Antimicrobial Resistance in the Food Chain: A Review
Abstract
:1. Introduction
2. Antimicrobial Resistance
2.1. Definition
2.2. Mechanisms of Antimicrobial Resistance
2.3. Mechanisms of Horizontal Gene Transfer

2.3.1. Conjugation
2.3.2. Transformation
2.3.3. Transduction
3. Antimicrobial Resistance in Food
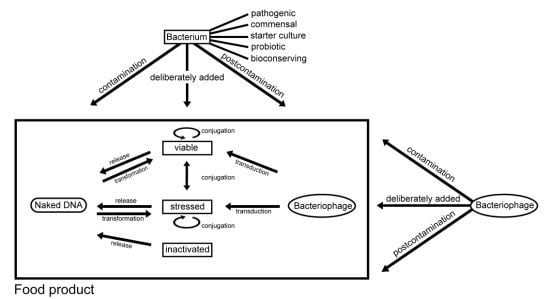
3.1. Contamination of Food with Antimicrobial Resistant Bacteria and Antimicrobial Resistance Genes
3.2. Intentional Addition of Microorganisms (with Antimicrobial Resistance Properties) to Food as Auxiliary Technical Substances
4. Transfer of Antimicrobial Resistance in the Food Processing Environment
4.1. Influence of Food Processing and Preservation Techniques
4.2. Influence of Biofilms
4.3. Cross-Resistance to Antibiotics and Chemical Biocides
5. Consequences of Foodborne Antimicrobial Resistance for the Consumer
6. Discussion and Conclusions
Acknowledgments
Conflict of Interest
References
- Carattoli, A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infec. 2008, 14, 117–123. [Google Scholar] [CrossRef]
- Depoorter, P.; Persoons, D.; Uyttendaele, M.; Butaye, P.; De Zutter, L.; Dierick, K.; Herman, L.; Imberechts, H.; Van Huffel, X.; Dewulf, J. Assessment of human exposure to 3rd generation cephalosporin resistant E. coli (CREC) through consumption of broiler meat in Belgium. Int. J. Food Microbiol. 2012, 159, 30–38. [Google Scholar] [CrossRef]
- Mayrhofer, S.; Paulsen, P.; Smulders, F.J.M.; Hilbert, F. Antimicrobial resistance in commensal Escherichia coli isolated from muscle foods as related to the veterinary use of antimicrobial agents in food-producing animals in Austria. Microb. Drug. Resist. 2006, 12, 278–283. [Google Scholar] [CrossRef]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial food animal production, antimicrobial resistance and human health. Annu. Rev. Publ. Health 2008, 29, 151–169. [Google Scholar] [CrossRef]
- Srinivasan, V.; Nam, H.-M.; Sawant, A.A.; Headrick, S.I.; Nguyen, L.T.; Oliver, S.P. Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 2008, 55, 184–193. [Google Scholar] [CrossRef]
- Stine, O.C.; Johnson, J.A.; Keefer-Norris, A.K.; Perry, K.L.; Tigno, J.; Qaiyumi, S.; Stine, M.S.; Morris, J.G., Jr. Widespread distribution of tetracycline resistance genes in a confined animal feeding facility. Int. J. Antmicrob. Ag. 2007, 29, 348–352. [Google Scholar] [CrossRef]
- Van Boxstael, S.; Dierick, K.; Van Huffel, X.; Uyttendaele, M.; Berkvens, D.; Herman, L.; Bertrand, S.; Wildemauwe, C.; Catry, B.; Butaye, P.; et al. Comparison of antimicrobial resistance patterns and phage types of Salmonella Typhimurium isolated from pigs, pork and humans in Belgium between 2001 and 2006. Food Res. Int. 2012, 45, 913–918. [Google Scholar] [CrossRef]
- Zirakzadeh, A.; Patel, R. Epidemiology and mechanisms of glycopeptide resistance in enterococci. Curr. Opin. Infect. Dis. 2005, 18, 507–512. [Google Scholar] [CrossRef]
- Zou, S.; Xu, W.; Zhang, R.; Tang, J.; Chen, Y.; Zhang, G. Occurrence and distribution of antibiotics in coastal water of the Bohai Bay, China: Impacts of river discharge and aquaculture activities. Environ. Pollut. 2011, 159, 2913–2920. [Google Scholar] [CrossRef]
- Miranda, C.D.; Kehrenberg, C.; Ulep, C.; Schwarz, S.; Roberts, M.C. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 2003, 47, 883–888. [Google Scholar] [CrossRef]
- FASFC. Advice 2003/07 of the Scientific Committee of the FASFC on Streptomycin Residues in Honey by Using the Product Fructocin on Apple Trees and Pear Trees. (in Dutch)(in French). Available online: www.favv-afsca.fgov.be/home/com-sci/avis03_fr.asp#07 (accessed on 27 August 2012).
- Acar, J.; Röstel, B. Antimicrobial resistance: An overview. Rev. Sci. Tech. OIE 2001, 20, 797–810. [Google Scholar]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef]
- McDonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar]
- Van Eldere, J. The Significance of in vitro Antibiotic Resistance. Available online: www.sbimc.org/2005/spring/slides/Vaneldere/Vaneldere.pdf (accessed on 30 August 2012).
- Maher, M.C.; Alemayehu, W.; Lakew, T.; Gaynor, B.D.; Haug, S.; Cevallos, V.; Keenan, J.D.; Lietman, T.M.; Porce, T.C. The fitness cost of antibiotic resistance in Streptococcus pneumoniae: Insight from the field. PLoS One 2012, 7, e29407:1–e29407:5. [Google Scholar] [CrossRef]
- Kang, Y.S.; Park, W. Trade-off between antibiotic resistance and biological fitness in Acinetobacter sp. strain DR1. Environ. Microbiol. 2010, 12, 1304–1318. [Google Scholar] [CrossRef]
- Rudolf, D.; Michaylov, N.; van der Linden, M.; Hoy, L.; Klugman, K.P.; Welte, T.; Pletz, M.W. International pneumococcal clones match or exceed the fitness of other strains despite the accumulation of antibiotic resistance. Antimicrob. Agents Chemother. 2011, 55, 4915–4917. [Google Scholar] [CrossRef]
- Livermore, D.M.; Woodford, N. The β-lactamase threat in Enterobacteriacea, Pseudomonas and Acinetobacter. Trends Microbiol. 2006, 14, 413–420. [Google Scholar] [CrossRef]
- Wright, G.D. Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 1999, 2, 499–503. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. R. 1997, 61, 1092–2172. [Google Scholar]
- Pinho, M.G.; Filipe, S.R.; de Lencastres, H.; Tomasz, A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001, 183, 6525–6531. [Google Scholar] [CrossRef]
- McMurry, L.; Petrucci, R.E., Jr.; Levy, S.B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Nat. Acad. Sci. USA 1980, 77, 3974–3977. [Google Scholar] [CrossRef]
- Pattishall, K.H.; Acar, J.; Burchall, J.J.; Goldstein, F.W.; Harvey, R.J. Two distinct types of trimethoprim-resistant dihydrofolate reductase specified by R-plasmids of different compatibility groups. J. Biol. Chem. 1977, 252, 2319–2323. [Google Scholar]
- Aarestrup, F.M. Antimicrobial Resistance in Bacteria of Animal Origin, 1st ed; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Viseur, N.; Lambert, M.L.; Delmée, M.; Van Broeck, J.; Catry, B. Nosocomial and non-nosocomial Clostridium difficile infections hospitalised patients in Belgium—Compulsory surveillance data from 2008 to 2010. Euro Surveill. 2011, 16, 5:1–5:5. [Google Scholar]
- Catry, B.; Croubel, S.; Schwarz, S.; Deprez, P.; Cox, B.; Kehrenberg, C.; Opsomer, G.; Decostere, A.; Haesebrouck, F. Influence of systemic fluoroquinolone administration on the presence of Pasteurella multocida in the upper respiratory tract of clinically healthy calves. Acta. Vet. Scand. 2008, 50, 36–39. [Google Scholar] [CrossRef]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Brit. J. Pharmacol. 2008, 153, 347–357. [Google Scholar] [CrossRef]
- Revilla, C.M.; Garcillán-Barcia, P.; Fernández-López, R.; Thomson, N.R.; Sanders, M.; Cheung, M.; Thomas, C.M.; de la Cruz, F. Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob. Agents Chemother. 2008, 52, 1472–1480. [Google Scholar] [CrossRef]
- Salyers, A.A.; Moon, K.; Schlesinger, D. The human intestinal tract—A hotbed of resistance gene transfer. Clin. Microbiol. Newsletter 2007, 29, 17–21. [Google Scholar] [CrossRef]
- Keese, P. Risks from GMOs due to horizontal gene transfer. Environ. Biosafety Res. 2008, 7, 123–149. [Google Scholar] [CrossRef]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guedon, G. Conjugative transposons: The tip of the iceberg. Mol. Microbiol. 2002, 46, 601–610. [Google Scholar] [CrossRef]
- Burrus, V.; Waldor, M.K. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004, 155, 376–386. [Google Scholar] [CrossRef]
- Wozniak, R.A.F.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar]
- Lyras, D.; Adams, V.; Lucet, I.; Rood, J.I. The large resolvase TnpX is the only transposon-encoded protein required for transposition of the Tn4451/3 family of integrative mobilizable elements. Mol. Microbiol. 2004, 51, 1787–1800. [Google Scholar] [CrossRef]
- Brochet, M.; Couve, E.; Glaser, P.; Guedon, G.; Payot, S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 2008, 190, 6913–6917. [Google Scholar] [CrossRef]
- Doublet, B.; Boyd, D.; Mulvey, M.R.; Cloeckaert, A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 2005, 55, 1911–1924. [Google Scholar] [CrossRef]
- Douard, G.; Praud, K.; Cloeckaert, A.; Doublet, B. The Salmonella genomic island 1 is specifically mobilized in trans by the lncA/C multidrug resistance plasmid family. PLoS One 2010, 5, e15302:1–e15302:8. [Google Scholar]
- Hall, R.M. Salmonella genomic islands and antibiotic resistance in Salmonella enterica. Future Microbiol. 2010, 5, 1525–1538. [Google Scholar] [CrossRef]
- Doublet, B.; Weill, F.X.; Fabre, L.; Chaslus-Dancla, E.; Cloeckaert, A. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar newport. Antimicrob. Agents Chemother. 2004, 48, 3806–3812. [Google Scholar] [CrossRef]
- Stokes, H.W.; Hall, R.M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions—Integrons. Mol. Microbiol. 1989, 3, 1669–1683. [Google Scholar] [CrossRef]
- Cambray, G.; Guerout, A.M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Nagachinta, S.; Chen, J. Integron-mediated antibiotic resistance in Shiga toxin-producing Escherichia coli. J. Food Protect. 2009, 72, 21–27. [Google Scholar]
- Van Meervenne, E.; Boon, N.; Verstraete, K.; Devlieghere, F.; De Reu, K.; Herman, L.; Buvens, G.; Piérard, D.; Van Coillie, E. Integron characterization and typing of Shiga toxin-producing Escherichia coli isolates in Belgium. J. Med. Microbiol. 2013, 62, 712–719. [Google Scholar] [CrossRef]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar]
- Andrup, L.; Damgaard, J.; Wassermann, K. Mobilization of small plasmids in Bacillus thuringiensis subsp. israelensis is accompanied by specific aggregation. J. Bacteriol. 1993, 175, 6530–6536. [Google Scholar]
- Kelly, B.G.; Verspermann, A.; Bolton, D.J. Horizonal gene transfer of virulence determinants in selected bacterial foodborne pathogens. Food Chem. Toxicol. 2008, 47, 969–977. [Google Scholar]
- Lorenz, M.G.; Wackernagel, W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994, 58, 563–602. [Google Scholar]
- Matsui, K.; Ishii, N.; Kawabata, Z. Release of extracellular transformable plasmid DNA from Escherchia coli cocultivated with algae. Appl. Environ. Microbiol. 2003, 69, 2399–2404. [Google Scholar] [CrossRef]
- Seitz, P.; Blokesch, M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microb. Rev. 2013, 37, 336–363. [Google Scholar] [CrossRef]
- Johnsborg, O.; Eldholm, V.; Håvarstein, L.S. Natural genetic transformation: Prevalence, mechanisms and function. Res. Microbiol. 2007, 158, 767–778. [Google Scholar] [CrossRef]
- Chen, I.; Christie, P.J.; Dubnau, D. The ins and outs of DNA transfer in bacteria. Science 2005, 310, 1456–1460. [Google Scholar] [CrossRef]
- Cérémonie, H.; Buret, F.; Simonet, P.; Vogel, T.M. Isolation of lightning-competent soil bacteria. Appl. Environ. Microb. 2004, 70, 6342–6346. [Google Scholar] [CrossRef]
- Cérémonie, H.; Buret, F.; Simonet, P.; Vogel, T.M. Natural Pseudomonas sp. strain N3 in artificial soil microscosms. Appl. Environ. Microb. 2006, 72, 2385–2389. [Google Scholar] [CrossRef]
- Davison, J. Genetic exchange between bacteria in the environment. Plasmid 1999, 42, 73–91. [Google Scholar] [CrossRef]
- Weiss, J.; Ros-Chumillas, M.; Pena, L.; Egea-Cortines, M. Effect of storage and processing on plasmid, yeast and plant genomic DNA stability in juice from genetically modified oranges. J. Biotechnol. 2007, 128, 194–203. [Google Scholar] [CrossRef]
- Van den Eede, G.; Aarts, A.; Buhk, H.-J.; Corthier, G.; Flint, H.J.; Hammes, J.; Jacobsen, B.; Midtvedt, T.; van der Vossen, J.; von Wright, A.; et al. The relevance of gene transfer to the safety of food and feed derived from genetically modified (GM) plants. Food Chem. Toxicol. 2004, 42, 1127–1156. [Google Scholar] [CrossRef]
- Straub, J.A.; Hertel, C.; Hammes, W.P. A 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J. Food Protect. 1999, 62, 1150–1156. [Google Scholar]
- Bauer, T.; Hammes, W.P.; Haase, N.U.; Hertel, C. Effect of food components and processing parameters on DNA degradation in food. Environ. Biosaf. Res. 2004, 3, 215–223. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Ge, Y.; Xu, B. Degradation of endogenous and exogenous genes of roundup ready soybean during food processing. J. Agr. Food Chem. 2005, 53, 10239–10243. [Google Scholar] [CrossRef]
- Kharazmi, M.; Bauer, T.; Hammes, W.P.; Hertel, C. Effect of food processing on the fate of DNA with regard to degradation and transformation capability in Bacillus subtilis. Syst. Appl. Microbiol. 2003, 26, 495–501. [Google Scholar] [CrossRef]
- De Vries, J.; Meier, P.; Wackernagel, W. The natural transformation of the soil bacterium Pseudomonas stutzeri and Acinetobacter sp. by transgenic plant DNA strictly depends on homologous sequences in recipient cells. FEMS Microbiol. Lett. 2001, 195, 211–215. [Google Scholar] [CrossRef]
- Simpson, D.J.; Dawson, L.F.; Fry, J.C.; Rogers, H.J.; Day, M.J. Influence of flanking homology insert size on the transformation frequency of Acinetobacter baylyi BD413. Environ. Biosaf. Res. 2007, 6, 55–69. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Holmfeldt, K.; Middelboe, M.; Nybroe, O.; Riemann, L. Large variabilities in host strains susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl. Environ. Microb. 2007, 73, 6703–6709. [Google Scholar]
- Jensen, E.C.; Schrader, H.S.; Rieland, B.; Thompson, T.L.; Lee, K.W.; Nickerson, K.W.; Kokjohn, T.A. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microb. 1998, 64, 575–580. [Google Scholar]
- Nakaminami, H.; Noguchi, N.; Nishijma, S.; Kurokawa, I.; So, H.; Sasatsu, M. Transduction of the plasmid encoding antiseptic resistance gene qacB in Staphylococcus aureus. Biol. Pharm. Bull. 2007, 30, 1412–1415. [Google Scholar] [CrossRef]
- Varga, M.; Kun Kuntová, L.; Pantůček, R.; Mašlaňová, I.; Růžičková, V.; Doškař, J. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 2012, 332, 146–152. [Google Scholar] [CrossRef]
- Bergogne-Bérézin, E. Who or what is the source of antibiotic resistance? J. Med. Microbiol. 1997, 46, 461–470. [Google Scholar]
- Walsh, C.; Duffy, G.; Nally, P.; O’Mahony, R.; McDowell, D.A.; Fanning, S. Transfer of ampicillin resistance from Salmonella Typhimurium DT104 to Escherichia coli K12 in food. Lett. Appl. Microbiol. 2008, 46, 210–215. [Google Scholar]
- Toomey, N.; Monaghan, A.; Fanning, S.; Bolton, D.J. Assessment of antimicrobial resistance transfer between lactic acid bacteria and potential foodborne pathogens using in vitro methods and mating in a food matrix. Foodborne Pathog. Dis. 2009, 6, 925–933. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Timmery, S.; Hoton, F.; Mahillon, J. Plasmid exchanges among members of the Bacillus cereus group in foodstuffs. Int. J. Food Microbiol. 2007, 113, 164–172. [Google Scholar] [CrossRef]
- Bezanson, G.S.; MacInnis, R.; Potter, G.; Hughes, T. Presence and potential for horizontal transfer of antibiotic resistance in oxidase-positive bacteria populating raw salad vegetables. Int. J. Food Microbiol. 2008, 127, 37–42. [Google Scholar] [CrossRef]
- Kharazmi, M.; Hammes, W.P.; Hertel, C. Construction of a marker rescue system in Bacillus subtilis for detection of horizontal gene transfer in food. Syst. Appl. Microbiol. 2002, 25, 471–477. [Google Scholar] [CrossRef]
- Zenz, K.I.; Neve, H.; Geis, A.; Heller, K.J. Bacillus subtilis develops competence for uptake of plasmid DNA when growing in milk products. Syst. Appl. Microbiol. 1998, 21, 28–32. [Google Scholar] [CrossRef]
- Jeon, B.; Muraoka, W.; Sahin, O.; Zhang, Q. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacer jejuni. Antimicrob. Agents Chemother. 2008, 52, 2699–2708. [Google Scholar] [CrossRef]
- Jeon, B.; Muraoka, W.; Scupham, A.; Zhang, Q. Roles of lipooligosaccharide and capsular polysaccharide in antimicrobial resistance and natural transformation of Campylobacter jejuni. J. Antimicrob. Chemother. 2009, 63, 462–468. [Google Scholar] [CrossRef]
- Hugas, M. Bacteriocinogenic lactic acid bacteria for the biopreservation of meat and meat products. Meat Sci. 1998, 49, 139–150. [Google Scholar] [CrossRef]
- Jacobsen, T.; Budde, B.B.; Koch, A.G. Application of Leuconostoc carnosum for bioperservation of cooked meat products. J. Appl. Microbiol. 2003, 95, 242–249. [Google Scholar] [CrossRef]
- Vermeiren, L.; Devlieghere, F.; Debevere, J. Evalutation of meat born lactic acid bacteria as protective cultures for the biopreservation of cooked meat products. Int. J. Food Microbiol. 2004, 96, 149–164. [Google Scholar] [CrossRef]
- Chahad, O.B.; Bour, M.E.; Calo-Mata, P.; Boudabous, A.; Barros-Velàzquez, J. Discovery of novel biopreservation agents with inhibitory effects on growth of food-borne pathogens and their application to seafood products. Res. Microbiol. 2012, 163, 44–45. [Google Scholar] [CrossRef]
- Olstorpe, M.; Passoth, V. Pichia anomala in grain biopreservation. A. Van Leeuw. J. Microb. 2011, 99, 57–62. [Google Scholar]
- Schnürer, J.; Jonsson, A. Pichia anomala J121: A 30-year overnight near success bioperservation story. A. Van Leeuw. J. Microb. 2011, 99, 5–12. [Google Scholar] [CrossRef]
- Sundh, I.; Melin, P. Safety and regulation of yeasts used for biocontrol or biopreservation in the food or feed chain. A. Van Leeuw. J. Microb. 2011, 99, 113–119. [Google Scholar] [CrossRef]
- El Bassi, L.; Hassouna, M.; Shinzato, N.; Matsui, T. Biopreservation of refrigerated and vacuum-pached Dicentrarchus labrax by lactic acid bacteria. J. Food Sci 2009, 74, 335–339. [Google Scholar] [CrossRef]
- EFSA. Foodborne antimicrobial resistance as a biological hazard. EFSA J. 2008, 765, 1–87.
- Teuber, M.; Meile, L.; Schwartz, F. Acquired antibiotic resistance in lactic acid bacteria from foods. A. Van Leeuw. J. Microb. 1999, 76, 115–137. [Google Scholar] [CrossRef]
- Masco, L.; van Hoorde, K.; de Brandt, E.; Swings, J.; Huys, G. Antimicrobial susceptibility of Bifidobacterium strains from humans, animals and probiotic products. J. Antimicrob. Chemother. 2006, 58, 85–94. [Google Scholar] [CrossRef]
- Gfeller, K.Y.; Roth, M.; Meile, L.; Teuber, M. Sequence and genetic organization of the 19.3-kb erythromycin and dalfopristin resistance plasmid pLME300 from Lactobacillus fermentum ROT1. Plasmid 2003, 50, 190–201. [Google Scholar] [CrossRef]
- Cataloluk, O.; Gogebakan, B. Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol. Lett. 2004, 236, 7–12. [Google Scholar]
- Gevers, D.; Masco, L.; Baert, L.; Huys, G.; Debevere, J.; Swings, J. Prevalence and diversity of tetracycline resistant lactic acid bacteria and their tet genes along the process line of fermented dry sausages. Syst. Appl. Microbiol. 2003, 26, 277–283. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Werner, G.; Huys, G.; Vankerckhoven, V.; Kahlmeter, G.; Hildebrandt, B.; Müller-Bertling, S.; Witte, W.; Goossens, H. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 2007, 59, 900–912. [Google Scholar] [CrossRef]
- Wang, H.H.; Manuzon, M.; Lehman, M.; Wan, K.; Luo, H.; Wittum, T.E.; Yousef, A.; Backaletz, L. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 2006, 254, 226–231. [Google Scholar] [CrossRef]
- Katla, A.-K.; Kruse, H.; Johnsen, G.; Herikstad, H. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 2001, 67, 147–152. [Google Scholar] [CrossRef]
- Kastner, S.; Perreten, V.; Bleuler, H.; Hugenschmidt, G.; Lacroix, C.; Meile, L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 2005, 29, 145–155. [Google Scholar]
- Resch, M.; Nagel, V.; Hertel, C. Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int. J. Food Microbiol. 2008, 127, 99–104. [Google Scholar] [CrossRef]
- Carlton, R.M.; Noordman, W.H.; Biswas, B.; De Meester, E.D.; Loessner, M.J. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharm. 2005, 43, 301–312. [Google Scholar] [CrossRef]
- Leverentz, B.; Conway, W.S.; Camp, M.J.; Janisiewicz, W.J.; Abuladze, T.; Yang, M.; Saftner, R.; Sulakvelidze, A. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 2003, 69, 4519–4526. [Google Scholar] [CrossRef]
- Leverentz, B.; Conway, W.S.; Janisiewicz, W.; Camp, M.J. Optimizing concentration and timing of a phage spray application to reduce Listeria monocytogenes on honeydew melon tissue. J. Food Protect. 2004, 67, 1682–1686. [Google Scholar]
- Atterbury, R.J.; Connerton, P.L.; Dodd, C.E.R.; Rees, C.E.D.; Connerton, I.F. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl. Environ. Microb. 2003, 69, 6302–6306. [Google Scholar] [CrossRef]
- Goode, D.; Allen, V.M.; Barrow, P.A. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microb. 2003, 69, 5032–5036. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 2011, 6, e17549:1–e17549:11. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Imamovic, L.; Jofre, J.; Muniesa, M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 2011, 55, 4908–4911. [Google Scholar] [CrossRef]
- Toomey, N.; Bolton, D.; Fanning, S. Characterisation and transferability of antibiotic resistance genes from lactic acid bacteria isolated from Irish pork and beef abattoirs. Res. Microbiol. 2010, 161, 127–135. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Chaofeng, M.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef]
- Vogel, R.F.; Becke-Schmid, M.; Entgens, P.; Gaier, W.; Hames, W.P. Plasmid transfer and segregation in Lactobacillus curvatus LTH1432 in vitro and during sausage fermentations. Syst. Appl. Microbiol. 1992, 15, 129–136. [Google Scholar]
- Cocconcelli, P.S.; Cattivelli, D.; Gazzola, S. Gene transfer of vancomycin and tetracycline resistances among Enterococcus faecalis during cheese and sausage fermentations. Int. J. Food Microbiol. 2003, 88, 315–323. [Google Scholar] [CrossRef]
- Chen, J.; Novick, R.P. Phage-mediated intergeneric transfer of toxin genes. Science 2009, 323, 139–141. [Google Scholar] [CrossRef]
- Brabban, A.D.; Hite, E.; Callaway, T.R. Evolution of foodborne pathogens via temperate bacteriophage-mediated gene transfer. Foodborne Pathog. Dis. 2005, 2, 287–303. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, X.M.; Zhang, Q.X.; Shen, Z.; Tian, F.W.; Zhang, H.; Sun, Z.H.; Zhang, H.P.; Chen, W. Influence of consumption of probiotics on the plasma lipid profile: A meta-anlysis of randomized controlled trials. Nutr. Metab. Cardiovas. 2011, 21, 844–850. [Google Scholar] [CrossRef]
- EFSA. Scientific Opinion on the evaluation of the safety and efficacy of ListexTM P100 for the removal of Listeria monocytogenes surface contamination of raw fish. EFSA J. 2012, 10, 2615:1–2615:43.
- Mattia, A.; Merker, R. Regulation of probiotic substances as ingredients in foods: Premarket approval of “Generally Recognized as Safe” notification. Clin. Infect. Dis. 2008, 46, S115–S118. [Google Scholar] [CrossRef]
- EFSA. Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA. EFSA J. 2007, 587, 1–16.
- Deák, T.; Farkas, J. Microbiology of Thermally Preserved Foods: Canning and Novel Physical Methods; DEStech Publications, Inc: Lancaster, PA, USA, 2013. [Google Scholar]
- Smigic, N.; Rajkovic, A.; Nielsen, D.S.; Arneborg, N.; Siegumfeldt, H.; Devlieghere, F. Survival of lactic acid and chlorine dioxide treated Campylobacter jejuni under suboptimal conditions of pH, temperature and modified atmosphere. Int. J. Food Microbiol. 2010, 141, S140–S146. [Google Scholar] [CrossRef]
- Allende, A.; Tomas-Barberan, F.A.; Gil, M.I. Minimal processing for healthy traditional foods. Trends Food Sci. Tech. 2006, 17, 513–519. [Google Scholar] [CrossRef]
- Farrell, H.P.; Garvey, M.; Cormican, M.; Laffey, J.G.; Rowan, N.J. Investigation of critical inter-related factors affecting the efficacy of pulsed light for inactivating clinically relevant bacterial pathogens. J. Appl. Microbiol. 2010, 108, 1494–1508. [Google Scholar] [CrossRef]
- Sivertsvik, M.; Jeksrud, W.K.; Rosnes, J.T. A review of modified atmosphere packaging of fish and fishery products—Significance of microbial growth, activities and safety. Int. J. Food Sci. Tech. 2002, 37, 107–127. [Google Scholar] [CrossRef]
- Van der Steen, C.; Jacxsens, L.; Devlieghere, F.; Debevere, J. Combining high oxygen atmospheres with low oxygen modified atmosphere packaging to improve the keeping quality of strawberries and raspberries. Postharvest Biol. Technol. 2002, 26, 49–58. [Google Scholar] [CrossRef]
- Rajkovic, A.; Smigic, N.; Devlieghere, F. Contemporary strategies in combating microbial contamination in food chain. Int. J. Food Microbiol. 2010, 141, S29–S42. [Google Scholar] [CrossRef]
- Wesche, A.M.; Gurtler, J.B.; Marks, B.P.; Ryser, E.T. Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J. Food Protect. 2009, 72, 1121–1138. [Google Scholar]
- McMahon, M.A.S.; Xu, J.; Moore, J.E.; Blair, I.S.; McDowell, D.A. Environmental stress and antibiotic resistance in food-related pathogens. Appl. Environ. Microbiol. 2007, 73, 211–217. [Google Scholar] [CrossRef]
- Ganjian, H.; Nikokar, I.; Tieshayar, A.; Mostafaei, A.; Amirmozafari, N.; Kiani, S. Effects of salt stress on the antimicrobial drug resistance and protein profile of Staphylococcus aureus. Jundishapur. J. Microbiol. 2012, 5, 328–331. [Google Scholar]
- Al-Nabulsi, A.A.; Osaili, T.M.; Elabedeen, N.A.; Jaradat, Z.W.; Shaker, R.R.; Kheirallah, K.A.; Tarazi, Y.H.; Holley, R.A. Impact of environmental stress desiccation, acidity, alkalinity, heat or cold on antibiotic susceptibility of Cronobacter sakazakii. Int. J. Food Microbiol. 2011, 146, 137–143. [Google Scholar] [CrossRef]
- McMahon, M.A.S.; Blair, I.S.; Moore, J.E.; McDowell, D.A. The rate of horizontal transmission of antibiotic resistance plasmids is increased in food preservation-stressed bacteria. J. Appl. Microbiol. 2007, 103, 1883–1888. [Google Scholar] [CrossRef]
- Capozzi, V.; Fiocco, D.; Amodio, M.L.; Gallone, A.; Spano, G. Bacterial stressors in minimally processed food. Int. J. Mol. Sci. 2009, 10, 3076–3105. [Google Scholar] [CrossRef]
- Van der Veen, S.; Abee, T. Bacterial SOS response: A food safety perspective. Curr. Opin. Biotech. 2011, 22, 136–142. [Google Scholar] [CrossRef]
- Cirz, R.T.; Chin, J.K.; Andes, D.R.; de Crecy-Lagard, V.; Craig, W.A.; Romesberg, F.E. Inhibition of mutation and combating the evolution of antibiotic resistance. PloS Biol. 2005, 3, 1024–1033. [Google Scholar]
- Leistner, L. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 2000, 55, 181–186. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Sofos, J.N.; Geornaras, I. Overview of current meat hygiene and safety risks and summary of recent studies on biofilms, and control of Escherichia coli O157:H7 in nonintact, and Listeria monocytogenes in ready-to-eat, meat products. Meat Sci. 2010, 86, 2–14. [Google Scholar] [CrossRef]
- Marchand, S.; De Block, J.; De Jonghe, V.; Coorevits, A.; Heyndrickx, M.; Herman, L. Biofilm formation in milk production and processing environments; influence on milk quality and safety. Compr. Rev. Food Sci. F. 2012, 11, 133–147. [Google Scholar] [CrossRef]
- Jahid, I.K.; Ha, S.D. A review of microbial biofilms of produce: Future challenge to food safety. Food Sci. Biotechnol. 2012, 21, 299–316. [Google Scholar] [CrossRef]
- Mah, T.F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Drenkard, E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 2003, 5, 1213–1219. [Google Scholar] [CrossRef]
- Król, J.E.; Nguyen, H.D.; Rogers, L.M.; Beyenal, H.; Krone, S.M.; Top, E.M. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl. Environ. Microbiol. 2011, 77, 5079–5088. [Google Scholar] [CrossRef]
- Molin, S.; Tolker-Nielsen, T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotech. 2003, 14, 255–261. [Google Scholar] [CrossRef]
- Reisner, A.; Höller, B.M.; Molin, S.; Zechner, E.L. Synergistic effects in mixed Escherichia coli biofilms: Conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 2006, 188, 3582–3588. [Google Scholar] [CrossRef]
- Hannan, S.; Ready, D.; Jasni, A.S.; Rogers, M.; Pratten, J.; Roberts, A.P. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Mic. 2010, 59, 345–349. [Google Scholar]
- Luo, H.L.; Wan, K.; Wang, H.H. High-frequency conjugation system facilitates biofilm formation and pAM beta 1 transmission by Lactococcus lactis. Appl. Environ. Microb. 2005, 71, 2970–2978. [Google Scholar] [CrossRef]
- Tumah, H.N. Bacterial biocide resistance. J. Chemother. 2009, 21, 5–15. [Google Scholar]
- Meyer, B. Does microbial resistance to biocides create a hazard to food hygiene? Int. J. Food Microbiol. 2006, 112, 275–279. [Google Scholar] [CrossRef]
- Buffet-Bataillon, S.; Branger, B.; Cormier, M.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Effect of higher minimum inhibitory concentrations of quaternary ammonium compounds in clinical E. coli isolates on antibiotic susceptibilities. J. Hosp. Infect. 2011, 79, 141–146. [Google Scholar] [CrossRef]
- Whitehead, R.N.; Overton, T.W.; Kemp, C.L.; Webber, M.A. Exposure of Salmonella enterica serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS One 2011, 6, e22833:1–e22833:9. [Google Scholar]
- Maseda, H.; Hashida, Y.; Konaka, R.; Shirai, A.; Kourai, H. Mutational upregulation of a resistance-nodulation-cell-division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob. Agents Chemother. 2009, 53, 5230–5235. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Webber, M.A.; Jorgensen, F.; Woodward, M.J.; Piddock, L.J.V.; Humphrey, T.J. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J. Antimicrob. Chemother. 2007, 60, 947–955. [Google Scholar] [CrossRef]
- Sidhu, M.S.; Heir, E.; Sørum, H.; Holck, A. Genetic linkage between resistance to quaternary ammonium compounds and beta-lactam antibiotics in food-related Staphylococcus spp. Microb. Drug Resist. 2001, 7, 363–371. [Google Scholar] [CrossRef]
- Sidhu, M.S.; Heir, E.; Leegaard, T.; Wiger, K.; Holck, A. Frequency of disinfectant resistance genes and genetic linkage with beta-lactamase transposon Tn552 among clinical staphylococci. Antimicrob. Agents Chemother. 2002, 46, 2797–2803. [Google Scholar] [CrossRef]
- IFH. Microbial Resistance and Biocides. A review by the International Scientific Forum on Home Hygiene. September 2000. Available online: www.ifh-homehygiene.org/sites/default/files/publications/antresFINAL.pdf (accessed on 27 August 2012).
- Mølbak, K. Spread of resistant bacteria and resistance genes from animals to humans—The public health consequences. J. Vet. Med. B 2004, 51, 364–369. [Google Scholar] [CrossRef]
- Mølbak, K. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 2005, 41, 1613–1620. [Google Scholar] [CrossRef]
- Streit, J.M.; Jones, R.N.; Toleman, M.A.; Stratchounski, L.S.; Fritsche, T.R. Prevalence and antimicrobial susceptibility patterns among gastroenteritis-causing pathogens recovered in Europe and Latin America and Salmonella isolates recovered from bloodstream infections in North America and Latin America: Report from the SENTRY antimicrobial surveillance program (2003). Int. J. Antmicrob. Ag. 2006, 27, 367–375. [Google Scholar]
- Varma, J.K.; Greene, K.D.; Ovitt, J.; Barrett, T.J.; Medalla, F.; Angulo, F.J. Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984–2002. Emerg. Infect. Dis. 2005, 11, 943–946. [Google Scholar] [CrossRef]
- Doucet-Populaire, F.; Trieu-Cuot, P.; Dosbaa, I.; Andremont, A.; Courvalin, P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 1991, 35, 185–187. [Google Scholar] [CrossRef]
- Fluit, A.C. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 2005, 43, 1–11. [Google Scholar] [CrossRef]
- Guerra, B.; Junker, E.; Miko, A.; Helmuth, R.; Mendoza, M.C. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 2004, 10, 83–91. [Google Scholar] [CrossRef]
- Gooderham, W.J.; Hancock, R.E.W. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2009, 33, 279–294. [Google Scholar] [CrossRef]
- Thomson, K.S.; Moland, E.S. Version 2000: The new β-lactamases of Gram-negative bacteria at the dawn of the new millennium. Microbes Infect. 2000, 2, 1225–1235. [Google Scholar] [CrossRef]
- Smet, A.; Rasschaert, G.; Martel, A.; Persoons, D.; Dewulf, J.; Butaye, P.; Catry, B.; Haesebrouck, F.; Herman, L.; Heyndrickx, M. In situ ESBL conjugation from avian to human Escherichia coli during cefotaxime administration. J. Appl. Microbiol. 2010, 110, 541–549. [Google Scholar]
- Leverstein-van Hall, M.A.; Dierickx, C.M.; Stuart, J.C.; Voets, G.M.; van den Munckhof, M.P.; van Essen-Zandbergen, A.; Platteel, T.; Fluit, A.C.; van de Sande-Bruinsma, N.; Scharinga, J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef]
- Septimus, E.J.; Kuper, K.M. Clinical challenges in addressing resistance to antimicrobial drugs in the twenty-first century. Clin. Pharmacol. Ther. 2009, 86, 336–339. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643-2669. https://doi.org/10.3390/ijerph10072643
Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, De Schaetzen M-A, Van Huffel X, Imberechts H, Dierick K, et al. Antimicrobial Resistance in the Food Chain: A Review. International Journal of Environmental Research and Public Health. 2013; 10(7):2643-2669. https://doi.org/10.3390/ijerph10072643
Chicago/Turabian StyleVerraes, Claire, Sigrid Van Boxstael, Eva Van Meervenne, Els Van Coillie, Patrick Butaye, Boudewijn Catry, Marie-Athénaïs De Schaetzen, Xavier Van Huffel, Hein Imberechts, Katelijne Dierick, and et al. 2013. "Antimicrobial Resistance in the Food Chain: A Review" International Journal of Environmental Research and Public Health 10, no. 7: 2643-2669. https://doi.org/10.3390/ijerph10072643
APA StyleVerraes, C., Van Boxstael, S., Van Meervenne, E., Van Coillie, E., Butaye, P., Catry, B., De Schaetzen, M.-A., Van Huffel, X., Imberechts, H., Dierick, K., Daube, G., Saegerman, C., De Block, J., Dewulf, J., & Herman, L. (2013). Antimicrobial Resistance in the Food Chain: A Review. International Journal of Environmental Research and Public Health, 10(7), 2643-2669. https://doi.org/10.3390/ijerph10072643