Abstract
Emiliania huxleyi is a single celled, marine phytoplankton with global distribution. As a key species for global biogeochemical cycling, a variety of strains have been amassed in various culture collections. Using a library consisting of 52 strains of E. huxleyi and an ‘in house’ enzyme screening program, we have assessed the functional biodiversity within this species of fundamental importance to global biogeochemical cycling, whilst at the same time determining their potential for exploitation in biocatalytic applications. Here, we describe the screening of E. huxleyi strains, as well as a coccolithovirus infected strain, for commercially relevant biocatalytic enzymes such as acid/alkali phosphodiesterase, acid/alkali phosphomonoesterase, EC1.1.1-type dehydrogenase, EC1.3.1-type dehydrogenase and carboxylesterase.
1. Introduction
Without doubt the oceanic environment represents a hotbed of microbial diversity. With an extra billion years of evolution over their terrestrial counterparts, the oceans contain some of the most ancient and diverse life forms in existence [1]. Attention was initially drawn to this potential metabolic treasure trove largely through the efforts of researchers to catalogue and assess marine biodiversity, as an academic exercise, through intense profiling of common markers such as ribosomal DNA sequence [2,3]. Yet, as our databases began to fill with newly identified permutations of well characterised marker genes, little real functional metabolic information was garnered in the process. Large scale metagenomic projects have gone some way to address this imbalance, yet relevant information on functional activity remains a sparse commodity [4,5]. This causes significant problems for both academic and applied researchers; indeed, without knowledge of the metabolic potential and activity of the individual components of complex ecosystems, the functional relevance of biodiversity remains poorly understood. This lack of understanding is particularly acute for microbial populations of similar strains which are considered as single closely-related groups with little or no attention paid to the variation contained within them which can be significant at the biochemical level.
With little functional information to hand, the first port of call for bioprospectors looking for novel metabolites, drugs and enzyme activities is often established strain libraries where the focus is often placed on screening as diverse a range of species as possible. With economics and efficiency in mind, intraspecies variation is overlooked despite the strong possibility that useful or more suitable properties may be found in “closely-related” strains to those screened. In particular, algal strains have generally been maintained within large collections, under long term continuous culture for many decades, and may therefore no longer be an accurate representation of natural activity levels, due to significant genetic drift and adaptation to artificial culture conditions.
Emiliania huxleyi, a single celled, lithed, marine-phytoplankton with global distribution, is the most abundant of the coccolithophores and is famous for its massive blooms which can be observed from space [6–8]. A species crucial to the study of processes including carbon and sulphur cycling in global marine systems [9], there are now over 450 known strains within culture collections around the world. Furthermore, it is host to one of the largest viruses ever discovered [10], with a genome of over 400,000 bp encoding largely novel genes [10–13]. We have assembled a diverse collection of E. huxleyi strains consisting of representatives established for over half a century in continuous culture as well as more recent isolates, geographically distinct strains and a virally infected strain, and assessed their biochemical diversity using a number of enzyme assays previously used to identify commercially-relevant enzyme activities from the marine environment. Enzyme activities tested for in this study were acid and alkali phosphodiesterase, acid and alkali phosphomonoesterase, EC1.1.1-type dehydrogenase, EC1.3.1-type dehydrogenase and carboxylesterase activity, respectively. Such activities could have applications in the synthesis of enantiomerically-pure chemicals for the pharmaceutical and fine chemical industry where the replacement of traditional synthetic chemistry methods is a rapidly-increasing multi-billion dollar market. We aimed to assess functional biodiversity within this species of fundamental importance to global biogeochemical cycling, whilst at the same time determining the exploitation potential of their enzymes for biocatalysis. This study demonstrates the value of screening similar strains in such biodiscovery programs.
2. Results and Discussion
Enzyme Activity Assays
Fifty two strains of Emiliania huxleyi, isolated from various geographical locations over a period of more than half a century were acquired from ‘in-house’ and external culture collections (Table 1). All strains were screened for acid and alkali phosphodiesterase, acid and alkali phosphomonoesterase, EC1.1.1-type dehydrogenase, EC1.3.1-type dehydrogenase and carboxylesterase activity. In addition, strain CCMP2090 (a confirmed axenic strain which provides a useful ‘clean’ system for studying viral infection dynamics) was infected with the coccolithovirus EhV-86, and following harvesting 72 h later (prior to mass viral induced cellular lysis) included with the other strains. All strains displayed at least residual enzymatic activity in all the screens performed, with all tested substrates (Tables 2–10).

Table 1.
Strains of Emiliania huxleyi used in this study.

Table 2.
Acid phosphomonoesterase (PPME) activity displayed by various E. huxleyi strains (arbitrary values).

Table 10.
Carboxylesterase activity (C16 substrate, CBXY-C16) displayed by various E. huxleyi strains (arbitrary values).
Permutational analysis of variance based on Euclidean distances among strains showed no significant main effects of, or interaction between, strains grouped according to the sea or ocean from which they originated, or the number of years strains had been maintained in culture (Pseudo-F < 1, p > 0.7). Principal Components Analysis (PCA) showed that 69% of variation in enzyme activity among the strains could be summarised by the first principal component (PC1). All subsequent principal components had eigenvalues below 1. All enzymes had similar coefficients (range −0.365 to −0.268) on PC1. Thus despite the differences in locations from which strains were originally collected, in the lengths of time strains had been maintained in culture, and in the range of enzyme activities screened, the overall pattern was a simple gradient in overall activity (Figure 1). A strain displaying high activity in one enzyme assay tended to have high activity in the other enzyme assays.
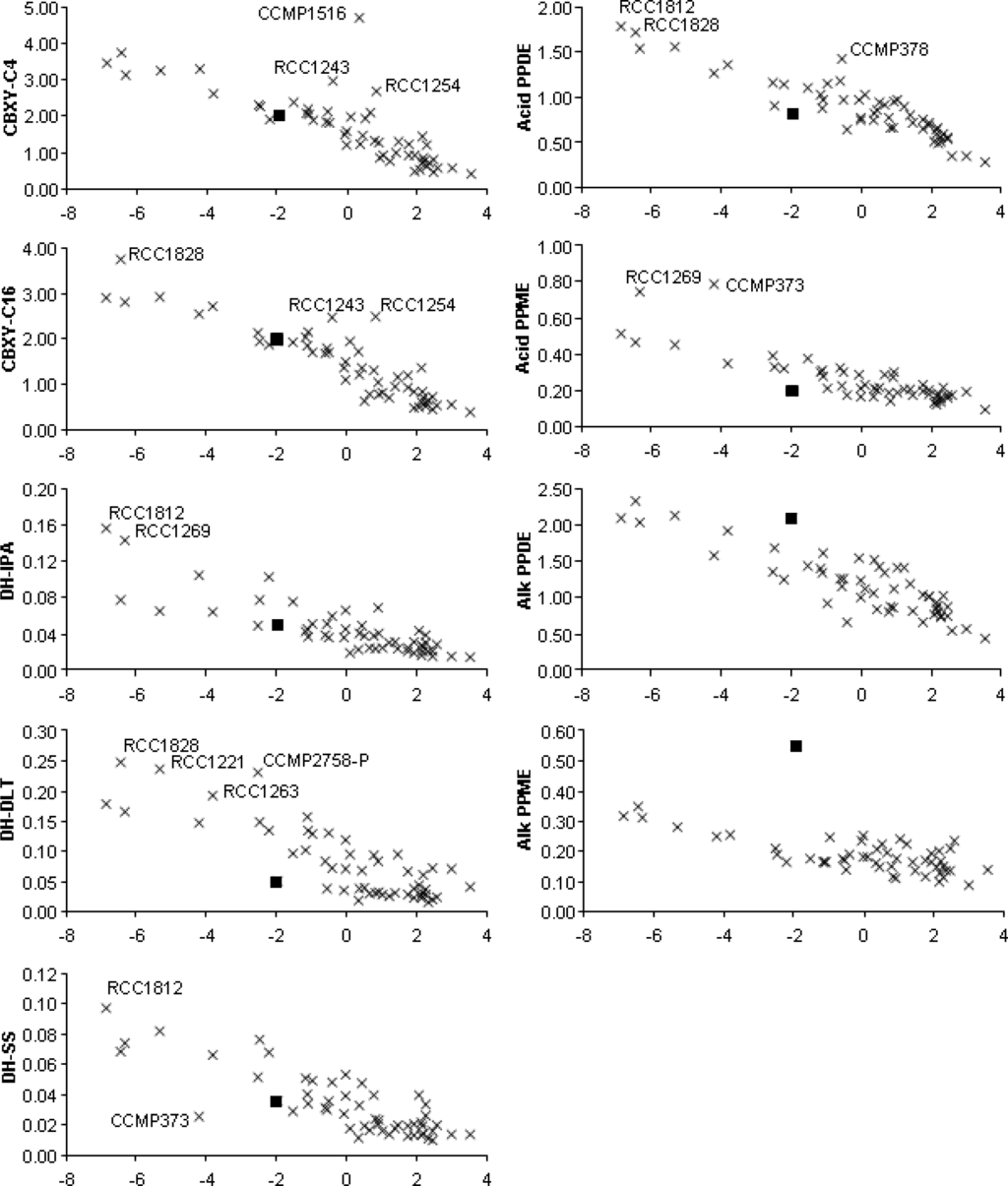
Figure 1.
Activities of each strain for each enzyme (arbitrary units) plotted against first principle component (PC1) scores for each strain (x axis) from a PCA of normalised activities for all enzymes (×). Selected individual strains are labelled. The virally infected strain (CCMP2090inf) is indicated by ▪. Enzyme activities are carboxylesterase with C4 substrate (CBXY-C4); carboxylesterase with C16 substrate (CBXY-C16); E.C.1.1.1-type dehydrogenase with isopropyl alcohol substrate (DH-IPA); E.C.1.1.1-type dehydrogenase with DL-threonine substrate (DH-DLT); E.C.1.3.1-type dehydrogenase with sodium succinate substrate (DH-SS); alkaline phosphodiesterase (Alk PPDE); acid phosphodiesterase (Acid PPDE); alkaline phosphomonoesterase (Alk PPME); and acid phosphomonoesterase (Acid PPME).
Although 69% of the variance among strains was explained by a simple gradient in activity, strains which differ from the overall pattern in terms of the activity of one or two enzymes could be of particular interest for novel enzyme discovery and biocatalysis. To explore this possibility, actual activities of each enzyme for each strain were plotted against the scores for each strain on PC1 (Tables 2–11, Figure 1). In each plot the overall gradient from high activity to low activity is apparent. Among strains which tended to have the highest activity (the most ‘active’ 6 strains on PC1 are RCC1812, RCC1828, RCC1269, RCC1221, CCMP373, RCC1263) none had the highest activity for all enzymes. As examples, RCC1828 had high activity in the carboxylesterase screen with the C4 substrate (Table 9), while for EC1.1.1-type dehydrogenase with isopropyl alcohol substrate it was RCC1812 and RCC1269 (Table 6), for EC1.1.1-type dehydrogenase with DL-threonine substrate RCC1828, RCC1221, RCC1263 and CCMP2758 (Table 7), and so on. Even among these strains of ‘high’ overall activity some had relatively low activity for some enzymes, such as a range of strains for EC1.1.1-type dehydrogenase with isopropyl alcohol substrate (Table 6) and CCMP373 for EC1.3.1-type dehydrogenase (Table 7). Some strains which generally had mid-range activity for most enzymes (i.e., have PC1 scores between −2 and +2) had relatively high activity for individual enzymes (Figure 1), such as CCMP1516 for carboxylesterase activity with C4 substrate; RCC1243 and RCC1254 for carboxylesterase activity with both C4 and C16 substrates; and CCMP378 for acid phosphodiesterase activity.

Table 11.
Principle Component scores (to 2 d.p.) for E. huxleyi strains in the enzyme activity screens.

Table 9.
Carboxylesterase activity (C4 substrate, CBXY-C4) displayed by various E. huxleyi strains (arbitrary values).

Table 6.
E.C.1.1.1-type dehydrogenase activity (isopropyl alcohol substrate, DH-IPA) displayed by various E. huxleyi strains (arbitrary values).

Table 7.
E.C.1.1.1-type dehydrogenase activity (DL-threonine substrate, DH-DLT) displayed by various E. huxleyi strains (arbitrary values).
Among the strains screened are some that might be expected to be highly similar in terms of their enzyme activity. CCMP373 and CCMP88E are thought to be the same strain, but they clearly differ in terms of their activity, as CCMP373 is identified as having relatively high overall activity (low PC1 score) with low dehydrogenase activity (sodium succinate substrate) and high acid phosphomonoesterase activity (Figure 1). Likewise CCMP2090 and CCMP1516 are also thought to be synonyms, but CCMP1516 is identified as having high activity for carboxylesterase (C4 substrate) whereas CCMP2090 is not. Although synonyms, CCMP2090 is an axenic version of CCMP1516 which fails to calcify. The physiological differences between CCMP1516 and CCMP2090 may account for the difference in carboxylesterase activity displayed. CCMP2758-P and CCMP2758-B are definitely the same strain, cultured separately in different collections for approximately 7 years, yet CCMP2758-P displayed a higher overall activity (lower PC1 score), especially in the dehydrogenase assay (DL-threonine substrate) (Figure 1). CCMP376-B and CCMP376-P, also cultured separately for 7 years, display no evidence of differences in activity. Moreover, despite the significant changes in cellular physiology between the haploid (motile) and diploid (lithed) state in E. huxleyi, RCC1217 and RCC1216 (haploid and diploid manifestations of the same strain) displayed no significant evidence of differences in activity in the assays tested. Previous studies have shown significant overlap (approximately 50%) exists between the transcriptional profiles of RCC1216 and RCC1217 with a core set of 3,519 EST clusters identified as common to both life stages [14]. Furthermore, 22 of these EST clusters display database homology to known esterases (including phosphomonoesterases, phosphodiesterases and carboxylesterases), while 94 display homology to known dehydrogenases (including succinate and threonine dehydrogenases) (see supplementary material of [14]). That is not to say that further investigation will not reveal significant metabolic differences between RCC1217 and RCC1216, however. These limited examples raise several crucial issues for further research, such as the repeatability of screening results, the reliability of strain-identification methods, and the relationships between function and taxonomy.
Of particular note is the difference in alkaline phosphomonoesterase and phosphodiesterase activity displayed by the EhV-86 infected strain of CCMP2090 in comparison with the uninfected CCMP2090, and other E. huxleyi strains. With a PC1 score of 1.93 for CCMP2090 and −1.96 for CCMP2090inf, the infected strain generally displayed higher overall activities in all enzyme assays than its uninfected counterpart. The reason for this is, as yet, unclear, but could be a physical effect of the infection process (e.g., variation in cellular integrity or segregation) or a biochemical effect (e.g., variation in metabolism). The infected strain, CCMP2090inf, is highlighted in each plot in Figure 1. Viral infection had little effect on relative carboxylesterase activity (with either C4 or C16 substrate); reduced E.C.1.1.1-type dehydrogenase with isopropyl alcohol substrate and E.C.1.3.1-type dehydrogenase activity slightly; reduced E.C.1.1.1-type dehydrogenase with DL-threonine substrate markedly; and reduced acid phosphodiesterase and phosphomonoesterase activity. However, viral infection had the effect of increasing both alkaline phosphomonoesterase and phosphodiesterase activity, especially the former. Indeed, EhV-86 infected CCMP2090 displayed a higher alkaline phosphomonoesterase activity than all the tested strains of E. huxleyi.
The higher activity observed in this assay may be due to the upregulation or increased activity of E. huxleyi phosphonomonoesterase function in response to viral infection. However, the increased activity could also be a direct consequence of infection through the action of virally encoded enzymes. Indeed, the EhV-86 genome has revealed two such candidates (ehv028 and ehv363) for this activity in the form of coding sequences which have homology to known esterases [10]. Whilst transcripts for ehv363 have so far not been detected during global transcriptional analysis of the infection cycle, transcripts for ehv028 have been detected within two hours of infection by EhV-86 [15].
3. Experimental Section
3.1. Strain Culture and Harvesting
The strains used in this study are shown in Table 1. For each strain of E. huxleyi, 500 mL of F/2 (Guillard 1975) was seeded with 25 mL of mid exponential starter culture [16]. The cultures were grown at 15 °C with a photoperiod of 16 h:8 h L:D. Culture flasks were gently shaken once per day until mid-exponential growth (4 × 106 cells mL−1) was reached. Biomass was harvested by centrifugation at 8000 g for 30 min at 15 °C. CCMP2090-B was infected 72 h prior to harvesting with 0.5 mL Emiliania huxleyi Virus 86 (EhV-86) giving MOI of 1:1.
3.2. Enzyme Activity Assays
Cell pellets were resuspended in 2.5 mL of 50 mM potassium phosphate buffer (pH 7.0) containing 5 mg/mL polyethylenimine and disrupted by sonication on ice. Cell debris was removed by centrifugation at 4,000 g for 10 mins at 4 °C and the protein concentration of extracts determined using Bradford’s assay. Enzyme assays were carried out in triplicate in 96 well, flat bottom microplates using 50 μL of cell extract per reaction in a total assay volume of 250 μL. Reaction mixes were incubated at room temperature for 60 min and absorbance changes (due to colour development) were monitored using a Molecular Devices Versamax platereader at 415 nm.
Acid or alkaline phosphodiesterase activity was measured by incubating extract plus bis-(4-nitrophenyl) phosphate (20 mM) in the presence of either 11.5 mM HCl or 7mM NaOH, respectively. Similarly, for acid or alkali phosphomonoesterase activity, extract plus 4-nitrophenyl phosphate (20 mM) was incubated with either 11.5 mM HCl or 7 mM NaOH, respectively. Carboxylesterase activity was detected by incubating extract in the presence of either 4-nitrophenyl butyrate (C4) or 4-nitrophenyl palmitate (C16) at a final concentration of 20 mM, respectively. EC.1.1.1-type dehydrogenase activity was detected incubating extract as follows: isopropyl alcohol or DL-threonine (20 mM), NaOH (7 mM), NAD (1 mM), XTT (0.5 mM), and 10.25 Units of Diaphorase solution. EC.1.3.1-type dehydrogenase activity was detected in an identical assay mix except that sodium succinate (20 mM) replaced the isopropyl alcohol or DL-threonine as substrate.
3.3. Statistical Analysis
Data were normalised to protein content. To account for differences in average activity among enzymes data were standardised across all strains by subtracting the mean activity and dividing through by the standard deviation. This placed the variation in activity for each enzyme across all strains on a scale of standard deviations centered on zero. The standardised dataset was analysed using multivariate methods in Primer v6 [17,18] with the Permanova+ add-in [19].
4. Conclusions
All E. huxleyi strains under study displayed acid and alkali phosphodiesterase, acid and alkali phosphomonoesterase, EC1.1.1-type dehydrogenase, EC1.3.1-type dehydrogenase and carboxylesterase activity with all variants of the substrates tested. Strains displaying higher activities for one enzyme function tended also to have higher activities for the other enzyme functions tested. Consequently, we observed a simple gradient in enzyme activity, from low activity strains to high activity strains. Along this gradient, we identified six strains displaying significantly higher enzymatic activities than their relatives. On the whole, strains of E. huxleyi displayed similar metabolic potentials, yet variations did occur within some strains which exhibited marked increases or decreases in particular enzyme activities relative to their “expected” activity (i.e., the gradual changes in enzymatic activity observed in the general population). These variations could have profound effects on ecosystem productivity and form the basis of functional biodiversity. Crucially, the activity gradient was skewed only on a few occasions, notably by viral infection. The display of increased phosphomonoesterase activity in virally infected cells is a particularly noteworthy example of this departure from the norm. As arguably the largest reservoir of genetic novelty on the planet, the metabolic potential of viruses is enormous. As we have shown here, viruses have much to offer the field of biocatalysis. Moreover, with their relatively small genomes, gene identification is not as arduous a task as it can be with the larger genomes found within their hosts. However, despite the massive potential for viruses in biocatalysis, the problem of identifying suitable hosts for culture-dependent enzyme screening of the nature undertaken in this study remains significant. Of further interest to biodiscovery programs, enzyme activity was not associated with geographic location or the length of time strains had been in culture, suggesting that, for preliminary screens, established culture collections are indeed a useful and valid starting point. A high degree of genetic diversity has previously been observed among E. huxleyi strains [20], as well as for other algal species [21], yet the ecological and functional relevance of this diversity has so far remained unassessed. The results here demonstrate that once a specific enzyme functional activity is identified in any particular strain under study, the screening of related strains (both close and distant relatives) for altered activity levels is a prudent and worthwhile approach for both ecological and biotechnological applications.
Acknowledgments
The authors would like to acknowledge Andrew Willets for establishing the enzyme screening program at PML Applications Ltd as part of the NERC follow on fund NE/F014406/1 awarded to STA, also supported by i-GPeninsula, a Public Sector Research Exploitation project funded by the Department of Business, Innovation, and Skills (HM Government, UK). MJA, PJS and STA are supported by NERC’s Oceans 2025 program. ER and CW are supported by BBSRC Industrial CASE studentships, sponsored by PML Applications Ltd. MJA, IP and CW were supported by funds from the Interreg program, Marinexus. JL and JN are supported by the BBSRC. Strains from the RCC were obtained through the ASSEMBLE program (FP7-227799).
- Samples Availability: Strains with CCMP prefix are available for purchase from The Provasoli-Guillard National Center for Culture of Marine Phytoplankton at Bigelow, USA ( https://ccmp.bigelow.org/). All other Emiliania huxleyi strains are available from the Roscoff Culture Collection ( http://www.sb-roscoff.fr/Phyto/RCC/). Coccolithovirus strain EhV-86 is available from the PML Virus Collection, contact Mike Allen (mija@pml.ac.uk).
References
- Watanabe, Y; Martini, JEJ; Ohmoto, H. Geochemical evidence for terrestrial ecosystems 2.6 billion years ago. Nature 2000, 408, 574–578. [Google Scholar]
- Caporaso, JG; Kuczynski, J; Stombaugh, J; Bittinger, K; Bushman, FD; Costello, EK; Fierer, N; Pena, AG; Goodrich, JK; Gordon, JI; Huttley, GA; Kelley, ST; Knights, D; Koenig, JE; Ley, RE; Lozupone, CA; McDonald, D; Muegge, BD; Pirrung, M; Reeder, J; Sevinsky, JR; Tumbaugh, PJ; Walters, WA; Widmann, J; Yatsunenko, T; Zaneveld, J; Knight, R. Qiime allows analysis of high-throughput community sequencing data. Nat Methods 2010, 7, 335–336. [Google Scholar]
- Hamady, M; Walker, JJ; Harris, JK; Gold, NJ; Knight, R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 2008, 5, 235–237. [Google Scholar]
- Sharon, I; Tzahor, S; Williamson, S; Shmoish, M; Man-Aharonovich, D; Rusch, DB; Yooseph, S; Zeidner, G; Golden, SS; Mackey, SR; Adir, N; Weingart, U; Horn, D; Venter, JC; Mandel-Gutfreund, Y; Beja, O. Viral photosynthetic reaction center genes and transcripts in the marine environment. Isme J 2007, 1, 492–501. [Google Scholar]
- Nealson, KH; Venter, JC. Metagenomics and the global ocean survey: What’s in it for us, and why should we care. Isme J 2007, 1, 185–187. [Google Scholar]
- Holligan, PM; Fernandez, E; Aiken, J; Balch, WM; Boyd, P; Burkill, PH; Finch, M; Groom, SB; Malin, G; Muller, K; Purdie, DA; Robinson, C; Trees, CC; Turner, SM; Vanderwal, P. A biogeochemical study of the coccolithophore, emiliania huxleyi, in the north-atlantic. Glob Biogeochem Cycle 1993, 7, 879–900. [Google Scholar]
- Holligan, PM; Viollier, M; Harbour, DS; Camus, P; Champagnephilippe, M. Satellite and ship studies of coccolithophore production along a continental-shelf edge. Nature 1983, 304, 339–342. [Google Scholar]
- Brown, CW; Yoder, JA. Coccolithophorid blooms in the global ocean. J Geophys Res-Oceans 1994, 99, 7467–7482. [Google Scholar]
- Westbroek, P; Brown, CW; Vanbleijswijk, J; Brownlee, C; Brummer, GJ; Conte, M; Egge, J; Fernandez, E; Jordan, R; Knappertsbusch, M; Stefels, J; Veldhuis, M; Vanderwal, P; Young, J. A model system approach to biological climate forcing—the example of emiliania huxleyi. Glob Planet Change 1993, 8, 27–46. [Google Scholar]
- Wilson, WH; Schroeder, DC; Allen, MJ; Holden, MTG; Parkhill, J; Barrell, BG; Churcher, C; Hamlin, N; Mungall, K; Norbertczak, H; Quail, MA; Price, C; Rabbinowitsch, E; Walker, D; Craigon, M; Roy, D; Ghazal, P. Complete genome sequence and lytic phase transcription profile of a coccolithovirus. Science 2005, 309, 1090–1092. [Google Scholar]
- Suttle, CA. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar]
- Suttle, CA. Marine viruses--major players in the global ecosystem. Nat Rev 2007, 5, 801–812. [Google Scholar]
- Allen, MJ; Martinez-Martinez, J; Schroeder, DC; Somerfield, PJ; Wilson, WH. Use of microarrays to assess viral diversity: From genotype to phenotype. Environ Microbiol 2007, 9, 971–982. [Google Scholar]
- von Dassow, P; Ogata, H; Probert, I; Wincker, P; Da Silva, C; Audic, S; Claverie, JM; de Vargas, C. Transcriptome analysis of functional differentiation between haploid and diploid cells of emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol 2009, 10. [Google Scholar] [CrossRef] [Green Version]
- Allen, MJ; Forster, T; Schroeder, DC; Hall, M; Roy, D; Ghazal, P; Wilson, WH. Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus. J Virol 2006, 80, 7699–7705. [Google Scholar]
- Guillard, RRL. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Smith, WL, Chanley, MH, Eds.; Plenum Press: New York, NY, USA, 1975; pp. 29–60. [Google Scholar]
- Clarke, K. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 1993, 18, 117–143. [Google Scholar]
- Clarke, KR; Gorley, RN. Primer v6: User Manual/Tutorial; Primer-E Ltd: Plymouth, UK, 2006. [Google Scholar]
- Anderson, MJ; Gorley, RN; Clarke, KR. Permanova+ for Primer: Guide to Software and Statistical Methods; Primer-E Ltd: Plymouth, UK, 2008. [Google Scholar]
- Iglesias-Rodriguez, MD; Schofield, OM; Batley, J; Medlin, LK; Hayes, PK. Intraspecific genetic diversity in the marine coccolithophore emiliania huxleyi (prymnesiophyceae): The use of microsatellite analysis in marine phytoplankton population studies. J Phycol 2006, 42, 526–536. [Google Scholar]
- Rynearson, TA; Armbrust, EV. DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom ditylum brightwellii. Limnol Oceanogr 2000, 45, 1329–1340. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).