Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075
Abstract
:1. Introduction
2. Results
2.1. Isolation of Actinomycetes from Deep-Sea Water
2.2. Genome Analysis of Microbacterium sp. B1075
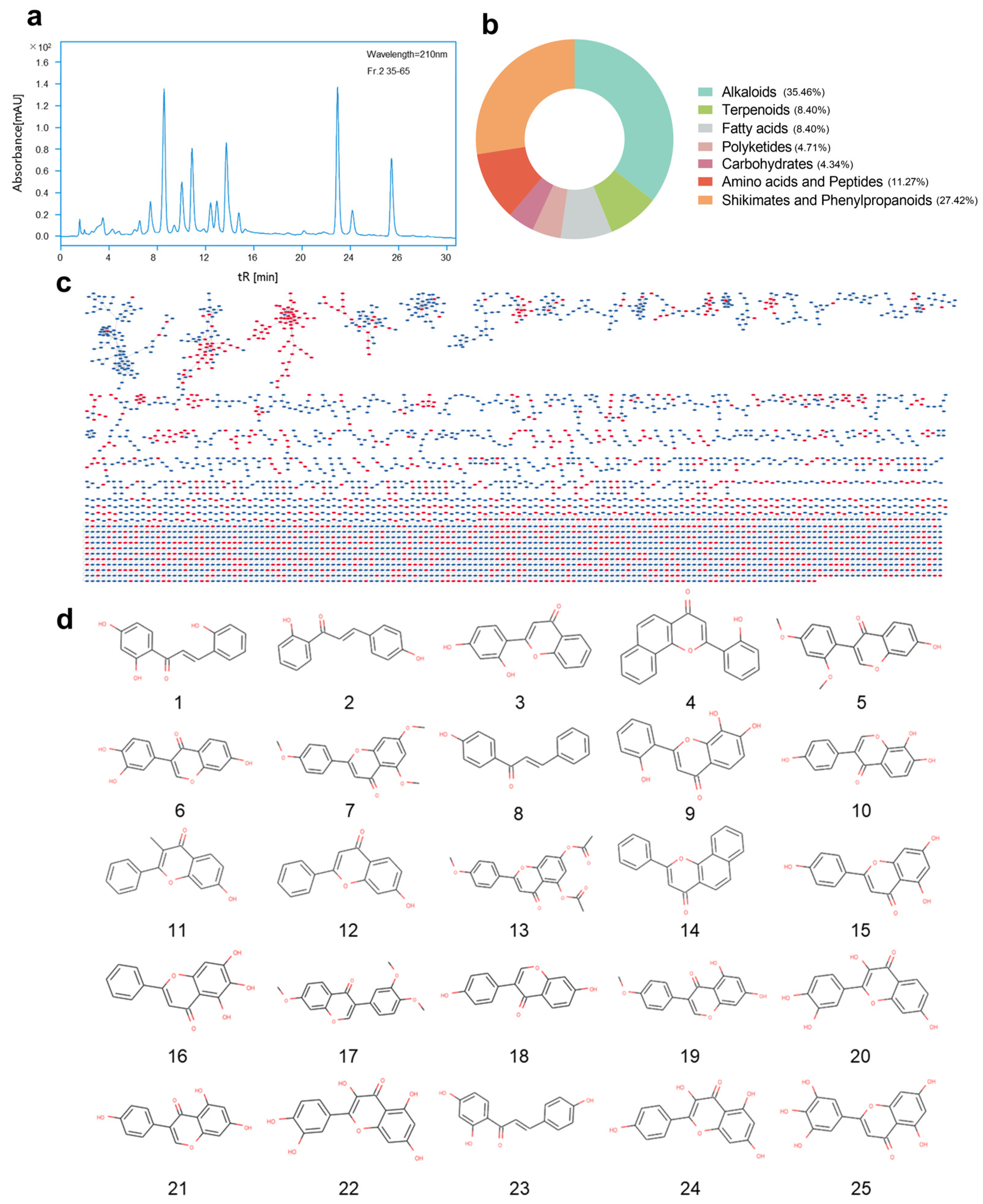
2.3. GNPS Molecular Network Analysis of Secondary Metabolites
2.4. Fractionation and Purification of Secondary Metabolites Guided by Antibacterial Activity
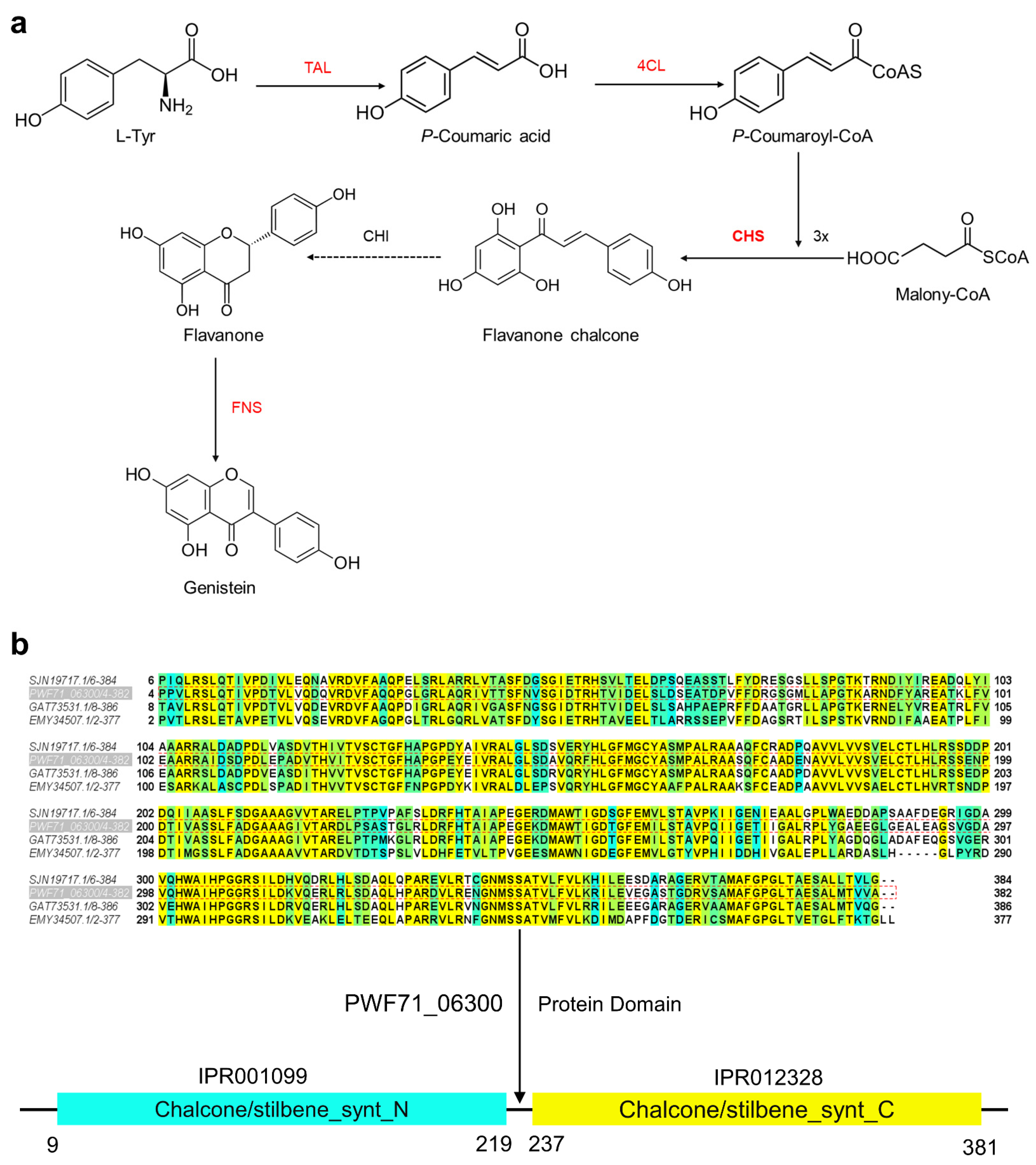
2.5. Proposed Genistein Biosynthesis Pathway in Microbacterium sp. B1075
2.6. Regulation of Key Genes of Flavonoids under High Pressure
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Strain Isolation
4.2. 16S rDNA Cloning and Sequencing for Bacterial Identification
4.3. Genomic DNA Preparation and Whole-Genome Sequencing
4.4. RNA Extraction and Quantitative Real-Time PCR
4.5. Extraction of Secondary Metabolites
4.6. UPLC-MS/MS Analysis of Secondary Metabolites
4.7. Isolation and Identification of Compounds
4.8. Antimicrobial Activity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ribeiro, I.; Antunes, J.T.; Alexandrino, D.A.M.; Tomasino, M.P.; Almeida, E.; Hilário, A.; Urbatzka, R.; Leão, P.N.; Mucha, A.P.; Carvalho, M.F. Actinobacteria from Arctic and Atlantic deep-sea sediments—Biodiversity and bioactive potential. Front. Microbiol. 2023, 14, 1158441. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Z.; Wang, L.; Li, Z.; Fang, J.; Wei, X.; Wei, W.; Cao, J.; Wei, Y.; Xie, Z. Bulk and Active Sediment Prokaryotic Communities in the Mariana and Mussau Trenches. Front. Microbiol. 2020, 11, 1521. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lu, Y.; Cao, S. Antimicrobial compounds from marine actinomycetes. Arch. Pharm. Res. 2020, 43, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Aalbersberg, W. Culturable rare Actinomycetes: Diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 2013, 97, 9291–9321. [Google Scholar] [CrossRef] [PubMed]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep Sea Actinomycetes and Their Secondary Metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef]
- Al-Fadhli, A.A.; Threadgill, M.D.; Mohammed, F.; Sibley, P.; Al-Ariqi, W.; Parveen, I. Macrolides from rare actinomycetes: Structures and bioactivities. Int. J. Antimicrob. Agents 2022, 59, 106523. [Google Scholar] [CrossRef]
- Ding, T.; Yang, L.-J.; Zhang, W.-D.; Shen, Y.-H. The secondary metabolites of rare actinomycetes: Chemistry and bioactivity. RSC Adv. 2019, 9, 21964–21988. [Google Scholar] [CrossRef]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.; Xie, Z.; Zhang, L.; Wang, J.; Fang, J. Influence of Extremely High Pressure and Oxygen on Hydrocarbon-Enriched Microbial Communities in Sediments from the Challenger Deep, Mariana Trench. Microorganisms 2023, 11, 630. [Google Scholar] [CrossRef]
- Corretto, E.; Antonielli, L.; Sessitsch, A.; Höfer, C.; Puschenreiter, M.; Widhalm, S.; Swarnalakshmi, K.; Brader, G. Comparative Genomics of Microbacterium Species to Reveal Diversity, Potential for Secondary Metabolites and Heavy Metal Resistance. Front. Microbiol. 2020, 11, 1869. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, D.; Pan, L.; Su, C.; Li, Z.; Liu, C.; He, Q. Characterization and removal mechanism of a novel enrofloxacin-degrading microorganism, Microbacterium proteolyticum GJEE142 capable of simultaneous removal of enrofloxacin, nitrogen and phosphorus. J. Hazard. Mater. 2023, 454, 131452. [Google Scholar] [CrossRef]
- Sohn, S.I.; Pandian, S.; Oh, Y.J.; Kang, H.J.; Cho, W.S.; Cho, Y.S. Metabolic Engineering of Isoflavones: An Updated Overview. Front. Plant Sci. 2021, 12, 670103. [Google Scholar] [CrossRef]
- Goh, Y.X.; Jalil, J.; Lam, K.W.; Husain, K.; Premakumar, C.M. Genistein: A Review on its Anti-Inflammatory Properties. Front. Pharmacol. 2022, 13, 820969. [Google Scholar] [CrossRef]
- Das, S.; Rosazza, J.P.N. Microbial and Enzymatic Transformations of Flavonoids. J. Nat. Prod. 2006, 69, 499–508. [Google Scholar] [CrossRef]
- Wang, J.-F.; Liu, S.-S.; Song, Z.-Q.; Xu, T.-C.; Liu, C.-S.; Hou, Y.-G.; Huang, R.; Wu, S.-H. Naturally Occurring Flavonoids and Isoflavonoids and Their Microbial Transformation: A Review. Molecules 2020, 25, 5112. [Google Scholar] [CrossRef]
- Meng, Y.; Liu, X.; Zhang, L.; Zhao, G.-R. Modular Engineering of Saccharomyces cerevisiae for De Novo Biosynthesis of Genistein. Microorganisms 2022, 10, 1402. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Reimold, U.; Kröger, M.; Kreuzaler, F.; Hahlbrock, K. Coding and 3′ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J. 1983, 2, 1801–1805. [Google Scholar] [CrossRef]
- Mazurek, A.P.; Kozerski, L.; Sadlej, J.; Kawę, R.; Bednarek, E.; Sitkowski, J.; Dobrowolski, J.C.; Maurin, J.K.; Biniecki, K.; Witowska, J.; et al. Genistein complexes with amines: Structure and properties. J. Chem. Soc. Perkin Trans. 2 1998, 1223–1230. [Google Scholar] [CrossRef]
- Álvarez-Álvarez, R.; Botas, A.; Albillos, S.M.; Rumbero, A.; Martín, J.F.; Liras, P. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Fact. 2015, 14, 178. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Sheng, H.; Sun, X.; Yan, Y.; Yuan, Q.; Wang, J.; Shen, X. Metabolic Engineering of Microorganisms for the Production of Flavonoids. Front. Bioeng. Biotechnol. 2020, 8, 589069. [Google Scholar] [CrossRef]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Qiu, X.; Cao, X.; Jian, H.; Wu, H.; Xu, G.; Tang, X. Transcriptomic Analysis Reveals that Changes in Gene Expression Contribute to Microbacterium sediminis YLB-01 Adaptation at Low Temperature Under High Hydrostatic Pressure. Curr. Microbiol. 2022, 79, 95. [Google Scholar] [CrossRef]
- Xia, J.-M.; Hu, X.-M.; Huang, C.-H.; Yu, L.-B.; Xu, R.-F.; Tang, X.-X.; Lin, D.-H. Metabolic profiling of cold adaptation of a deep-sea psychrotolerant Microbacterium sediminis to prolonged low temperature under high hydrostatic pressure. Appl. Microbiol. Biotechnol. 2020, 104, 277–289. [Google Scholar] [CrossRef]
- Liu, D.; Lin, H.; Proksch, P.; Tang, X.; Shao, Z.; Lin, W. Microbacterins A and B, New Peptaibols from the Deep Sea Actinomycete Microbacterium sediminis sp. nov. YLB-01(T). Org. Lett. 2015, 17, 1220–1223. [Google Scholar] [CrossRef]
- Vitale, G.A.; Scarpato, S.; Mangoni, A.; D’Auria, M.V.; Della Sala, G.; De Pascale, D. Enhanced Molecular Networking Shows Microbacterium sp. V1 as a Factory of Antioxidant Proline-Rich Peptides. Mar. Drugs 2023, 21, 256. [Google Scholar] [CrossRef]
- Jung, W.; Yu, O.; Lau, S.-M.C.; O’Keefe, D.P.; Odell, J.; Fader, G.; McGonigle, B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 2000, 18, 208–212. [Google Scholar] [CrossRef]
- Liu, R.; Hu, Y.; Li, J.; Lin, Z. Production of soybean isoflavone genistein in non-legume plants via genetically modified secondary metabolism pathway. Metab. Eng. 2007, 9, 1–7. [Google Scholar] [CrossRef]
- Jrumg, Z.-D.; Fenieal, P.J.W. Actinellavosiae, A Novel Flavonoid-Like Glycoside Produced by a Marine Becterium of the Genus Streptomyces. Tetrahedron Lett. 1997, 38, 5065–5068. [Google Scholar]
- Savi, D.C.; Shaaban, K.A.; Gos, F.M.W.; Thorson, J.S.; Glienke, C.; Rohr, J. Secondary metabolites produced by Microbacterium sp. LGMB471 with antifungal activity against the phytopathogen Phyllosticta citricarpa. Folia Microbiol. 2019, 64, 453–460. [Google Scholar] [CrossRef]
- Bartlett, D.H.; Lauro, F.M.; Eloe, E.A. Microbial Adaptation to High Pressure. In Physiology and Biochemistry of Extremophiles; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2007; pp. 331–348. ISBN 978-1-68367-165-7. [Google Scholar]
- Xie, Z.; Jian, H.; Jin, Z.; Xiao, X. Enhancing the Adaptability of the Deep-Sea Bacterium Shewanella piezotolerans WP3 to High Pressure and Low Temperature by Experimental Evolution under H2O2 Stress. Appl. Environ. Microbiol. 2018, 84, e02342-17. [Google Scholar] [CrossRef]
- Yang, S.; Lv, Y.; Liu, X.; Wang, Y.; Fan, Q.; Yang, Z.; Boon, N.; Wang, F.; Xiao, X.; Zhang, Y. Genomic and enzymatic evidence of acetogenesis by anaerobic methanotrophic archaea. Nat. Commun. 2020, 11, 3941. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, Y.; Wang, F. Hydrostatic pressure is the universal key driver of microbial evolution in the deep ocean and beyond. Environ. Microbiol. Rep. 2021, 13, 68–72. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Imran, M.; Rauf, A.; Nadeem, M.; Gondal, T.A.; Ahmad, B.; Atif, M.; Mubarak, M.S.; Sytar, O.; et al. Genistein: An Integrative Overview of Its Mode of Action, Pharmacological Properties, and Health Benefits. Oxidative Med. Cell. Longev. 2021, 2021, 3268136. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Ye, X.; Yu, S.; Lian, X.-Y.; Zhang, Z. New Capoamycin-Type Antibiotics and Polyene Acids from Marine Streptomyces fradiae PTZ0025. Mar. Drugs 2012, 10, 2388–2402. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Cui, Y.; Wu, W.; Zhang, Z.; Fang, J.; Yu, X.; Cao, J. Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075. Mar. Drugs 2024, 22, 276. https://doi.org/10.3390/md22060276
Li X, Cui Y, Wu W, Zhang Z, Fang J, Yu X, Cao J. Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075. Marine Drugs. 2024; 22(6):276. https://doi.org/10.3390/md22060276
Chicago/Turabian StyleLi, Xin, Yukun Cui, Weichao Wu, Zhizhen Zhang, Jiasong Fang, Xi Yu, and Junwei Cao. 2024. "Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075" Marine Drugs 22, no. 6: 276. https://doi.org/10.3390/md22060276




