 | 795.4 ± 98.84 |
| oHG-11 | 11 | 3417 | 23.00 ± 0.06 | 237.0 ± 30.41 | ||
| oHG-14 | 14 | 4373 | 9.31 ± 0.05 | 136.6 ± 20.87 | ||
| oHG-17 | 17 | 5328 | 5.87 ± 0.08 | 14.75 ± 1.31 | ||
| HfFG | 42,600 b | 3.19 ± 0.02 | 15.74 ± 0.29 | |||
| LMWH e | 4500 | 7.46 ± 0.02 | 84.44 ± 7.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Sun, H.; Shang, X.; Wen, J.; Li, P.; Yang, S.; Chen, L.; Huang, X.; Li, H.; Yin, R.; et al. Purification and Structural Analyses of Sulfated Polysaccharides from Low-Value Sea Cucumber Stichopus naso and Anticoagulant Activities of Its Oligosaccharides. Mar. Drugs 2024, 22, 265. https://doi.org/10.3390/md22060265
Cui L, Sun H, Shang X, Wen J, Li P, Yang S, Chen L, Huang X, Li H, Yin R, et al. Purification and Structural Analyses of Sulfated Polysaccharides from Low-Value Sea Cucumber Stichopus naso and Anticoagulant Activities of Its Oligosaccharides. Marine Drugs. 2024; 22(6):265. https://doi.org/10.3390/md22060265
Chicago/Turabian StyleCui, Lige, Huifang Sun, Xiaolei Shang, Jing Wen, Pengfei Li, Shengtao Yang, Linxia Chen, Xiangyang Huang, Haoyang Li, Ronghua Yin, and et al. 2024. "Purification and Structural Analyses of Sulfated Polysaccharides from Low-Value Sea Cucumber Stichopus naso and Anticoagulant Activities of Its Oligosaccharides" Marine Drugs 22, no. 6: 265. https://doi.org/10.3390/md22060265




