The Impact of Human Activities on Microbial Quality of Rivers in the Vhembe District, South Africa
Abstract
:1. Introduction
2. Experimental Section
2.1. Study Area and Sample Collection
2.2. Detecting the Presence of Indicator Organisms in River Samples
2.3. Identification of Pathogenic E. coli Strains
2.3.1. Growth and Maintenance of Bacterial Strains
2.3.2. Characterization of E. coli Communities with Multiplex PCR (m-PCR)
2.3.3. Visualization of PCR Products
2.4. Risk Assessment Analysis
2.5. Data and Statistical Analyses
3. Results and Discussion
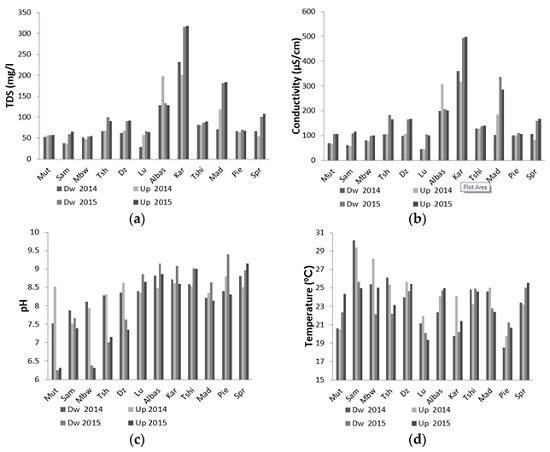
3.1. Physical Parameters
3.2. Microbiological Results
3.3. Pathogens Identified in Water Samples
3.4. Risk Activities in Rivers and Dams
3.5. Risk Assessment and Seasonal Variation of Microbiological Water Quality Monitoring
3.6. Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kurup, R.; Persuad, R.; Ceaser, J.; Raja, V. Microbial and Physiochemical Analysis in Georgetown, Guyana. Nat. Sci. 2010, 8, 261–265. [Google Scholar]
- Zamxala, M.; Pironcheva, G.; Muyima, N.Y.O. Microbiological and physico-chemical assessment of the quality of domestic water sources in selected rural communities of the Eastern Cape Province, South Africa. Water SA 2004, 30, 333–340. [Google Scholar]
- Obi, C.L.; Green, E.; Bessong, P.O.; Matsaung, G. Assessment of the microbial quality of river water sources in rural Venda communities in South Africa. Water SA 2002, 28, 287–291. [Google Scholar] [CrossRef]
- Obi, C.L.; Green, E.; Bessong, P.O.; de Villiers, B.; Hoosen, A.A.; Igumbor, E.O.; Potgieter, N. Gene encoding virulence markers among Escherichia coli isolates from diarrhoeagenic stool samples and river sources in rural Venda communities of South Africa. Water SA 2004, 3, 37–42. [Google Scholar]
- Nyairo, W.N.; Owuor, P.O.; Kengara, F.O. Effect of anthropogenic activities on the water quality of Amala and Nyangores tributaries of River Mara in Kenya. Environ. Monit. Assess. 2015, 187, 4913–4918. [Google Scholar] [CrossRef] [PubMed]
- Chigor, V.N.; Sibanda, T.; Okoh, A.I. Studies on the bacteriological qualities of the Buffalo River and three source water dams along its course in the Eastern Cape Province of South Africa. Environ. Sci. Pollut. Res. 2013, 20, 4125–4136. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Water, Sanitation and Health Protection and the Human Environment. Available online: htttp://www.who.int/water_sanitation_health/gdwqrevision/desalination (accessed on 22 April 2014).
- Nikaeen, M.; Pehjan, A.; Jalali, M. Rapid Monitoring of Indicator Coliforms in Drinking Water by an Enzymatic Assay. Iran J. Environ. Health Sci. Engl. 2009, 6, 7–10. [Google Scholar]
- Dundas, S.; Todd, W.T.A. Acute diarrhoea and fever. J. R. Coll. Phys. Edinb. 2006, 36, 236–240. [Google Scholar]
- Ballester, F.; Sunyer, J. Drinking water and gastrointestinal disease: Need of better understanding and an improvement in public health surveillance. J. Epidermiol. Community Health 2000, 54, 3–5. [Google Scholar] [CrossRef]
- Smeets, P.W.M.H.; Rietveld, L.C.; van Dijik, J.C.; Medema, G.J. Practical applications of quantitative microbial risk assessment (QMRA) for water safety plans. Water Sci. Technol. 2010, 61, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Ganesh, A. Water quality indicators: Bacteria, coliphages, enteric viruses. Int. J. Environ. Health. Res. 2013, 23, 484–506. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, J.H. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 2004, 198, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Omar, K.B.; Barnard, T.G. Detection of diarrhoeagenic Escherichia coli in clinical and environmental water sources in South Africa using single-step 11-gene m-PCR. World J. Microbiol. Biotechnol. 2014, 30, 2663–2671. [Google Scholar] [CrossRef] [PubMed]
- Omar, K.B.; Barnard, T.G.; Jagals, P. Development of a competitive PCR assay for the quantification of total Escherichia coli DNA in water. Afr. J. Biotechnol. 2010, 9, 564–572. [Google Scholar]
- South African Bureau of Standard. South African National Standard (SANS): Drinking Water Standard; SABS: Pretoria, South Africa, 2011; pp. 1–23. [Google Scholar]
- World Health Organization (WHO). A Toolkit for Monitoring and Evaluating Household Water Treatment and Safe Storage Programmes: 1–20, Geneva, Switzerland, 2012. Available online: http://www.who.int/household_water/resources/ (accessed on 10 December 2015).
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Langauge and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2013. [Google Scholar]
- Barton, K. MuMIn: Multi-Model Inference. Available online: https://cran.r-project.org/web/packages/MuMIn/index.html (accessed on 7 January 2016).
- Burnham, K.P.; Anderson, D.R. Multimodel inference understanding AIC and BIC in model selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Department of Water Affairs and Forestry (DWAF). Quality of Domestic Water Supplies, 2nd ed.; DWAF: Pretoria, South Africa, 1998; pp. 48–59. [Google Scholar]
- World Health Organization (WHO). Guidelines for Drinking Water Quality, 4th ed.; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Abd El Gawad, H.S. Aquatic environmental monitoring and removal efficiency of detergents. Water Sci. 2014, 28, 51–64. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Guidelines for Drinking Water Quality; WHO: Geneva, Switzerland, 1993. [Google Scholar]
- Karikari, A.Y.; Ansa-Asare, O.D. Physico-chemical water quality of Densu River of Ghana. West Afr. J. Appl. Ecol. 2006, 10, 1–12. [Google Scholar]
- Ferrar, A.A. Ecological Flow Requirements for South African Rivers; Report No.162; South African National Scientific Programmes: Pretoria, South Africa, 1989. [Google Scholar]
- Koning, N.; Roos, J.C. The Continued Influence of Organic Pollution on the Water Quality of the Turbid Modder River. Water S. Afr. 1999, 25, 285–292. [Google Scholar]
- Obi, C.L.; Potgieter, N.; Bessong, P.O.; Matsaung, G. Scope of potential bacterial agents of diarrhoea and microbial assessment of quality of river water sources in rural Venda communities in South Africa. Water Sci. Technol. 2003, 47, 59–64. [Google Scholar] [PubMed]
- Grabow, W.O.K. Water borne diseases: Update on water quality assessment and control. Water SA 1996, 22, 193–202. [Google Scholar]
- Jagals, P.; Barnard, T.G.; Mokoena, M.M.; Ashbolt, N.; Roser, D.J. Pathogenic Escherichia coli in rural household container waters. Water Technol. 2013, 67, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Momba, M.N.B.; Kaleni, P. Regrowth and survival of indicator microorganisms on the surfaces of household containers used in the storage of drinking water in rural South African communities. Water Res. 2002, 36, 3023–3028. [Google Scholar] [CrossRef]
- Griesel, M.; Jagals, P. Fecal indicator organisms in the Renoster Spruit system of the Modder-Reit River catchment area and implications for human users of the water. Water SA 2002, 28, 227–234. [Google Scholar] [CrossRef]
- Howard, G.; Pedley, S.; Barret, M.; Nalubega, M.; Johal, K. Risk factors contributing to microbial contamination of shallow groundwater in Kampala, Uganda. Water Res. 2003, 37, 3421–3429. [Google Scholar] [CrossRef]
- Wilkes, G.; Brassard, J.; Edge, T.A.; Gannon, V.; Jokinen, C.C.; Jones, T.H.; Neumann, N.; Pintar, K.D.; Ruecker, N.; Schmidt, P.J.; et al. Bacteria, viruses, and parasites in an intermittent stream protected from and exposed to pasturing cattle: Prevalence, densities, and quantitative microbial risk assessment. Water Res. 2013, 47, 6244–6257. [Google Scholar] [CrossRef] [PubMed]
- Aström, J.; Pettersson, T.J.; Strenström, T.A.; Bergstedt, O. Variability analysis of pathogen and indicator loads from urban sewer systems along a river. Water Sci. Technol. 2009, 59, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Franke, C.; Rechenburg, A.; Baumanns, S.; Willkomm, M.; Christoffels, E.; Exner, M.; Kistermann, T. The emission potential of different land use patterns for the occurrence of coliphages in surface water. Int. J. Hyg. Environ. Health 2009, 212, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Bezuidenhout, C.C. A Large Scale Study of Microbial and Physio-Chemical Quality of Selected Groundwater and Surface Water in North West Province, South Africa; WRC Report no. 1966/1/3; Water Resources Commission: Pretoria, South Africa, 2011; pp. 44–178. [Google Scholar]


| Observed Risk Activities | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sampling Sites | Car Wash | Animal Grazing | Drinking Water | Laundry | Farming | Body Washing | Sewage Discharge | Fecal Matter | Baby Diaper Washing | % of Risks Activities per Site |
| Mutale | X | X | X | - | X | X | X | - | - | 66.7 |
| Sambandou | - | X | X | X | - | - | - | X | - | 44.4 |
| Mbwedi | X | - | - | X | - | X | - | X | - | 44.4 |
| Tshinane | - | - | - | X | - | X | - | X | X | 44.4 |
| Dzindi | X | - | - | - | X | - | - | X | X | 44.4 |
| Lutanandwa | - | - | - | - | - | - | - | X | X | 22.2 |
| Albasin Dam | - | - | - | - | X | - | - | X | - | 22.2 |
| Karringmelkspruit | - | - | - | - | X | - | - | X | X | 33.3 |
| Tshino | X | - | - | X | - | X | - | X | - | 44.4 |
| Madadzhe | - | - | - | - | X | - | X | X | - | 33.3 |
| Piesanghoek | - | - | - | - | X | - | - | X | - | 22.2 |
| Springfield | - | - | - | - | X | - | - | X | - | 22.2 |
| Cumulative percent per risk activity | 33.3 | 16.7 | 16.7 | 33.3 | 58.3 | 33.3 | 16.7 | 91.7 | 33.3 | |
| Bacterial Strain | Reference | Use | Genes Present |
|---|---|---|---|
| Escherichia coli (Commensal) a | PCR | mdh | |
| Enterohaemorrhagic (EHEC) | ESCCO 21 b | PCR | mdh, stx1, stx2 and eaeA |
| Enteroinvasive (EIEC) | ESCCOS ATCC 43893 b | PCR | mdh and ial |
| Enterotoxigenic (ETEC) | ESCCO 22 b | PCR | mdh, lt and st |
| Enteropathogenic (EPEC) | S-ESCCO 16 Pl b | PCR | mdh, eaeA, bfp |
| Enteroaggregative (EAEC) | ESCCO 14 b | PCR | mdh and eagg |
| E. coli | ATCC 25922 | Microbiology | |
| Pseudomonas aeruginosa | ATCC 31488 | Microbiology | |
| Klebsiella pneumoniae | ATCC 10145 | Microbiology |
| Sampling Sites | Total Coliforms | Escherichia coli | ||||||
|---|---|---|---|---|---|---|---|---|
| Up 2014 | Dw 2014 | Up 2015 | Dw 2015 | Up 2014 | Dw 2014 | Up 2015 | Dw 2015 | |
| Mut | 2420 | 2420 | 2420 | 2420 | 163 | 643 | 1203.3 | 345 |
| Samb | 2420 | 2420 | 2420 | 2420 | 528 | 241 | 66 | 355 |
| Mbwe | 2420 | 1716 | 2420 | 2420 | 863 | 609 | 203 | 228 |
| Tshin | 2420 | 2420 | 2203 | 1595 | 241 | 247 | 114 | 85 |
| Dzin | 2420 | 2420 | 2203 | 2420 | 540 | 1517 | 214 | 242 |
| Lutan | 2420 | 1733 | 1917 | 2420 | 805 | 942 | 35 | 30 |
| Albas | 1812 | 1770 | 721 | 1700 | 40 | 12 | 6 | 6 |
| Karr | 2420 | 2420 | 2420 | 2420 | 227 | 141 | 157 | 206 |
| Tshino | 2420 | 2420 | 2420 | 2076 | 704 | 1250 | 275 | 178 |
| Mada | 2420 | 2420 | 2420 | 2420 | 2420 | 2420 | 2420 | 2420 |
| Pies | 2420 | 2420 | 1553 | 1378 | 212 | 143 | 125 | 126 |
| Spring | 2420 | 2420 | 1553 | 1393 | 331 | 372 | 21 | 19 |
| Year | Com | aEPEC | tEPEC | EAEC | EHEC | EIEC | ETEC |
|---|---|---|---|---|---|---|---|
| 2014 | 11 (22%) | 14 (26%) | 9 (18%) | 17 (35%) | 13 (27%) | 3 (6%) | 21 (43%) |
| 2015 | 3 (6%) | 31 (60%) | 8 (15%) | 27 (52%) | 14 (27%) | 8 (15%) | 21 (40%) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traoré, A.N.; Mulaudzi, K.; Chari, G.J.E.; Foord, S.H.; Mudau, L.S.; Barnard, T.G.; Potgieter, N. The Impact of Human Activities on Microbial Quality of Rivers in the Vhembe District, South Africa. Int. J. Environ. Res. Public Health 2016, 13, 817. https://doi.org/10.3390/ijerph13080817
Traoré AN, Mulaudzi K, Chari GJE, Foord SH, Mudau LS, Barnard TG, Potgieter N. The Impact of Human Activities on Microbial Quality of Rivers in the Vhembe District, South Africa. International Journal of Environmental Research and Public Health. 2016; 13(8):817. https://doi.org/10.3390/ijerph13080817
Chicago/Turabian StyleTraoré, Afsatou N., Khodani Mulaudzi, Gamuchirai J.E. Chari, Stefan H. Foord, Lutendo S. Mudau, Tobias G. Barnard, and Natasha Potgieter. 2016. "The Impact of Human Activities on Microbial Quality of Rivers in the Vhembe District, South Africa" International Journal of Environmental Research and Public Health 13, no. 8: 817. https://doi.org/10.3390/ijerph13080817
APA StyleTraoré, A. N., Mulaudzi, K., Chari, G. J. E., Foord, S. H., Mudau, L. S., Barnard, T. G., & Potgieter, N. (2016). The Impact of Human Activities on Microbial Quality of Rivers in the Vhembe District, South Africa. International Journal of Environmental Research and Public Health, 13(8), 817. https://doi.org/10.3390/ijerph13080817