Isolation and Characterization of a Bacterial Strain Capable of Efficient Berberine Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enrichment and Isolation of a BBR−Degrading Culture
2.1.1. Source of Inoculation
2.1.2. Isolation of Pure Cultures and Initial Screening
2.2. Characterization of the Isolated Strain
2.2.1. Colony and Cell Morphology
2.2.2. Identification of Biochemical Characteristics
2.2.3. DNA Extraction, PCR, Sequencing and Phylogenetic Analysis
2.3. Tests on Degradation Characteristics
2.3.1. Operational Conditions
2.3.2. Growth Characterization of the Isolated Strain
2.3.3. Kinetic Model
2.4. Analytical Methods
2.4.1. BBR
2.4.2. Biomass Abundance
2.4.3. TOC
3. Results
3.1. Morphology and Physical Characteristics of Strain B16
3.2. Biochemical Properties of Strain B16
3.3. Microbial Identification and Phylogenetic Analysis
3.4. Degradation of Organic Substances by Strain B16
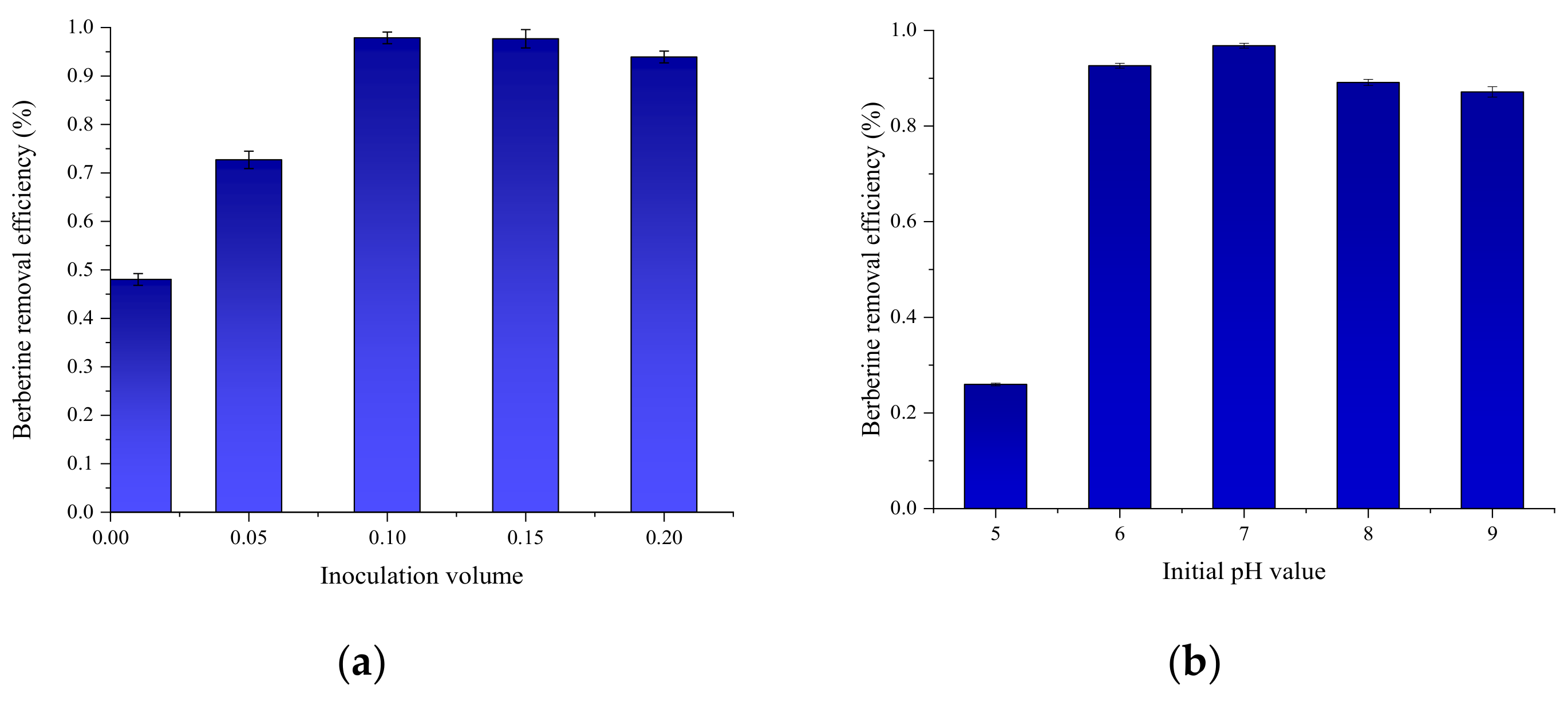
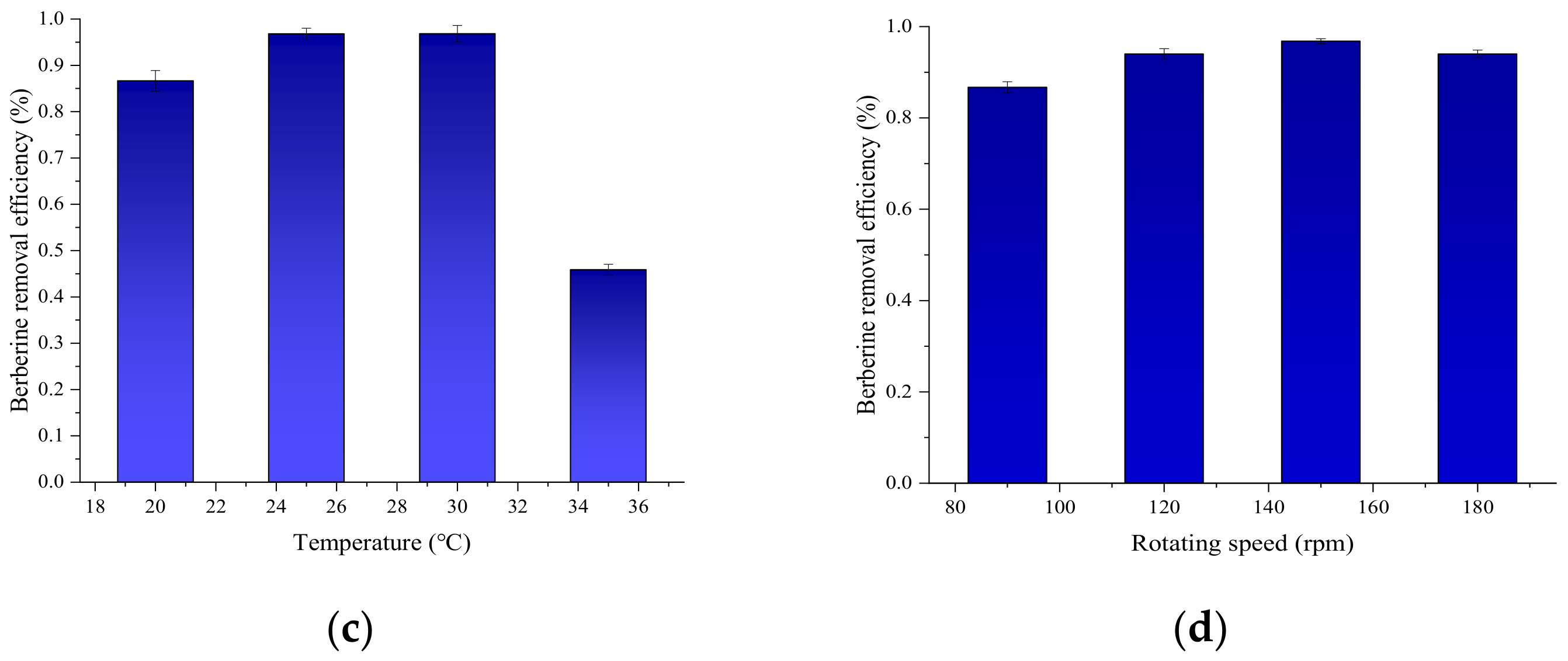
3.5. Optimization of Growth Conditions
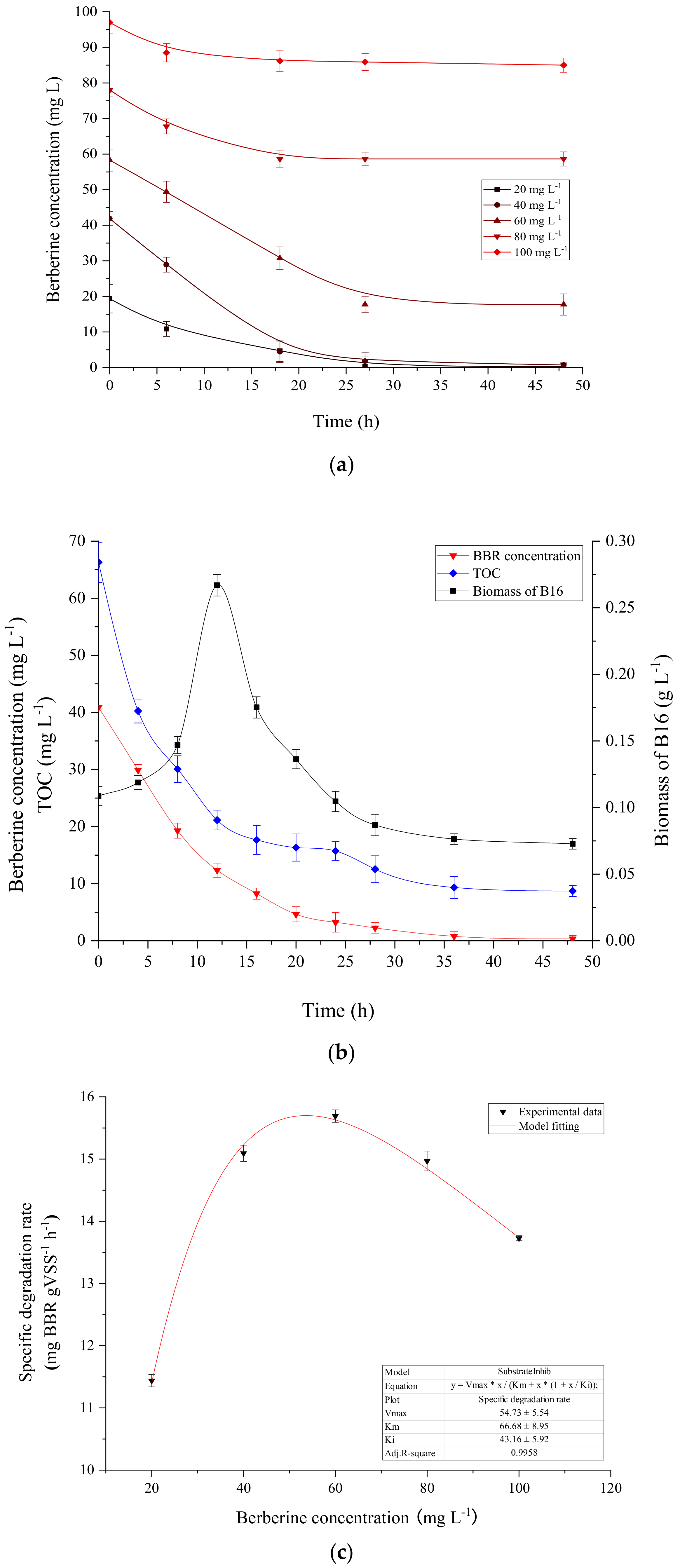
3.6. The Growth of Strain B16 and Degradation Kinetics of BBR
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wojtyczka, R.D.; Arkadiusz, D.; MalGorzata, K.; Robert, K.; Agata, K.-D.; Tomasz, M.; Danuta, I. Berberine enhances the antibacterial activity of selected antibiotics against coagulase−negative Staphylococcus strains in vitro. Molecules 2014, 19, 6583–6596. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yin, Y.M.; Wang, X.Y.; Wu, H.; Ge, X.Z. Evaluation of berberine as a natural fungicide: Biodegradation and antimicrobial mechanism. J. Asian Nat. Prod. Res. 2017, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Othman, M.S.; Safwat, G.; Aboulkhair, M.; Abdel Moneim, A.E. The potential effect of berberine in mercury−induced hepatorenal toxicity in albino rats. Food Chem. Toxicol. 2014, 69, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Song, Y.; Dai, Y.; Qiu, G.; Ren, M.; Zeng, P. Treatment of berberine hydrochloride pharmaceutical wastewater by O3/UV/H2O2 advanced oxidation process. Environ. Earth Sci. 2015, 73, 4939–4946. [Google Scholar] [CrossRef]
- Ren, M.; Song, Y.; Xiao, S.; Ping, Z.; Peng, J. Treatment of berberine hydrochloride wastewater by using pulse electro−coagulation process with Fe electrode. Chem. Eng. J. 2011, 169, 84–90. [Google Scholar] [CrossRef]
- Qiu, G.; Song, Y.H.; Zeng, P.; Duan, L.; Xiao, S. Characterization of bacterial communities in hybrid upflow anaerobic sludge blanket (UASB)−membrane bioreactor (MBR) process for berberine antibiotic wastewater treatment. Bioresour. Technol. 2013, 142, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Song, Y.; Zeng, P.; Wang, Q.; Han, S.; Song, B.; Lin, H.U. Cultivation of Aerobic Granules with Effluent from ABR and Analysis of Their Microbial Diversity of Microbial Communities. Environ. Sci. Technol. (Chin.) 2014, 37, 70–74. [Google Scholar]
- Takeda, H.; Ishikawa, K.; Wakana, D.; Fukuda, M.; Sato, F.; Hosoe, T. 11−Hydroxylation of Protoberberine by the Novel Berberine−Utilizing Aerobic Bacterium Sphingobium sp. Strain BD3100. J. Nat. Prod. 2015, 78, 2880–2886. [Google Scholar] [CrossRef] [PubMed]
- Guillaumegentil, O.; Sonnard, V.; Kandhai, M.C.; Marugg, J.D.; Joosten, H. A simple and rapid cultural method for detection of Enterobacter sakazakii in environmental samples. J. Food Prot. 2005, 68, 64. [Google Scholar] [CrossRef]
- Rojas, I.; Bernardi, L.; Ratsch, E.; Kania, R.; Wittig, U.; Saric, J. A database system for the analysis of biochemical pathways. Silico Biol. 2002, 2, 75–86. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI−BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position−specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor−joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406. [Google Scholar] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor−joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G. Hybrid UASB−MBR Process for Berberine Pharmaceutical Wastewater Treatment and the Process Mechanisms. Ph.D. Thesis, Beijing Normal University, Beijing, China, 2011. [Google Scholar]
- Yi, L.; Xu, X. Study on the precipitation reaction between baicalin and berberine by HPLC. J. Chromatogr. B 2004, 810, 165–168. [Google Scholar] [CrossRef]
- Ruan, B.; Wu, P.; Chen, M.; Lai, X.; Chen, L.; Yu, L.; Gong, B.; Kang, C.; Dang, Z.; Shi, Z.; et al. Immobilization of Sphingomonas sp. GY2B in polyvinyl alcohol–alginate–kaolin beads for efficient degradation of phenol against unfavorable environmental factors. Ecotoxicol. Environ. Saf. 2018, 162, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Moy, B.; Song, Y.; Tay, J. Biodegradation of dimethyl phthalate by Sphingomonas sp. isolated from phthalic−acid−degrading aerobic granules. Appl. Microbiol. Biotechnol. 2008, 80, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Iwasa, K.; Kamigauchi, M.; Sugiura, M.; Nanba, H. Antimicrobial activity of some 13−alkyl substituted protoberberinium salts. Planta Med. 1997, 63, 196–198. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.L.; Hua, G.Q.; Meng, Z. Antibacterial Mechanisms of Berberine and Reasons for Little Resistance of Bacteria. Chin. Herb. Med. 2011, 3, 27–35. [Google Scholar]
- Mohsen, I.; Hossein, H. Pharmacological and therapeutic effects of Berberis vulgaris and its active constituent, berberine. Phytother. Res. 2010, 22, 999–1012. [Google Scholar]
- Wei, G.X.; Xu, X.; Wu, C.D. In vitro synergism between berberine and miconazole against planktonic and biofilm Candida cultures. Arch. Oral Biol. 2011, 56, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Han, Y.M.; Wei, L.; Xiao, X.H. Effect of berberine alkaloids on Bifidobacterium adolescentis growth by microcalorimetry. J. Therm. Anal. Calorim. 2009, 95, 495–499. [Google Scholar] [CrossRef]
- Takeuchi, M.; Hamana, K.; Hiraishi, A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2001, 51, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tay, J. Toxic and inhibitory effects of trichloroethylene aerobic co−metabolism on phenol−grown aerobic granules. J. Hazard. Mater. 2015, 286, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Habe, H.; Yamane, H.; Omori, T.; Nojiri, H. Diversity of carbazole−degrading bacteria having the car gene cluster: Isolation of a novel gram−positive carbazole−degrading bacterium. FEMS Microbiol. Lett. 2005, 245, 145–153. [Google Scholar] [CrossRef] [PubMed]
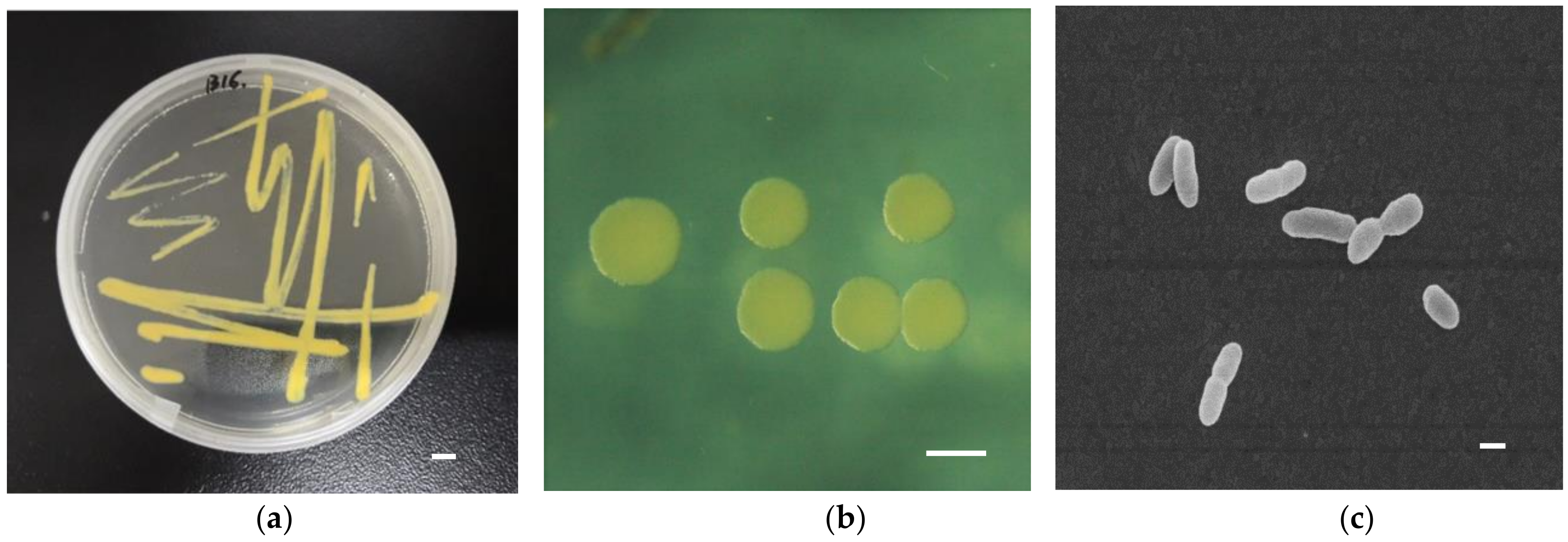
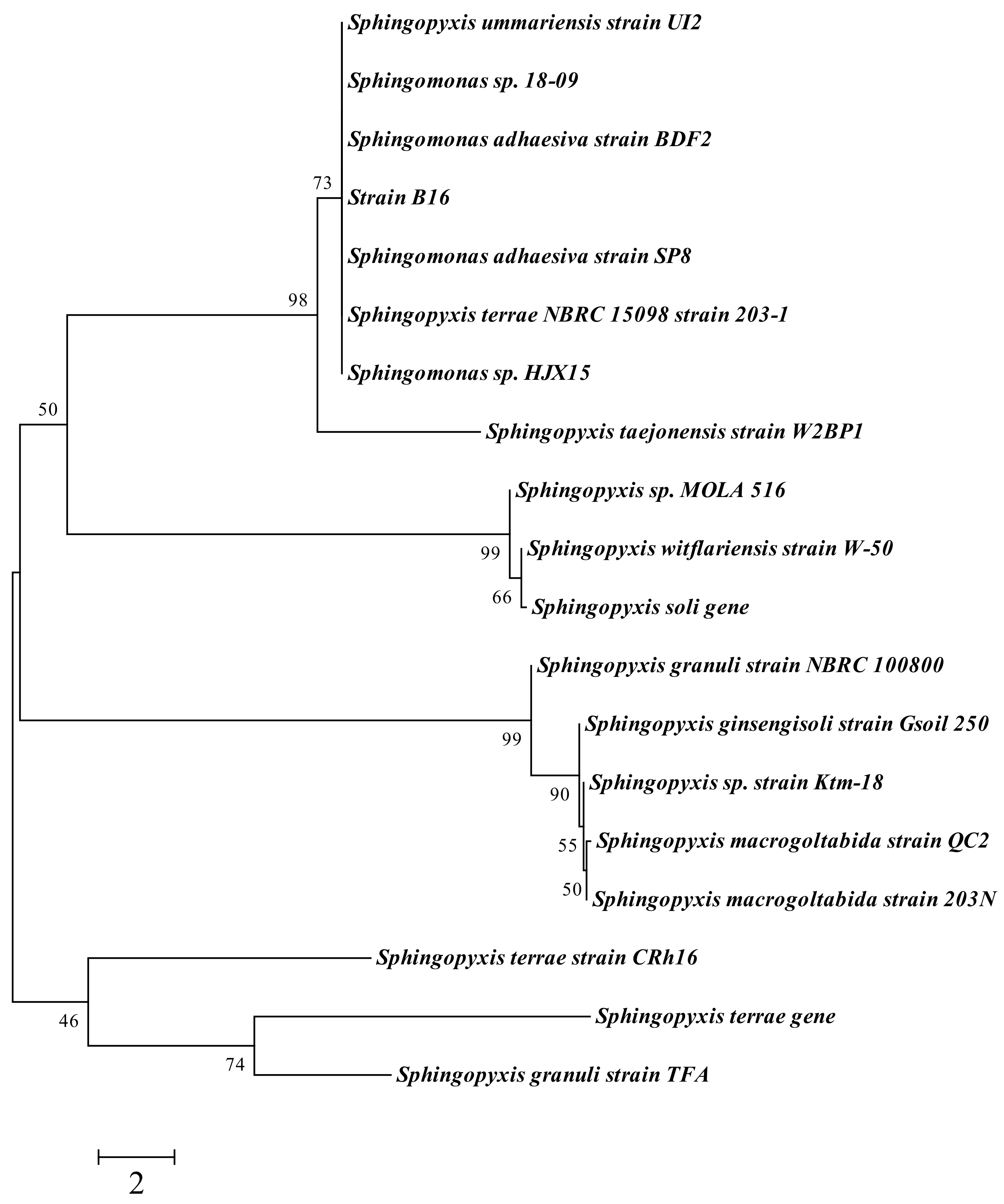
| Accession | Description | Max Score | Total Score | Query Coverage | Evalue | Max Ident |
|---|---|---|---|---|---|---|
| CP013342.1 | Sphingopyxis terrae; NBRC 15098 | 2318 | 2318 | 100% | 0.0 | 99% |
| KP979546.1 | Sphingopyxis sp.; HJX15 | 2318 | 2318 | 100% | 0.0 | 99% |
| AB680906.1 | Sphingopyxis macrogoltabida; NBRC 15593; | 2318 | 2318 | 100% | 0.0 | 99% |
| MK217492.1 | Sphingopyxis sp.; YC−JH3 | 2318 | 2318 | 100% | 0.0 | 99% |
| Substrate | Growth | Substrate | Growth |
|---|---|---|---|
| Benzene | + | Tetrabutylammonium bromide | ++ |
| Chlorobenzene | + | Sodium acetate | ++ |
| Catechol | + | Urea | ++ |
| Formaldehyde | − | Phenol | + |
| Benzaldehyde | ++ | P−methylphenol | − |
| Glyoxal | + | 3,5−dimethylphenol | ++ |
| Vanillin | − | Acetophenone | + |
| Acetic acid | + | Benzothiazole | ++ |
| Methanol | ++ | Benzyl benzoate | ++ |
| Ethanol | + | Dimethyl phthalate | ++ |
| Sorbitol | ++ | Di−n−butyl phthalate | ++ |
| Isopropanol | ++ | Diisobutyl phthalate | ++ |
| Dimethyl sulfate | − | Chloramphenicol | + |
| Methyl dichloroacetate | − |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zhang, Y.; Zeng, P.; Wang, H.; Song, Y.; Li, J. Isolation and Characterization of a Bacterial Strain Capable of Efficient Berberine Degradation. Int. J. Environ. Res. Public Health 2019, 16, 646. https://doi.org/10.3390/ijerph16040646
Liu S, Zhang Y, Zeng P, Wang H, Song Y, Li J. Isolation and Characterization of a Bacterial Strain Capable of Efficient Berberine Degradation. International Journal of Environmental Research and Public Health. 2019; 16(4):646. https://doi.org/10.3390/ijerph16040646
Chicago/Turabian StyleLiu, Shiyue, Yi Zhang, Ping Zeng, Heli Wang, Yonghui Song, and Juan Li. 2019. "Isolation and Characterization of a Bacterial Strain Capable of Efficient Berberine Degradation" International Journal of Environmental Research and Public Health 16, no. 4: 646. https://doi.org/10.3390/ijerph16040646
APA StyleLiu, S., Zhang, Y., Zeng, P., Wang, H., Song, Y., & Li, J. (2019). Isolation and Characterization of a Bacterial Strain Capable of Efficient Berberine Degradation. International Journal of Environmental Research and Public Health, 16(4), 646. https://doi.org/10.3390/ijerph16040646







