Spatiotemporal Variation and Hotspot Detection of the Avian Influenza A(H7N9) Virus in China, 2013–2017
Abstract
:1. Introduction
2. Data and Methods
2.1. Data Acquisition
2.2. Methodology
3. Results
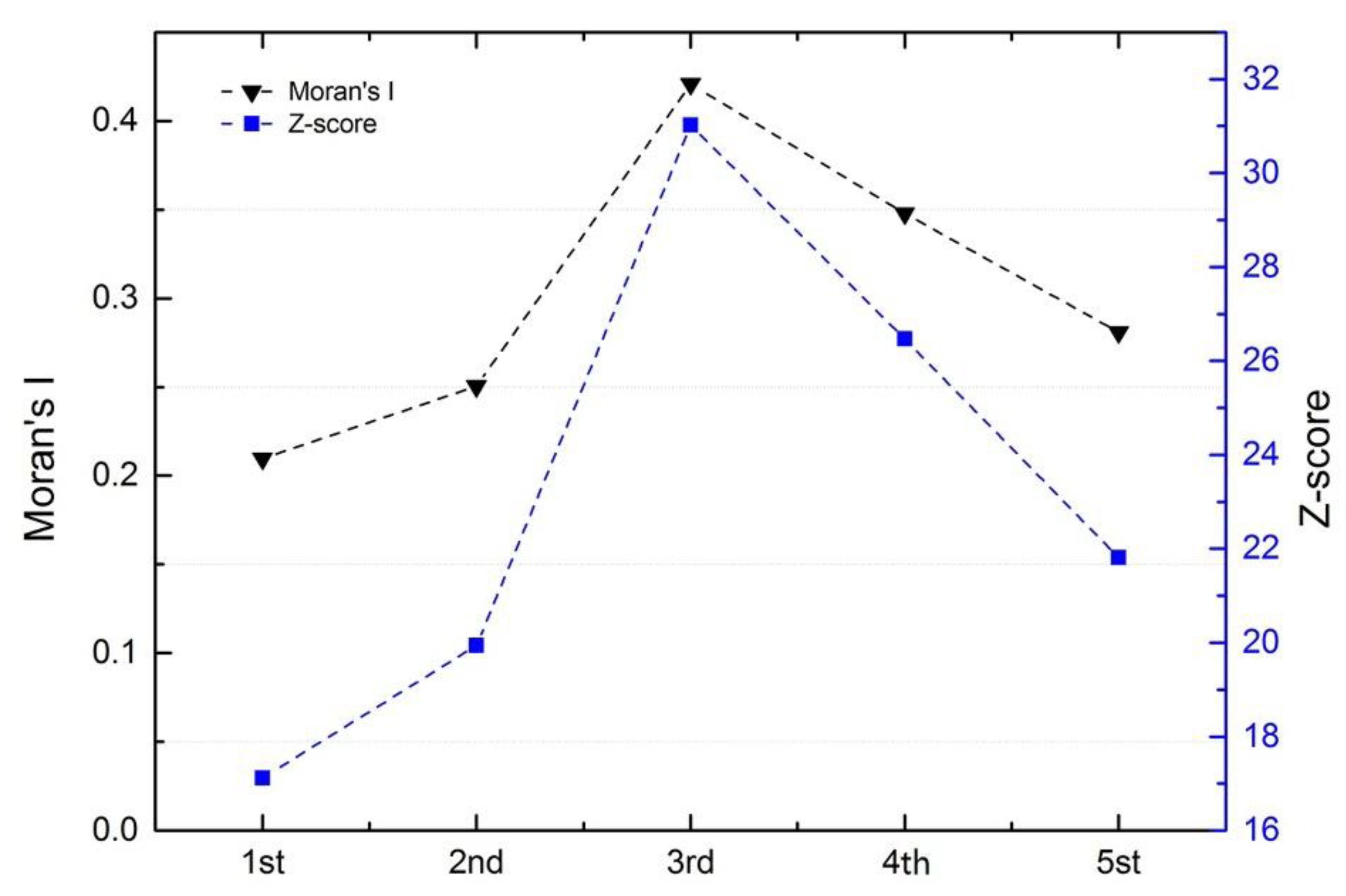
3.1. Characterization in Time
3.2. Characterization in Space
3.3. Characterization of Hotspots
3.4. Characterization of Time-Space Clustering
4. Discussion
4.1. Analysis of The Epidemiological Characteristics of Human H7N9 Cases
4.2. Limitations
5. Conclusions
- (1)
- The fifth epidemic started earlier in the year and affected more individuals and more districts than did the previous four epidemics based on analyses of the timeline and geographical distribution. Cases shifted from coastline areas to more inland areas over time.
- (2)
- The cases exhibited local spatial aggregation, with high-risk areas most found in the southeast coastal regions of China. Shanghai, Jiangsu, Zhejiang, and Guangdong were the high-risk epidemic areas, which should arouse the attention of the local governments.
- (3)
- Combining GIS and SaTScan methods increased our understanding of the development of A(H7N9) virus epidemics. A strong cluster from 9 April to 24 June 2017 was identified in Northern China, and there were many secondary clusters in Eastern and Southern China, especially in Zhejiang, Fujian, Jiangsu, and Guangdong Provinces.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yu, H.; Cowling, B.J.; Feng, L.; Lau, E.H.Y.; Liao, Q.; Tsang, T.K.; Peng, Z.; Wu, P.; Liu, F.; Fang, V.J.; et al. Human infection with avian influenza A(H7N9) virus: An assessment of clinical severity. Lancet 2013, 382, 138–145. [Google Scholar] [CrossRef]
- WHO. Avian Influenza A(H7N9) Virus. 2017. Available online: http://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_10_30_2017.pdf (accessed on 10 November 2017).
- Van Riel, D.; Leijten, L.M.; de Graaf, M.; Siegers, J.Y.; Short, K.R.; Spronken, M.I.; Schrauwen, E.J.; Fouchier, R.A.; Osterhaus, A.D.; Kuiken, T. Novel Avian-Origin Influenza A(H7N9) Virus Attaches to Epithelium in Both Upper and Lower Respiratory Tract of Humans. Am. J. Pathol. 2013, 183, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Chan, R.W.; Chan, L.L.; Mok, C.K.; Hui, K.P.; Fong, J.H.; Tao, K.P.; Poon, L.L.; Nicholls, J.M.; Guan, Y.; et al. Tropism and innate host responses of a novel avian influenza A(H7N9) virus: An analysis of ex-vivo and in-vitro cultures of the human respiratory tract. Lancet Respir. Med. 2013, 1, 534–542. [Google Scholar] [CrossRef]
- Chinese Center for Disease Control and Prevention. Mutations on H7N9 Virus Have Been Identified in H7N9 Patients in China. 2017. Available online: http://www.chinacdc.cn/gwswxx/yjzx/201702/t20170219_138185.html (accessed on 19 February 2017).
- Shi, J.; Deng, G.; Kong, H.; Gu, C.; Ma, S.; Yin, X.; Zeng, X.; Cui, P.; Chen, Y.; Yang, H.; et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 2017, 27, 1409. [Google Scholar] [CrossRef] [PubMed]
- Xiang, N.; Li, X.; Ren, R. Assessing Change in Avian Influenza A(H7N9) Virus Infections During the Fourth Epidemic—China, September 2015–August 2016. Morb. Mortal. Wkly. Rep. MMWR 2016, 65, 1390–1394. [Google Scholar] [CrossRef] [PubMed]
- Kapczynski, D. Identification of viral epitopes recognized by the immune system following vaccination and challenge with the H7N9 avian influenza virus from China. Virology 2011, 417, 369–378. [Google Scholar]
- Zhu, H.; Lam, T.T.; Smith, D.K.; Guan, Y. Emergence and development of H7N9 influenza viruses in China. Curr. Opin. Virol. 2016, 16, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Guinat, C.; Nicolas, G.; Vergne, T.; Bronner, A.; Durand, B.; Courcoul, A.; Gilbert, M.; Guérin, J.; Paul, M. Spatio-temporal patterns of highly pathogenic avian influenza virus subtype H5N8 spread, France, 2016 to 2017. Euro. Surveill. 2018, 23, 1–10. [Google Scholar] [CrossRef]
- Li, J.; Yu, X.; Pu, X.; Yang, X.; Kou, Y.; Zhou, Y.; Qian, X.; Xie, L.; Pan, J. The diversity of avian influenza virus subtypes in live poultry markets before and during the second wave of A(H7N9) infections in Hangzhou, China. Emerg. Microbes Infect. 2015, 4, e14. [Google Scholar] [CrossRef]
- Lin, Q.; Lin, Z.; Chiu, A.P.; He, D. Seasonality of Influenza A(H7N9) Virus in China—Fitting Simple Epidemic Models to Human Cases. PLoS ONE 2016, 11, e0151333. [Google Scholar] [CrossRef]
- World Health Organization. Human Infection with Avian Influenza A(H7N9) Virus—China. Available online: www.who.int/csr/don/17-january-2017-ah7n9-china/en/ (accessed on 16 June 2017).
- Zhang, Y.; Shen, Z.; Ma, C.; Jiang, C.; Feng, C.; Shankar, N.; Yang, P.; Sun, W.; Wang, Q. Cluster of Human Infections with Avian Influenza A(H7N9) Cases: A Temporal and Spatial Analysis. Int. J. Environ. Res. Public Health 2015, 12, 816–828. [Google Scholar] [CrossRef]
- Dong, W.; Yang, K.; Xu, Q.L.; Yang, Y.L. A Predictive Risk Model for A(H7N9) Human Infections Based on Spatial-Temporal Autocorrelation and Risk Factors: China, 2013–2014. Int. J. Environ. Res. Public Health 2015, 12, 15204–15221. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Yang, K.; Qi, X.; Xu, K.; Ji, H.; Ai, J.; Ge, A.; Wu, Y.; Li, Y.; Dai, Q.; et al. Spatial and temporal analysis of human infection with avian influenza A(H7N9) virus in China, 2013. Eur. Commun. Dis. Bull. 2013, 18, 3474–3476. [Google Scholar] [CrossRef]
- Songchitruksa, P.; Zeng, X. Getis–Ord Spatial Statistics to Identify Hot Spots by Using Incident Management Data. Transp. Res. Rec. J. Transp. Res. Board 2010, 2165, 42–51. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, X.; Gao, F.; Liu, Y. Impacts of changing scale on Getis-Ord Gi hotspots of CPUE: A case study of the neon flying squid (Ommastrephes bartramii) in the northwest Pacific Ocean. Acta Oceanol. Sin. 2018, 37, 67–76. [Google Scholar] [CrossRef]
- Dong, W.; Yang, K.; Xu, Q.; Liu, L.; Chen, J. Spatio-temporal pattern analysis for evaluation of the spread of human infections with avian influenza A(H7N9) virus in China, 2013–2014. BMC Infect. Dis. 2017, 17, 704. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, H.; Wu, P.; Uyeki, T.M.; Feng, L.; Lai, S.; Wang, L.; Huo, X.; Xu, K.; Chen, E.; et al. Epidemiology of avian influenza A(H7N9) virus in human beings across five epidemics in mainland China, 2013–2017: An epidemiological study of laboratory-confirmed case series. Lancet Infect. Dis. 2017, 17, 822–832. [Google Scholar] [CrossRef]
- Iuliano, A.D.; Jang, Y.; Jones, J.; Davis, C.T.; Wentworth, D.E.; Uyeki, T.M.; Roguski, K.; Thompson, M.G.; Gubareva, L.; Fry, A.M.; et al. Increase in Human Infections with Avian Influenza A(H7N9) Virus During the Fifth Epidemic—China, October 2016–February 2017. Morb. Mortal. Wkly. Rep. MMWR 2017, 66, 254. [Google Scholar] [CrossRef]
- Sokal, R.R.; Oden, N.L.; Thomson, B.A. Local spatial autocorrelation in biological variables. Biol. J. Linn. Soc. 2010, 65, 41–62. [Google Scholar] [CrossRef]
- Costa, M.A.; Kulldorff, M. Maximum linkage space-time permutation scan statistics for disease outbreak detection. Int. J. Health Geographic 2014, 13, 20. [Google Scholar] [CrossRef] [Green Version]
- Kulldorff, M.; Heffernan, R.; Hartman, J.; Assunçao, R.; Mostashari, F. A Space–Time Permutation Scan Statistic for Disease Outbreak Detection. PLoS Med. 2005, 2, e59. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, C.; Ma, C.; Yang, P.; Tang, S.; Lau, A.; Sun, W.; Wang, Q. The impact of temperature and humidity measures on influenza A(H7N9) outbreaks—Evidence from China. Int. J. Infect. Dis. 2015, 30, 122–124. [Google Scholar] [CrossRef]
- Ou, H.; Yao, H.; Yao, W.; Wu, N.; Wu, X.; Han, C.; Cheng, L.; Chen, K.; Chen, H.; Li, L. Analysis of the immunogenicity and bioactivities of a split influenza A/H7N9 vaccine mixed with MF59 adjuvant in BALB/c mice. Vaccine 2016, 34, 2362–2370. [Google Scholar] [CrossRef] [Green Version]
- Bart, S.A.; Hohenboken, M.; Della Cioppa, G.; Narasimhan, V.; Dormitzer, P.R.; Kanesa-thasan, N. A cell culture-derived MF59-adjuvanted pandemic A/H7N9 vaccine is immunogenic in adults. Sci. Transl. Med. 2014, 6, 234ra55. [Google Scholar] [CrossRef]
- Soltani, A.; Askari, S. Exploring spatial autocorrelation of traffic crashes based on severity. Injury 2017, 48, 637–647. [Google Scholar] [CrossRef]
- Valcour, J.E.; Charron, D.F.; Berke, O.; Wilson, J.B.; Edge, T.; Waltner-Toews, D. A descriptive analysis of the spatio-temporal distribution of enteric diseases in New Brunswick, Canada. BMC Public Health 2016, 16, 204. [Google Scholar] [CrossRef]
- Bui, C.; Bethmont, A.; Chughtai, A.A.; Gardner, L.; Sarkar, S.; Hassan, S.; Seale, H.; MacIntyre, C.R. A Systematic Review of the Comparative Epidemiology of Avian and Human Influenza A H5N1 and H7N9—Lessons and Unanswered Questions. Transbound. Emerg. Dis. 2016, 63, 602–620. [Google Scholar] [CrossRef]
- Yu, H.; Wu, J.T.; Cowling, B.J.; Liao, Q.; Fang, V.J.; Zhou, S.; Wu, P.; Zhou, H.; Lau, E.H.; Guo, D.; et al. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A(H7N9) virus: An ecological study. Lancet 2014, 383, 541–548. [Google Scholar] [CrossRef]
- Wu, P.; Jiang, H.; Wu, J.T.; Chen, E.; He, J.; Zhou, H.; Wei, L.; Yang, J.; Yang, B.; Qin, Y.; et al. Poultry market closures and human infection with influenza A(H7N9) virus, China, 2013–2014. Emerg. Infect. Dis. 2014, 20, 1891–1894. [Google Scholar] [CrossRef]
- Artois, J.; Lai, S.; Feng, L.; Jiang, H.; Zhou, H.; Li, X.; Dhingra, M.S.; Linard, C.; Nicolas, G.; Xiao, X.; et al. H7N9 and H5N1 avian influenza suitability models for China: Accounting for new poultry and live-poultry markets distribution data. Stoch. Environ. Res. Risk Assess. Res. J. 2017, 31, 393–402. [Google Scholar] [CrossRef]
- Artois, J.; Jiang, H.; Wang, X.; Qin, Y.; Pearcy, M.; Lai, S.; Shi, Y.; Zhang, J.; Peng, Z.; Zheng, J.; et al. Changing Geographic Patterns and Risk Factors for Avian Influenza A(H7N9) Infections in Humans, China. Emerg. Infect. Dis. J. 2018, 24, 87. [Google Scholar] [CrossRef]
- Offeddu, V.; Cowling, B.J.; Peiris, J.S.M. Interventions in live poultry markets for the control of avian influenza: A systematic review. One Health 2016, 2, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Lu, J.; Faria, N.R.; Zeng, X.; Song, Y.; Zou, L.; Yi, L.; Liang, L.; Ni, H.; Kang, M.; et al. Effect of Live Poultry Market Interventions on Influenza A(H7N9) Virus, Guangdong, China. Emerg. Infect. Dis. 2016, 22, 2104. [Google Scholar] [CrossRef]
- Bethmont, A.; Bui, C.M.; Gardner, L.; Sarkar, S.; Chughtai, A.A.; Macintyre, C.R. Quantified degree of poultry exposure differs for human cases of avian influenza H5N1 and H7N9. Epidemiol. Infect. 2016, 144, 2633–2640. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Fu, J.; Lin, G.; Jiang, D. Spatiotemporal Variation and Hotspot Detection of the Avian Influenza A(H7N9) Virus in China, 2013–2017. Int. J. Environ. Res. Public Health 2019, 16, 648. https://doi.org/10.3390/ijerph16040648
Li Z, Fu J, Lin G, Jiang D. Spatiotemporal Variation and Hotspot Detection of the Avian Influenza A(H7N9) Virus in China, 2013–2017. International Journal of Environmental Research and Public Health. 2019; 16(4):648. https://doi.org/10.3390/ijerph16040648
Chicago/Turabian StyleLi, Zeng, Jingying Fu, Gang Lin, and Dong Jiang. 2019. "Spatiotemporal Variation and Hotspot Detection of the Avian Influenza A(H7N9) Virus in China, 2013–2017" International Journal of Environmental Research and Public Health 16, no. 4: 648. https://doi.org/10.3390/ijerph16040648
APA StyleLi, Z., Fu, J., Lin, G., & Jiang, D. (2019). Spatiotemporal Variation and Hotspot Detection of the Avian Influenza A(H7N9) Virus in China, 2013–2017. International Journal of Environmental Research and Public Health, 16(4), 648. https://doi.org/10.3390/ijerph16040648







