Risk Assessment of the Schmutzdecke of Biosand Filters: Identification of an Opportunistic Pathogen in Schmutzdecke Developed by an Unsafe Water Source
Abstract
:1. Introduction
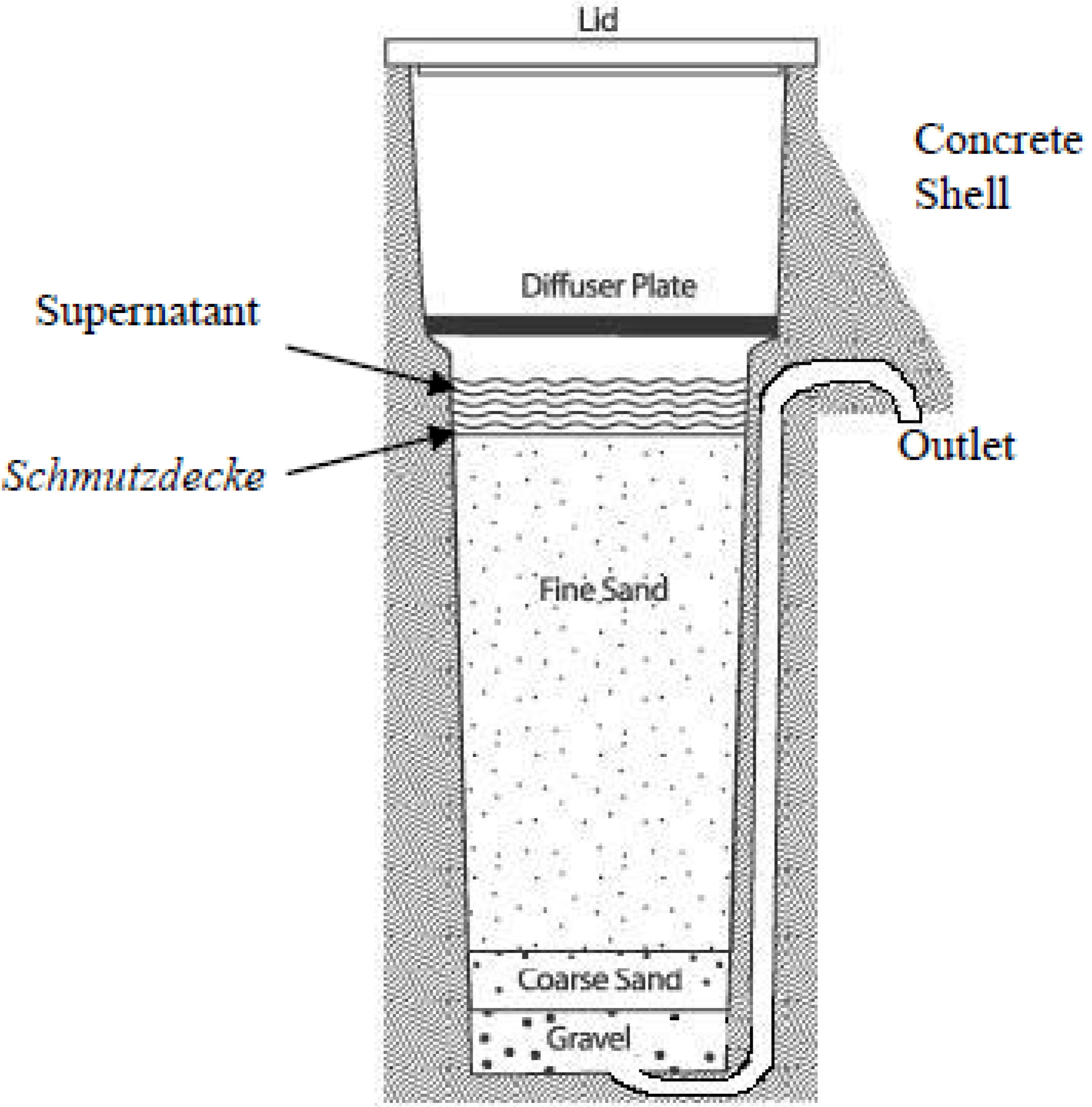
2. Experimental Section
2.1. Source of Samples
2.1.1. Sample Collection from Hyung-San River
2.1.2. Sample Collection from the Schmutzdecke (Biofilm) of Biosand Filters
2.2. DNA Isolation
2.3. PCR Amplification and Purification
2.4. 16S rRNA Gene Sequencing
2.5. Phylogenetic Analysis
3. Results
3.1. Comparing the Diversity and Proportions of Microorganisms from Two Different Sources
| Source | Labeling | Number of Isolates | Observed genus | Observed Phylum | % Strain of Opportunistic Pathogens |
|---|---|---|---|---|---|
| Hyung-San River | H | 17 | Novosphingobium, Catellibacterium, Aeromonas, Leclercia, Raoultella, Microbacterium, | Proteobacteria, Actinobacteria | 29% |
| Schmutzdecke (biofilm) of BSF | HB | 20 | Novosphingobium, Sphingomonas, Bradyrhizobium, Klebsiella, Enterobacter, Aeromonas, Pantoea, Cloacibacterium, Streptomyces, Arthrobacter Microbacterium, Brevibacillus. | Proteobacteria Actinobacteria, Fermicutes, Bacteroidetes | 55% |
3.2. Phylogenic Analysis of Isolated Strains from Samples
| Sl No. | Strain No. | Nearest Phylogenic Neighbor | 16S rRNA Gene Sequence Similarity % |
|---|---|---|---|
| Gram-negative bacterial strains | |||
| Proteobacteria | |||
| Alphaproteobacteria | |||
| 1 | H1 | Novosphingobium resinovorum strain SQ85 | 98.1 |
| 2 | H8 | Catellibacterium aquatile strain A1-9 | 98.0 |
| 3 | H16 | Novosphingobium subterraneum strain T4AR15 | 98.2 |
| 4 | HB12 | Novosphingobium sp. HU1-AH51 | 98.8 |
| 5 | HB13 | Sphingomonas sp. M16 | 99.7 |
| 6 | HB15 | Sphingomonas sp. M16 | 99.6 |
| 7 | HB17 | Novosphingobium aromaticivorans DSM_12444 | 97.7 |
| 8 | HB19 | Bradyrhizobium sp. CCBAU 7128301 | 99.0 |
| 9 | HB20 | Bradyrhizobium sp. CCBAU 7128301 | 98.6 |
| Gammaproteobacteria | |||
| 10 | H2 | Aeromonas hydrophila strain AN-3 | 99.8 |
| 11 | H4 | Leclercia adecarboxylata strain HPC21 | 99.6 |
| 12 | H5 | Raoultella ornithinolytica strain B18 | 99.7 |
| 13 | H10 | Aeromonas caviae strain T84 | 99.7 |
| 14 | H11 | Aeromonas hydrophila strain AN-3 | 99.6 |
| 15 | HB2 | Klebsiella oxytoca strain LF-1 | 99.5 |
| 16 | HB4 | Enterobacter aerogenes strain DCH-2 | 99.5 |
| 17 | HB5 | Aeromonas hydrophila strain AN-3 | 99.7 |
| 18 | HB6 | Pantoea agglomerans strain 1BJN10 | 99.2 |
| 19 | HB7 | Enterobacter cancerogenus strain KNUC5008 | 98.9 |
| 20 | HB14 | Pantoea agglomerans strain 1BJN10 | 99.7 |
| Bacteroidetes | |||
| Flavobacteria | |||
| 21 | HB8 | Cloacibacterium normanense strain tu29 | 98.2 |
| 22 | HB10 | Cloacibacterium rupense strain R2A-16 | 97.9 |
| 23 | HB18 | Cloacibacterium normanense strain tu29 | 98.0 |
| Gram-positive bacterial strains | |||
| Actinobacteria | |||
| Actinobacteridae | |||
| 24 | H3 | Microbacterium flavescens strain 173 | 99.0 |
| 25 | H6 | Microbacterium trichotecenolyticum strain 3370 | 99.8 |
| 26 | H7 | Microbacterium laevaniformans strain 1YJ19 | 99.6 |
| 27 | H9 | Microbacterium trichotecenolyticum strain 3370 | 99.7 |
| 28 | H12 | Microbacterium flavescens strain 173 | 99.3 |
| 29 | H13 | Microbacterium trichotecenolyticum strain 3370 | 99.6 |
| 30 | H14 | Microbacterium testaceum strain 343 | 99.7 |
| 31 | H15 | Microbacterium testaceum strain 343 | 99.5 |
| 32 | H17 | Microbacterium testaceum strain 343 | 99.6 |
| 33 | HB1 | Streptomyces sp. MJM3179 | 99.8 |
| 34 | HB3 | Arthrobacter oryzae strain T42 | 99.2 |
| 35 | HB9 | Microbacterium laevaniformans strain 1YJ19 | 99.5 |
| 36 | HB11 | Streptomyces sp. MJM3179 | 99.9 |
| Fermicutes | |||
| Bacilli | |||
| 37 | HB16 | Brevibacillus panacihumi strain C17 | 99.4 |


3.3. Identification of Opportunistic Pathogens and Their Associated Diseases
| Sl No. | Nearest Phylogenic Neighbor | Phylum | General Characteristics | Associated Human Disease | Reference |
|---|---|---|---|---|---|
| 1 | Raoultella ornithinolytica B6 | Proteobacteria | Gram-negative, aerobic/facultative anaerobic | Enteric fever-like syndrome and bacteremia | Victoria Pulian Morais et al., 2009 [15] |
| 2 | Aeromonas caviae, strain NCIMB 13016 | Proteobacteria | Gram-negative, facultative anaerobic | Gastrointestinal infectious disease | Meiyanti et al., 2010 [16] |
| 3 | Klebsiella oxytoca strain LF-1 | Proteobacteria | Gram-negative, anaerobic | Septic arthritis | Mendard A et al., 2010 [18] |
| 4 | Enterobacter aerogenes strain DCH-2 | Proteobacteria | Gram-negative, facultative aerobic | All kinds of infections | Irene G et al., 2007 [19] |
| 5 | Pantoea agglomerans strain 1BJN10 | Proteobacteria | Gram-negative, aerobic | soft tissue or bone/joint infections | Andrea T et al., 2007 [20] |
| 6 | Enterobacter cancerogenus strain KNUC5008 | Proteobacteria | Gram-negative, facultative anaerobic | Wound and urinary tract infection, sepsis, and osteomyelitis | I. Stock et al., 2002 [21] |
| 7 | Novosphingobium aromaticivorans DSM 12444 | Proteobacteria | Gram-negative, strictly aerobic | Autoimmune primary biliary cirrhosis induced by infection | Mohammed JP et al., 2011 [22] |
| 8 | Aeromonas hydrophila strain RB5-M1 | Proteobacteria | Gram-negative, facultative anaerobic | Mild diarrhea, life-threatening necrotizing fasciitis, septicemia, meningitis, cholera-like illness, and hemolytic-uremic syndrome | Grim CJ et al., 2013 [13] |
| 9 | Leclercia adecarboxylata strain HPC21 | Proteobacteria | Gram-negative, aerobic | Fever and leukocytosis | Zelalem Temesgen et al., 1997 [14] |
| 10 | Streptomyces sp. MJM3179 | Actinobacteria | Gram-positive, aerobic | Hypersensitivity | Monk et al., 2007 [17] |
| 11 | Sphingomonas sp. M16 | Proteobacteria | Gram-negative, aerobic | Infectious disease | David C White et al., 1996 [23] |
4. Discussion
5. Conclusions
Acknowledgments
Conflicts of Interest
Author Contributions
References
- Ashobolt, N.J. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 2004, 198, 229–238. [Google Scholar] [CrossRef]
- Sianipar, C.P.M.; Yudoko, G.; Dowaki, K.; Adhiutama, A. Design methodology for appropriate technology: Engineering as if people mattered. Sustainability 2013, 5, 3382–3425. [Google Scholar] [CrossRef]
- Clifford, M.J. Appropriate technology: The poetry of science. Sci. Christ. Belief. 2005, 17, 71–82. [Google Scholar]
- Centre for Affordable Water and Sanitation Technology (CAWST). 2012 Annual Report; CAWST: Alberta, AB, Canada, 2012; pp. 1–36. [Google Scholar]
- Biosand Filter Info. Projects. Available online: http://www.biosandfilters.info/projects (accessed on 26 December 2013).
- Stauber, C.E.; Elliott, M.A.; Ortiz, G.M.; DiGiano, F.A.; Sobsey, M.D. Charactersation of the biosand filter for E. coli reductions from housegold drinking water under controlled laboratory and filed use conditions. Water Sci. Technol. 2006, 54, 1–7. [Google Scholar]
- Palmateer, G.; Manz, D.; Jurkovic, A.; Mclnnis, R.; Unger, S.; Kwan, K.K.; Dutka, B.J. Toxicant and parasite challenge of manz intermittent slow sand filter. Environ. Toxicol. 1999, 14, 217–225. [Google Scholar] [CrossRef]
- Barnes, D.; Collin, C.; Ziff, S. The Biosand. Filter, Shiphon. Filter, and Rainwater Harvesting: Strategic Recommendation for New Water Treatment Technologies and Safe Water Storage to Pure Home Water; Massachusetts Institute of Technology: New York, NY, USA, 2009; pp. 1–61. [Google Scholar]
- World Health Organization (WHO). Slow Sand Filtration; WHO: Geneva, Switzerland, 1974; pp. 1–120. [Google Scholar]
- Centre for Affordable Water and Sanitation Technology (CAWST). Biosand Filter Manual; CAWST: Alberta, AB, Canada, 2009; pp. 1–129. [Google Scholar]
- Centre for Affordable Water and Sanitation Technology (CAWST). Household Water Treatment and Safe Storage Fact Sheet: Biosand Filter; CAWST: Alberta, AB, Canada, 2009; pp. 1–5. [Google Scholar]
- Basic Local Alignment Search Tool. Available online: http://blast.ncbi.nlm.nih.gov (accessed on 13 November 2013).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Gabriel, B. Microbial Indicators of Fecal Contamination: Application to Microbial Source Taking; University of Florida: Gainesville, FL, USA, 2005; pp. 1–71. [Google Scholar]
- Grim, C.J.; Kozlova, E.V.; Sha, J.; Fitts, E.C.; van Lier, C.J.; Kirtley, M.L.; Joseph, S.J.; Read, T.D.; Burd, E.M.; Tall, B.D.; et al. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. MBio 2013, 4, 1–13. [Google Scholar] [CrossRef]
- Temesgen, Z.; Douglas, R.T.; Franklin, R.C. Leclercia adecarboxylata infections: Case report and review. Clin. Infect. Dis. 1997, 25, 79–81. [Google Scholar]
- Morais, V.P.; Daporta, M.T.; Bao, A.F.; Campello, M.G. Enteric fever-like syndrome caused by Raoultella ornithinolytica (Klebsiella ornithinolytica). J. Clin. Microbiol. 2009, 47, 868–869. [Google Scholar] [CrossRef]
- Meiyanti; Salim, O.C.; Surjawidjaja, J.E.; Lesmana, M. Isolation and antibiotic sensitivity of Aeromonas from children with diarrhea. Univ. Med. 2010, 29, 14–20. [Google Scholar]
- Kapadia, M.; Rolston, K.V.; Han, X.Y. Invasive Streptomyces infections: Six cases and literature review. Am. J. Clin. Pathol. 2007, 127, 619–624. [Google Scholar] [CrossRef]
- Menard, A.; Harambat, J.; Pereyre, S.; Pontailler, J.R.; Megraud, F.; Richer, O. First report of septic arthritis caused by Klebsiela oxytoca. J. Clin. Microbiol. 2010, 48, 3021–3023. [Google Scholar] [CrossRef]
- Galani, I.; Souli, M.; Koratzanis, E.; Koratzanis, G.; Chryssouli, Z.; Glamerellou, H. Emerging bacterial pathogens: Escherichia coli, Enterobacter aerogenes and Proteus mirabilis clinical isolates harbouring the same transferable plasmid coding for metallo-beta-lactamase VIM-1 in Greece. J. Antimicrob. Chemoth. 2007, 59, 578–579. [Google Scholar] [CrossRef]
- Cruz, A.T.; Cazacu, A.C.; Allen, C.H. Pantoea agglomerans, a plant pathogen causing human disease. J. Clin. Microbiol. 2007, 45, 1989–1992. [Google Scholar] [CrossRef]
- Stock, I.; Wiedemann, B. Natural antibiotic susceptibility of Enterobacter amnigenus, Enterobacter cancerogenus, Enterobacter gergoviae and Enterobacter sakazakii strains. Clin. Microbiol. Infect. 2002, 8, 564–578. [Google Scholar] [CrossRef]
- Mohammed, J.P.; Fusakio, M.E.; Rainbow, D.B.; Moule, C.; Fraser, H.I.; Clark, J.; Todd, J.A.; Peterson, L.B.; Savage, P.B.; Wills-Karp, M.; et al. Identification of Cd101 as a susceptibility gene for Novosphingobium aromaticivorans-induced liver autoimmunity. J. Immunol. 2011, 187, 337–349. [Google Scholar]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Environ. Biotech. 1996, 7, 301–306. [Google Scholar]
- Gomez, V.B.; Calvo, C.; Vilchez, R.; Gonazlez, L.J.; Rodelas, B. TGGE analysis of the diversity of ammonia-oxidizing and denitrifying bacteria in submerged filter biofilms for the treatment of urban wastewater. Appl. Microbiol. Biot. 2006, 72, 393–400. [Google Scholar] [CrossRef]
- Feng, S.; Chen, C.; Wang, Q.; Yang, Z.; Zhang, X.; Xie, S. Microbial community in a full-scale drinking water biosand filter. J. Environ. Biol. 2013, 34, 321–324. [Google Scholar]
- Feng, S.; Chen, C.; Wang, Q.F.; Zhang, X.J.; Yang, Z.Y.; Xie, S.G. Characterization of microbial communities in a granular activated carbon-sand dual media filter for drinking water treatment. Int. J. Environ. Sci. Te. 2013, 10, 917–922. [Google Scholar] [CrossRef]
- Kolari, M. Attachment Mechanisms and Properties of Bacterial Biofilms on Non-Living Surfaces; University of Helsinki: Helsinki, Finland, 2003; pp. 1–79. [Google Scholar]
- Szewzyk, U.; Szewyk, R.; Manz, W.; Schleifer, K.H. Microbiological safety of drinking water. Annu. Rev. Microbiol. 2000, 54, 81–127. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hwang, H.G.; Kim, M.S.; Shin, S.M.; Hwang, C.W. Risk Assessment of the Schmutzdecke of Biosand Filters: Identification of an Opportunistic Pathogen in Schmutzdecke Developed by an Unsafe Water Source. Int. J. Environ. Res. Public Health 2014, 11, 2033-2048. https://doi.org/10.3390/ijerph110202033
Hwang HG, Kim MS, Shin SM, Hwang CW. Risk Assessment of the Schmutzdecke of Biosand Filters: Identification of an Opportunistic Pathogen in Schmutzdecke Developed by an Unsafe Water Source. International Journal of Environmental Research and Public Health. 2014; 11(2):2033-2048. https://doi.org/10.3390/ijerph110202033
Chicago/Turabian StyleHwang, Hyun Gyu, Min Seo Kim, Soo Min Shin, and Cher Won Hwang. 2014. "Risk Assessment of the Schmutzdecke of Biosand Filters: Identification of an Opportunistic Pathogen in Schmutzdecke Developed by an Unsafe Water Source" International Journal of Environmental Research and Public Health 11, no. 2: 2033-2048. https://doi.org/10.3390/ijerph110202033
APA StyleHwang, H. G., Kim, M. S., Shin, S. M., & Hwang, C. W. (2014). Risk Assessment of the Schmutzdecke of Biosand Filters: Identification of an Opportunistic Pathogen in Schmutzdecke Developed by an Unsafe Water Source. International Journal of Environmental Research and Public Health, 11(2), 2033-2048. https://doi.org/10.3390/ijerph110202033




