A Low-Cost Wheat Bran Medium for Biodegradation of the Benzidine-Based Carcinogenic Dye Trypan Blue Using a Microbial Consortium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Dye Used

2.2. Wheat Bran Medium
2.3. Enrichment of the Trypan Blue Decolorizing Microbial Consortium
2.4. Microbial Diversity Analysis
2.5. Pre-enrichment of the Microbial Consortium
2.6. Decolorization Studies
2.7. Optimization of Decolorization Parameters
2.8. Determination of Aromatic Amines, TOC and COD
2.9. Enzyme Studies
2.10. Biodegradation Studies
2.11. Toxicity Studies
2.12. Data Analysis
3. Results and Discussion
3.1. Microbial Diversity Analysis

3.2. Decolorization of Trypan Blue


3.3. Decolorization Parameters
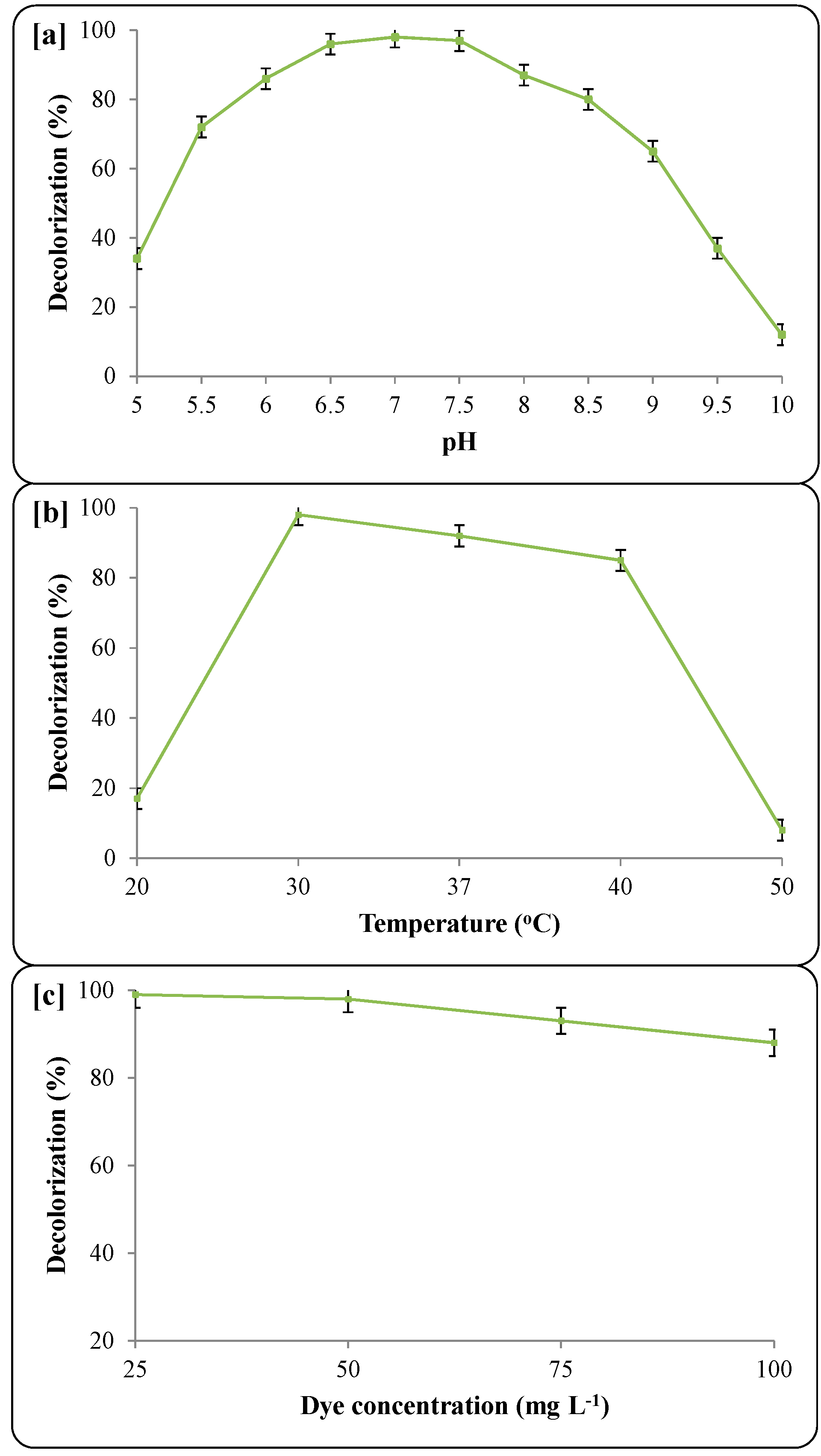
3.4. Mineralization of Dye
| Incubation Time (h) | Decolorization (%) | Amine Conc. (mM) |
|---|---|---|
| 0 | 0 ± 0.0 | n.d. |
| 8 | 34 ± 1.5 | 0.10 ± 0.01 |
| 16 | 70 ± 2.0 | 0.24 ± 0.02 |
| 24 | 98 ± 1.0 | n.d. |
| Environmental Parameters | Untreated Dye | After Decolorization (24 h) |
|---|---|---|
| TOC (mg∙L−1) | 1642 ± 3.0 | 591 ± 4.0 |
| COD (mg∙L−1) | 1265 ± 5.0 | 152 ± 3.0 |
| Color (%) | 100 ± 0.0 | 1 ± 1.0 |
3.5. Enzyme Studies
| Enzymes | Control (0 h) | Test (After Decolorization for 24 h) |
|---|---|---|
| Laccase 1 | 0.4 ± 0.01 | 1.5 ±0.03 *** |
| Tyrosinase 2 | 0.8 ± 0.02 | 0.97 ±0.03 ** |
| Veratryl alcohol oxidase 3 | 0.80 ± 0.02 | 1.3 ±0.05 *** |
| Azoreductase 4 | 0.6 ± 0.01 | 3.6 ±0.04 *** |
| NADH-DCIP reductase 5 | 12.2 ± 1.1 | 44.4 ±4.2 *** |
3.6. Biodegradation Studies

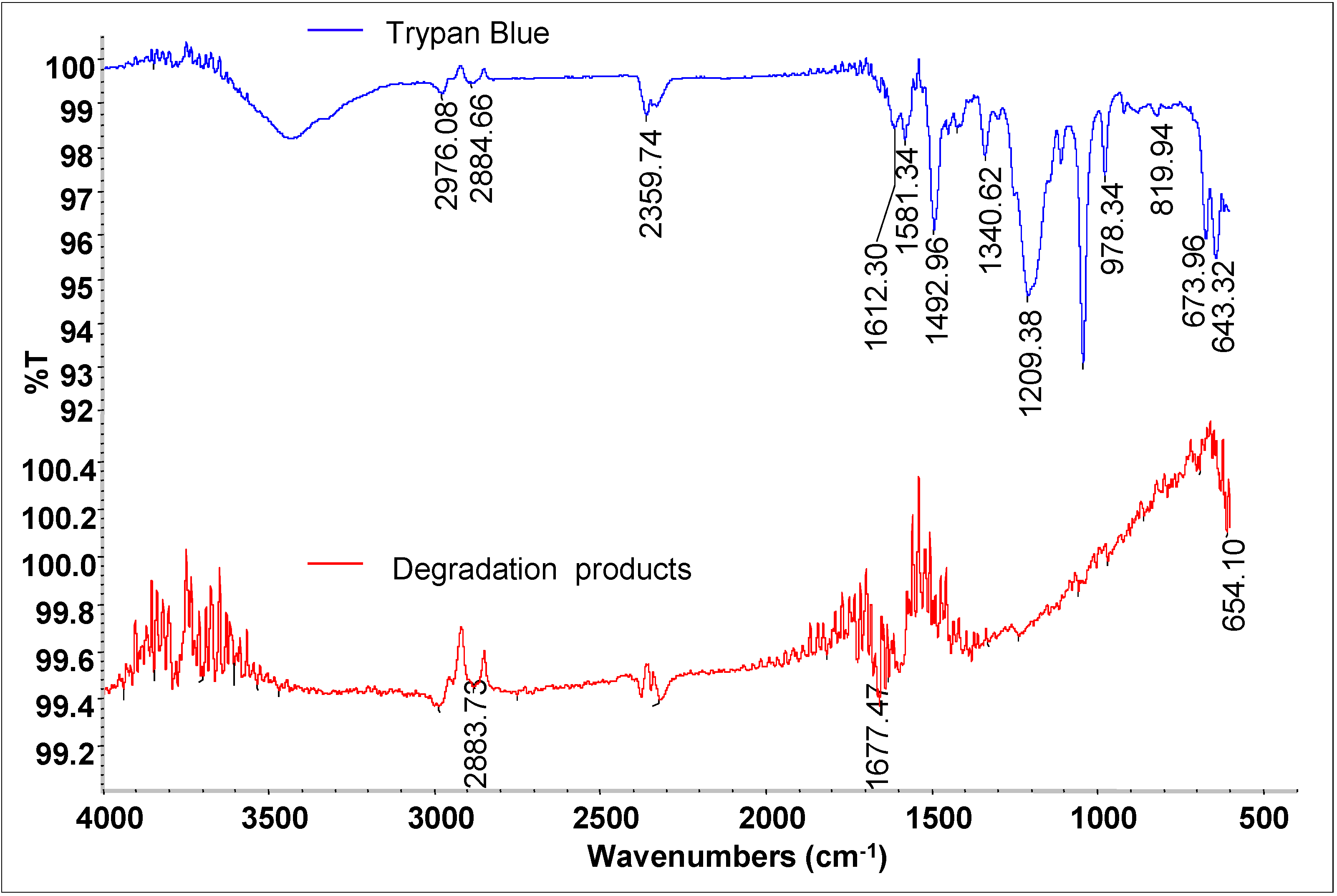
| Retention Time (min) | Mol. Weight (m/z) | Name of Metabolite | Mass Spectrum |
|---|---|---|---|
| 23.152 | 378 (m/z = 380) (+2) | Disodium 3,5-diamino-4-hydroxynaphthalene-2,7-disulfonate [I] |  |
| 17.102 | 144 (m/z = 143) (−1) | Naphthalen-1-ol [II] | 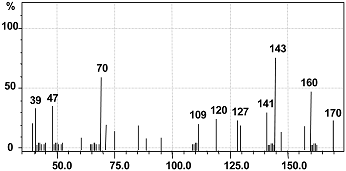 |

3.7. Toxicity Studies
| Samples | S. vulgare | P. mungo | ||||
|---|---|---|---|---|---|---|
| Germination (%) | Shoot Length (cm) | Root Length (cm) | Germination (%) | Shoot Length (cm) | Root Length (cm) | |
| Distilled water | 100 | 9.8 ± 0.2 | 4.1 ± 0.2 | 100 | 10.2 ± 0.3 | 4.8 ± 0.3 |
| Trypan Blue | 20 | 3.5 ± 0.2 * | 1.9 ± 0.1 * | 30 | 4.9 ± 0.2 * | 1.8 ± 0.2 * |
| Degradation products | 90 | 9.5 ± 0.3 | 3.8 ± 0.2 | 90 | 10.1 ± 0.2 | 4.6 ± 0.2 |
| Samples | Mortality (%) |
|---|---|
| Distilled water | 0 ± 0.0 |
| Trypan Blue | 60 ± 3.0 |
| Decolorized medium | 0 ± 0.0 |
4. Conclusions
Acknowledgments
Author Contributions
Conflict of Interests
References
- US EPA. Dyes Derived from Benzidine and Its Congeners. U.S. Environmental Protection Agency. 2010. Available online: http://www.epa.gov/oppt/existingchemicals/pubs/actionplans/benzidine.html (accessed on 2 March 2015). [Google Scholar]
- CEPA (Canadian Environmental Protection Act., 1999): Notice with Respect to Certain Aromatic Amines and Aromatic Azo- and Benzidine-Based Substances. Canada Gazette, Part I, Volume 145, No. 51, Supplement to the Canada Gazette. Available online: www.gazette.gc.ca/rp-pr/p1/2011/index-eng.html (accessed on 2 March 2015).
- Morton, K.C.; King, K.M.; Baetcke, K.P. Metabolism of benzidine and subsequent nucleic acid binding and mutagenicity. Cancer Res. 1979, 39, 3107–3113. [Google Scholar]
- Morgan, D.L.; Dunnick, J.K.; Goehl, T.; Jokinen, M.P.; Matthews, H.B.; Zeiger, E.; Mennear, J.H. Summary of the National Toxicology Program benzidine dye initiative. Environ. Health Perspect. 1994, 102, 63–78. [Google Scholar]
- Combes, R.D.; Haveland-Smith, R.B. A review of the genotoxicity of food, drug and cosmetic colours and other azo triphenylmethane and xanthene dyes. Mutat. Res. 1982, 98, 101–248. [Google Scholar]
- Chung, K.T.; Cerniglia, C.E. Mutagenicity of azo dyes: Structure-activity relationships. Mutat. Res. 1992, 277, 201–220. [Google Scholar]
- Turesky, R.J. Interspecies metabolism of heterocyclic aromatic amines and the uncertainties in extrapolation of animal toxicity data for human risk assessment. Mol. Nutr. Food Res. 2005, 49, 101–117. [Google Scholar]
- Dumont, J.; Josse, R.; Lambert, C.; Antherieu, S.; Le Hegarat, L.; Aninat, C.; Robin, M.A.; Guguen-Guillouzo, C.; Guillouzo, A. Differential toxicity of hetero- cyclic aromatic amines and their mixture in metabolically competent HepaRG cells. Toxicol. Appl. Pharmacol. 2010, 245, 256–263. [Google Scholar]
- Jager, I.; Hafner, C.; Schneider, K. Mutagenicity of different textile dye products in Salmonella typhimurium and mouse lymphoma cells. Mutat. Res. 2004, 561, 35–44. [Google Scholar]
- Brown, J.P.; Dietrich, P.S. Mutagenicity of selected sulfonated azo dyes in the Salmonella/microsome assay: Use of aerobic and anaerobic activation procedures. Mutat. Res. 1983, 116, 305–315. [Google Scholar]
- Gillman, T.; Kinns, A.M.; Hallowes, R.C.; Lloyd, J.B. Malignant lymphoreticular tumors induced by trypan blue and transplanted in inbred rats. J. Natl. Cancer Inst. 1973, 50, 1179–1193. [Google Scholar]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar]
- Kadam, A.A.; Lade, H.S.; Patil, S.M.; Govindwar, S.P. Low cost CaCl2 pretreatment of sugarcane bagasse for enhancement of textile dyes adsorption and subsequent biodegradation of adsorbed dyes under solid state fermentation. Bioresour. Technol. 2013, 132, 276–284. [Google Scholar]
- Dutta, A.; Banerjee, P.; Sarkar, D.; Bhattacharjee, S.; Chakrabarti, S. Degradation of Trypan Blue in wastewater by sunlight-assisted modified photo-Fenton reaction. Desalin. Water Treat. 2014, 8, 1–9. [Google Scholar]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Decolorization and biodegradation of textile dye Navy blue HER by Trichosporon beigelii NCIM-3326. J. Hazard. Mater. 2009, 166, 1421–1428. [Google Scholar]
- Lucas, M.S.; Peres, J.A. Decolorization of the azo dyeReactive Black 5 by Fenton and photo-Fenton oxidation. Dyes Pigm. 2006, 71, 236–244. [Google Scholar]
- Robinson, T.; Nigam, P. Remediation of textile dye waste water using a white rot fungus Bjerkandera adusta through solid-state fermentation (SSF). Appl. Biochem. Biotechnol. 2008, 151, 618–628. [Google Scholar]
- Muruganandham, M.; Swaminathan, M. Solar driven decolourisation of Reactive Yellow 14 by advanced oxidation processes in heterogeneous and homoge-neous media. Dyes Pigm. 2007, 72, 137–143. [Google Scholar]
- Muller, J.P.; Jekel, M. Comparison of advanced oxida-tion processes if flow-through pilot plants (Part I). Water Sci. Technol. 2001, 44, 303–309. [Google Scholar]
- Rosenfeldt, E.J.; Linden, K.G.; Canonica, S.; Von, G.U. Comparison of the efficiency of *OH radical formation during ozonation and the advanced oxidation processes O3/H2O2 and UV/H2O2. Water Res. 2006, 40, 3695–3704. [Google Scholar]
- Butani, N.; Jobanputra, J.; Bhatiya, P.; Patel, R. Recent biological technologies for textile effluent treatment. Int. Res. J. Biol. Sci. 2013, 2, 77–82. [Google Scholar]
- Kalyani, D.C.; Telke, A.A.; Dhanve, R.S.; Jadhav, J.P. Ecofriendly biodegradation and detoxification of Reactive Red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J. Hazard. Mater. 2009, 163, 735–742. [Google Scholar]
- Parshetti, G.K.; Kalme, S.D.; Gomare, S.S.; Govindwar, S.P. Biodegradation of reactive blue-25 by Aspergillus. ochraceus NCIM-1146. Bioresour. Technol. 2007, 98, 3638–3642. [Google Scholar]
- Khataee, A.R.; Zarei, M.; Pourhassan, M. Application of microalga Chlamydomonas sp. for biosorptive removal of a textile dye from contaminated water: Modelling by a neural network. Environ. Technol. 2009, 30, 1615–1623. [Google Scholar]
- Kulkarni, A.N.; Kadam, A.A.; Kachole, M.S.; Govindwar, S.P. Lichen Permelia perlata: A novel system for biodegradation and detoxification of disperse dye Solvent Red 24. J. Hazard. Mater. 2014, 276, 461–468. [Google Scholar]
- Khandare, R.V.; Kabra, A.N.; Kurade, M.B.; Govindwar, S.P. Phytoremediation potential of Portulaca grandiflora Hook. (Moss-Rose) in degrading a sulfonated diazo reactive dye Navy Blue HE2R (Reactive Blue 172). Bioresour. Technol. 2011, 102, 6774–6777. [Google Scholar]
- Lade, H.S.; Waghmode, T.R.; Kadam, A.A.; Govindwar, S.P. Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int. Biodeterior. Biodegrad. 2012, 72, 94–107. [Google Scholar]
- Kabra, A.N.; Khandare, R.V.; Govindwar, S.P. Development of a bioreactor for remediation of textile effluent and dye mixture: A plant-bacterial synergistic strategy. Water Res. 2013, 47, 1035–1048. [Google Scholar]
- Singh, R.; Kapoor, V.; Kumar, V. Utilization of agro-industrial wastes for the simultaneous production of amylase and xylanase by thermophilic Actinomycetes. Braz. J. Microbiol. 2012, 43, 1545–1552. [Google Scholar]
- Kadam, A.A.; Telke, A.A.; Jagtap, S.S.; Govindwar, S.P. Decolorization of adsorbed textile dyes by developed consortium of Pseudomonas sp. SUK1 and Aspergillus. ochraceus NCIM-1146 under solid state fermentation. J. Hazard. Mater. 2011, 189, 486–494. [Google Scholar]
- Kadam, A.A.; Kamatkar, J.D.; Khandare, R.V.; Jadhav, J.P.; Govindwar, S.P. Solid-state fermentation: Tool for bioremediation of adsorbed textile dyestuff on distillery industry waste-yeast biomass using isolated Bacillus cereus strain EBT1. Environ. Sci. Poll. Res. 2013, 20, 1009–1020. [Google Scholar]
- Waghmare, P.R.; Kadam, A.A.; Saratale, G.D.; Govindwar, S.P. Enzymatic hydrolysis and characterization of waste lignocellulosic biomass produced after dye bioremediation under solid state fermentation. Bioresour. Technol. 2014, 168, 136–141. [Google Scholar]
- Krishna, C. Solid-state fermentation systems-an overview. Crit. Rev. Biotechnol. 2005, 25, 1–30. [Google Scholar]
- Gomathi, D.; Muthulakshmi, C.; Kumar, D.G.; Ravikumar, G.; Kalaiselvi, M.; Uma, C. Submerged fermentation of wheat bran by Aspergillus. flavus for production and characterization of carboxy methyl cellulose. Asian Pac. J. Trop Biomed. 2012, 2, S67–S73. [Google Scholar]
- Liang, Y.L.; Zhang, Z.; Wu, M.; Wu, Y.; Feng, J.-X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus. terrae ME27–1. BioMed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- El-Shishtawy, R.M.; Mohamed, S.A.; Asiri, A.M.; Gomaa, A.B.; Ibrahim, I.H.; Al-Talhi, H.A. Solid fermentation of wheat bran for hydrolytic enzymes production and saccharification content by a local isolate Bacillus megatherium. BMC Biotechnol. 2014, 14, 1–19. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Preparation and analysis of DNA. In Current Protocols in Molecular Biology; John Wiley and Sons: New York, NY, USA, 1997; p. 66. [Google Scholar]
- Joshi, S.M.; Inamdar, S.A.; Patil, S.M.; Govindwar, S.P. Molecular assessment of shift in bacterial community in response to Congo Red. Int. Biodeterior. Biodegrad. 2013, 77, 18–21. [Google Scholar]
- Marik, J.; Song, A.; Lam, K.S. Detection of primary aromatic amines on solid phase. Tetrahedron Lett. 2003, 44, 4319–4320. [Google Scholar]
- APHA. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Wolfenden, B.S.; Willson, R.L. Radical-cations as reference chromogens in kinetic studies of ono-electron transfer reactions: pulse radiolysis studies of 2,2ʹ-azinobis-(3-ethylbenzthiazoline-6-sulphonate). J. Chem. Soc. Perkin Trans. 1982, 2, 805–812. [Google Scholar]
- Zhang, X.; Flurkey, W.H. Phenoloxidases in Portabella mushrooms. J. Food Sci. 1997, 62, 97–100. [Google Scholar]
- Jadhav, U.U.; Dawkar, V.V.; Tamboli, D.P.; Govindwar, S.P. Purification and characterization of veratryl alcohol oxidase from Comamonas sp. UVS and its role in decolorization of textile dyes. Biotechnol. Bioprocess. Eng. 2009, 14, 369–376. [Google Scholar]
- Chen, H.; Hopper, S.L.; Cerniglia, C.E. Biochemical and molecular characterization of an azoreductase from Staphylococcus aereus, a tetrameric NADPH-dependent flavoprotein. Microbiology 2005, 151, 1433–1441. [Google Scholar]
- Salokhe, M.D.; Govindwar, S.P. Effect of carbon source on the biotransformation enzyme in Serratia. marcescens. World J. Microbiol. Biotechnol. 1999, 15, 259–263. [Google Scholar]
- Balapure, K.H.; Jain, K.; Chattaraj, S.; Bhatt, N.S. Madamwar, D. Co-metabolic degradation of diazo dye- reactive blue 160 by enriched mixed cultures BDN. J. Hazard. Mater. 2014, 279, 85–95. [Google Scholar]
- Agrawal, S.; Tipreb, D.; Patelc, B.; Daveb, S. Optimization of triazo Acid Black 210 dye degradation by Providencia sp. SRS82 and elucidation of degradation pathway. Process. Biochem. 2014, 49, 110–119. [Google Scholar]
- Jain, K.; Shah, V.; Chapla, D.; Madamwar, D. Decolorization and degradation of azo dye—Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. J. Hazard. Mater. 2012, 213–214, 378–386. [Google Scholar]
- Franciscon, E.; Zille, A.; Dias, G.F.; Ragagnin, M.C.; Durrant, L.R.; Cavaco-Paulo, A. Biodegradation of textile azo dyes by a facultative Staphylococcus arlettae strain VN-11 using a sequential microaerophilic/aerobic process. Int. Biodeterior. Biodegrad. 2009, 63, 280–288. [Google Scholar]
- Franciscon, E.; Zille, A.; Durrant, L.R.; Fantinatti, G.F.; Cavaco-Paulo, A. Microaerophilic-aerobic sequential decolourization/biodegradation of textile azo dyes by a facultative Klebsiella sp. strain VN-31. Process. Biochem. 2009, 44, 446–452. [Google Scholar]
- Voordouw, G.; Niviere, V.; Ferris, F.G.; Fedorak, P.M.; Westlake, D.W.S. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ. Microbiol. 1990, 56, 3748–3754. [Google Scholar]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar]
- Annuar, M.S.M.; Adnan, S.; Vikineswary, S.; Chisti, Y. Kinetics and energetics of azo dye decolorization by Pycnoporus sanguineus. Water Air Soil Poll. 2009, 202, 179–188. [Google Scholar]
- Taha, M.; Adetutu, E.M.; Shahsavari, E.; Smith, A.T.; Ball, A.S. Azo and anthraquinone dye mixture decolourization at elevated temperature and concentration by a newly isolated thermophilic fungus, Thermomucor. indicae-seudaticae. J. Environ. Chem. Eng. 2014, 2, 415–423. [Google Scholar]
- Chung, K.T.; Stevens, S.E. Degradation of azo dyes by environmental microorganisms and helminthes. Environ. Toxicol. Chem. 1993, 12, 2121–2132. [Google Scholar]
- Hu, T.L. Degradation of azo dye RP2B by Pseudomonas luteola. Water Sci. Technol. 1998, 38, 299–306. [Google Scholar]
- Phugare, S.S.; Kalyani, D.C.; Patil, A.V.; Jadhav, J.P. Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J. Hazard. Mater. 2011, 186, 713–723. [Google Scholar]
- Lourenco, N.D.; Novais, J.M.; Pinheiro, H.M. Reactive textile dye colour removal in a sequencing batch reactor. Water Sci. Technol. 2000, 42, 321–328. [Google Scholar]
- Phugare, S.S.; Kalyani, D.C.; Surwase, S.N.; Jadhav, J.P. Ecofriendly degradation, decolorization and detoxification of textile effluent by a developed bacterial consortium. Ecotoxicol. Environ. Saf. 2011, 74, 1288–1296. [Google Scholar]
- Pandey, A.; Singh, P.; Iyengar, L. Bacterial decolorization and degradation of azo dyes. Int. Biodeterior. Biodegrad. 2007, 59, 73–84. [Google Scholar]
- Pearce, C.I.; Lloyd, J.R.; Guthrie, J.T. The removal of colour from textile wastewater using whole bacterial cells: A review. Dyes Pigm. 2003, 58, 179–196. [Google Scholar]
- Kadpan, I.K.; Kargi, F.; McMullan, G.; Marchant, R. Effect of environmental conditions on biological decolorization of textile dyestuff by C. versicolor. Enzyme Microb. Technol. 2000, 26, 381–387. [Google Scholar]
- Levine, W.G. Metabolism of azo dyes: Implication for detoxification and activation. Drug Metab. Rev. 1991, 23, 253–309. [Google Scholar]
- Singh, P.; Sanghi, R.; Pandey, A.; Iyengar, L. Decolorization and partial degradation of monoazo dyes in sequential fixed-filmed anaerobic batch reactor (SFABR). Bioresour. Technol. 2007, 98, 2053–2056. [Google Scholar]
- Telke, A.A.; Ghodake, G.S.; Kalyani, D.C.; Dhanve, R.S.; Govindwar, S.P. Biochemical characteristics of a textile dye degrading extracellular laccase from a Bacillus sp. ADR. Bioresour. Technol. 2011, 102, 1752–1756. [Google Scholar]
- Telke, A.; Kadam, A.; Govindwar, S. Bacterial enzymes and their role in decolorization of azo dyes. In Microbial Degradation of Synthetic Dyes in Wastewaters; Singh, S.N., Ed.; Springer International Publishing: Basel, Switzerland, 2015; Volume XIV, pp. 149–168. [Google Scholar]
- Zimmerman, T.; Kulla, H.G.; Leisinger, T. Properties of purified orange II azo reductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur. J. Biochem. 1982, 129, 197–203. [Google Scholar]
- Telke, A.; Kadam, A.; Jagtap, S.; Jadhav, J.; Govindwar, S. Biochemical characterization and potential for textile dye degradation of blue laccase from Aspergillus ochraceus NCIM-1146. Biotechnol. Bioprocess. Eng. 2010, 15, 696–703. [Google Scholar]
- Balapure, K.; Bhatt, N.; Madamwar, D. Mineralization of reactive azo dyes present in simulated textile waste water using down flow microaerophilic fixed film bioreactor. Bioresour. Technol. 2015, 175C, 1–7. [Google Scholar]
- Russ, R.; Rau, J.; Stolz, A. The function of cytoplasmic flavin reductases in the reduction of azo dyes by bacteria. Appl. Environ. Microbiol. 2000, 66, 1429–1434. [Google Scholar]
- Chang, J.S.; Lin, C.Y. Decolorization kinetics of a recombinant Escheria. coli strain harboring azo dye decolorizing determinants from Rhodococcus sp. Biotechnol. Lett. 2001, 23, 631–636. [Google Scholar]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Decolorization and biodegradation of reactive dyes and dye wastewater by a developed bacterial consortium. Biodegradation 2010, 21, 999–1015. [Google Scholar]
- Guilhermino, L.; Diamantino, T.; Silva, M.C.; Soares, A.M. Acute toxicity test with Daphnia magna: An alternative to mammals in the prescreening of chemical toxicity. Ecotoxicol. Environ. Saf. 2000, 46, 357–362. [Google Scholar]
- Villegas-Navarro, A.; González, M.C.R.; López, E.R.; Aguilar, R.D.; Marçal, W.S. Evaluation of Daphnia magna as an indicator of toxicity and treatment efficacy of textile wastewaters. Environ. Int. 1999, 25, 619–624. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lade, H.; Kadam, A.; Paul, D.; Govindwar, S. A Low-Cost Wheat Bran Medium for Biodegradation of the Benzidine-Based Carcinogenic Dye Trypan Blue Using a Microbial Consortium. Int. J. Environ. Res. Public Health 2015, 12, 3480-3505. https://doi.org/10.3390/ijerph120403480
Lade H, Kadam A, Paul D, Govindwar S. A Low-Cost Wheat Bran Medium for Biodegradation of the Benzidine-Based Carcinogenic Dye Trypan Blue Using a Microbial Consortium. International Journal of Environmental Research and Public Health. 2015; 12(4):3480-3505. https://doi.org/10.3390/ijerph120403480
Chicago/Turabian StyleLade, Harshad, Avinash Kadam, Diby Paul, and Sanjay Govindwar. 2015. "A Low-Cost Wheat Bran Medium for Biodegradation of the Benzidine-Based Carcinogenic Dye Trypan Blue Using a Microbial Consortium" International Journal of Environmental Research and Public Health 12, no. 4: 3480-3505. https://doi.org/10.3390/ijerph120403480









