Merging Data Diversity of Clinical Medical Records to Improve Effectiveness
Abstract
:1. Introduction
2. State of the Art
2.1. Unstructured Data
2.2. Mixed Approaches
- In Reference [31], topic modeling was used. It is a type of statistical modeling for discovering the abstract “topics” that occur in a collection of documents, and latent Dirichlet allocation (LDA) is an example of a topic model and is used to classify text in a document to a particular topic.
- In Reference [32], both techniques (Bayesian belief networks and decision trees (DT)) were used, and their results were compared. In other words, to detect early stages of dementia, the authors used two different techniques to assist specialists in the diagnosis of patients with clinical suspicion of dementia. It was possible to conclude that the model that used the structured data and the clustering of the texts written in free format by the physicians integrated, improved the accuracy of predictive models in all pathologies [32].
- In Reference [33], a prediction of patient admission was performed, applied to the logistic regression using five different iterations.
- In Reference [34], researchers had the objective of comparing the number of geriatric syndrome cases identified using structured claims and structured and unstructured EMR data to understand the added value of the latter. Conclusions were that results improved when combining both models. This type of fact led the authors to encourage “incorporating NLP methods to increase the sensitivity of identification of individuals with geriatric syndromes” [34].
2.3. Data Formats in Medicine
2.4. Data Processing in Medicine
3. Work Methodology Proposed
Data Integration Using ETL Process
- 1)
- The extract process is responsible for the data extraction from the source system or database and makes it accessible for further processing or management process. In health care, this process needs to deal with data privacy, and most of the extract process has an anonymization process associated. At this point, the researcher decides which data make sense to use.
- 2)
- The transform is a process based on a set of rules to transform the data from the source to the target. In the current research, a new approach using semantics is proposed for this phase. This can be a complex process taking into account different dimensionalities cases, and it needs to assure that all variables are in the same units so that they can later be joined and a clean process can be conducted. The transformation step also requires joining data from several sources, generating aggregations, surrogate keys, sorting, deriving newly calculated values, and applying advanced validation rules.
- 3)
- The load process merges all data into a target database (output data).
- (1)
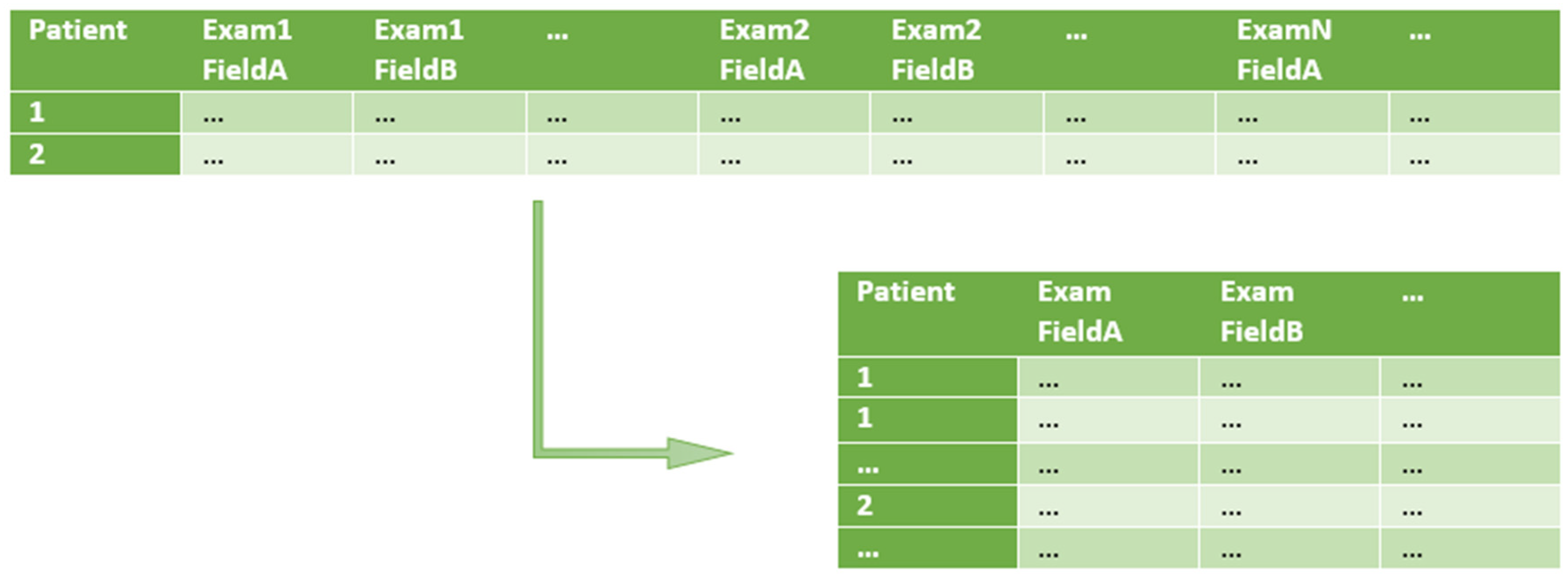
- Split one row into many (1 : m row split). Input rows are split in a 1 : m transformation, in order to have just one row per patient examination (instead of one row containing multiple patient examinations). At this point, only the relevant input fields are kept. The specification of row splitting and field selection is made through an auxiliary file called split rules. In this file, each selected field of the input CSV file is associated to a type and eventually to a structure (e.g., a specific date format to be used such as ‘YYYY-MM-DD’, for ‘2019-01-23’).
- (2)
- Semantic validation. At this step, the resulting data files of 1:m row split are submitted to semantic validation, where the rules defined in the auxiliary file semantic rules are applied. These rules are based on domain knowledge and aim at validating specific domain constraints, like thresholds or non-empty fields in specific columns;
- (3)
- Data join. At this point, data is joint by applying the rules defined in the last auxiliary file, named join rules. This file describes which fields should be present in the output files and which will be used as keys when joining the rows from each file. In addition to the output data files, a file named transparency report is generated. It includes statistical information about operations applied during the transformation process.
4. Description of the Case Study
4.1. E3TL First Step—The 1: m Row Split
- OutFile—The intermediary output file generated by the field mapping step;
- ExamFeatures—The number of fields per examination. This variable supports the indexing of each input field with the correct offset;
- ExamCountPerRow—The number of examinations per row. In the ALS-FRS.csv file, each row is associated with a specific patient. Therefore, it includes multiple examinations per row;
- OutputFeatures—The number of fields of the output table;
- FeatureMapping—The list of mappings between the column index of the selected input fields and the desired column output index;
- InputIdx—The index of a specified column in the input file;
- output is—The position of the field in the output intermediary file;
- StaticIdx—There are static and non-static based indexes. A static input column index is not dependent on the number of examinations per row. Differently, the “Date” field, with a non-static index, will be retrieved using the offset: InputColumnIndex = (exam number ∗ExamFeatures) + InputIdx where ExamNumber <= ExamCount;
- Name—The name to be used in the output file;
- Type—The data type to be used. Depending on the data type, different parsing routines will be used, either for multiple date formats, or different decimal notations, for example;
- Format—The output format of non-basic types to be used. For dates, for example, ‘YYYY-MM-DD’, for ‘2019-01-23’.
4.2. E3TL Second Step—The Semantic Validation
- OutFile—The intermediary output file generated by the semantic validation step;
- Semantic Rules—The list domain validations that should be applied to a specific field;
- InputIdx—The index of a specified column in the input file. NotNull requires a non-null value;
- RegEx—Verify if the value of the field maps to the specified regular expression;
- Threshold—Verify if the value of the field is over the specified threshold (only applicable to integer or double fields).
4.3. E3TL Third Step—Data Join
- OutFile—The final output filename;
- FeatureMapping—The list of fields that should be present in each row of the output file and their relation to the input fields (from the intermediary files). Typically, each row includes dynamic (exams) and static information (like patient date of the birth, for example);
- InputIdx—The index of the input field on the auxiliary input file.
- InputFile—The auxiliary input filename;
- Name—The final output field name;
- Type—The output field data type;
- Format—The output field format for non-basic types (like dates, for example);
- JoinKey—Indicates if the field is part of the Join key.
5. Outcomes from the Case Study
- (a)
- 1 : m row split execution example. The function receives two variables as arguments: One with the name of the CSV input data file and the second with the name of the JSON specification auxiliary file. As output, the function returns the name of the intermediary CSV output file.
6. Other Application Cases
7. Discussion
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Luo, L.; Li, L.; Hu, J.; Wang, X.; Hou, B.; Zhang, T.; Zhao, L.P. A hybrid solution for extracting structured medical information from unstructured data in medical records via a double-reading/entry system. BMC Med. Inform. Decis. Mak. 2016, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- McLane, S. Designing an EMR planning process based on staff attitudes toward and opinions about computers in healthcare. CIN Comput. Inform. Nurs. 2005, 23, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, L.G.; Khan, A.N.G.A. Challenges of Electronic Medical Record Implementation in the Emergency Department. Pediatr. Emerg. Care 2006, 22, 184–191. [Google Scholar] [PubMed]
- Yadav, P.; Steinbach, M.; Kumar, V.; Simon, G. Mining Electronic Health Records: A Survey. arXiv, 2017; arXiv:1702.03222. [Google Scholar] [CrossRef]
- Sun, W.; Cai, Z.; Li, Y.; Liu, F.; Fang, S.; Wang, G. Data processing and text mining technologies on electronic medical records: A review. J. Healthc. Eng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lamy, M.; Pereira, R.; Ferreira, J.C.; Vasconcelos, J.B.; Melo, F.; Velez, I. Extracting clinical information from electronic medical records. Adv. Intell. Syst. Comput. 2018, 806, 113–120. [Google Scholar] [CrossRef]
- Cooper, P. Data, information, knowledge and wisdom. Anaesth. Intensive Care Med. 2017, 18, 55–56. [Google Scholar] [CrossRef]
- Allen, G.D. Hierarchy of Knowledge—from Data to Wisdom. Int. J. Curr. Res. Multidiscip. 2004, 2, 15–23. [Google Scholar]
- Garets, D.; Davis, M. Electronic Medical Records vs. Electronic Health Records: Yes, There Is a Difference A HIMSS Analytics TM White Paper. Available online: https://s3.amazonaws.com/rdcms-himss/files/production/public/HIMSSorg/Content/files/WP_EMR_EHR.pdf (accessed on 27 February 2019).
- The Dorenfest Complete Integrated Healthcare Delivery System Plus (Ihds+) Database and Library. 2019. Available online: https://foundation.himss.org/Dorenfest/About (accessed on 27 February 2019).
- Koh, H.C.; Tan, G. Data Mining Applications in Healthcare. J. Healthc. Inf. Manag. 2011, 19, 64–72. [Google Scholar]
- Sun, W.; Cai, Z.; Liu, F.; Fang, S.; Wang, G. A survey of data mining technology on electronic medical records. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; pp. 1–6. [Google Scholar]
- Roy, S.B.; Teredesai, A.; Zolfaghar, K.; Liu, R.; Hazel, D.; Newman, S.; Marinez, A. Dynamic Hierarchical Classification for Patient Risk-of Readmission. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, NSW, Australia, 10–13 August 2015; pp. 1691–1700. [Google Scholar]
- Baba, Y.; Kashima, H.; Nohara, Y.; Kai, E.; Ghosh, P.; Islam, R.; Ahmed, A.; Kuroda, M.; Inoue, S.; Hiramatsu, T.; et al. Predictive Approaches for Low-Cost Preventive Medicine Program in Developing Countries. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, NSW, Australia, 10–13 August 2015; pp. 1681–1690. [Google Scholar]
- Somanchi, S.; Adhikari, S.; Lin, A.; Eneva, E.; Ghani, R. Early Prediction of Cardiac Arrest (Code Blue) using Electronic Medical Records. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, NSW, Australia, 10–13 August 2015; pp. 2119–2126. [Google Scholar]
- Ravindranath, K.R. Clinical Decision Support System for heart diseases using Extended sub tree. In Proceedings of the 2015 International Conference on Pervasive Computing (ICPC), Pune, India, 8–10 January 2015; pp. 1–5. [Google Scholar]
- Amin, S.U.; Agarwal, K.; Beg, R. Genetic neural network based data mining in prediction of heart disease using risk factors. In Proceedings of the 2013 IEEE Conference on Information & Communication Technologies, Hanoi, Vietnam, 20–22 March 2013; pp. 1227–1231. [Google Scholar]
- Chia, C.-C.; Syed, Z. Scalable noise mining in long-term electrocardiographic time-series to predict death following heart attacks. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 125–134. [Google Scholar]
- Toerper, M.F.; Flanagan, E.; Siddiqui, S.; Appelbaum, J.; Kasper, E.K.; Levin, S. Cardiac catheterization laboratory inpatient forecast tool: A prospective evaluation. J. Am. Med. Inform. Assoc. 2016, 23, e49–e57. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Phung, D.; Luo, W.; Venkatesh, S. Stabilized sparse ordinal regression for medical risk stratification. Knowl. Inf. Syst. 2015, 43, 555–582. [Google Scholar] [CrossRef]
- Kop, R.; Hoogendoorn, M.; Moons, L.M.G.; Numans, M.E.; Teije, A.T. On the advantage of using dedicated data mining techniques to predict colorectal cancer. In Proceedings of the Conference on Artificial Intelligence in Medicine in Europe, Pavia, Italy, 20 June 2015; Volume 9105, pp. 133–142. [Google Scholar]
- Simon, G.J.; Caraballo, P.J.; Therneau, T.M.; Cha, S.S.; Castro, M.R.; Li, P.W. Extending association rule summarization techniques to assess risk of diabetes mellitus. IEEE Trans. Knowl. Data Eng. 2015, 27, 130–141. [Google Scholar] [CrossRef]
- Rabbi, K.; Mamun, Q.; Islam, M.D.R. Dynamic feature selection (DFS) based Data clustering technique on sensory data streaming in eHealth record system. In Proceedings of the 2015 IEEE 10th Conference on Industrial Electronics and Applications (ICIEA), Auckland, New Zealand, 15–17 June 2015; pp. 661–665. [Google Scholar]
- Sumana, B.V.; Santhanam, T. Prediction of diseases by cascading clustering and classification. In Proceedings of the Advances in Electronics, Computers and Communications. International Conference (ICAECC 2014), Bangalore, India, 10–11 October 2014; pp. 1–8. [Google Scholar]
- Feldman, K.; Hazekamp, N.; Chawla, N.V. Mining the Clinical Narrative: All Text are Not Equal. In Proceedings of the 2016 IEEE International Conference on Healthcare Informatics (ICHI), Chicago, IL, USA, 4–7 October 2016; pp. 271–280. [Google Scholar]
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Pulmano, C.E.; Estuar, M.R.J.E. Towards Developing an Intelligent Agent to Assist in Patient Diagnosis Using Neural Networks on Unstructured Patient Clinical Notes: Initial Analysis and Models. Procedia Comput. Sci. 2016, 100, 263–270. [Google Scholar] [CrossRef]
- Araneo, R.; Celozzi, S. The Feasibility of Using Large-Scale Text Mining to Detect Adverse Childhood Experiences in a VA-Treated Population. Appl. Comput. Electromagn. Soc. J. 2015, 28, 505–514. [Google Scholar]
- Luther, S.L. Improving identification of fall-related injuries in ambulatory care using statistical text mining. Am. J. Public Health 2015, 105, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Jonnagaddala, J.; Liaw, S.T.; Ray, P.; Kumar, M.; Chang, N.W.; Dai, H.J. Coronary artery disease risk assessment from unstructured electronic health records using text mining. J. Biomed. Inform. 2015, 58, S203–S210. [Google Scholar] [CrossRef] [PubMed]
- Fong, A.; Hettinger, A.Z.; Ratwani, R.M. Exploring methods for identifying related patient safety events using structured and unstructured data. J. Biomed. Inform. 2015, 58, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.B.; Namen, A.A. A hybrid data mining model for diagnosis of patients with clinical suspicion of dementia. Comput. Methods Prog. Biomed. 2018, 165, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, A.; Ramanathan, S.V.; Thati, R. Novel Approach to Predict Hospital Readmissions Using Feature Selection from Unstructured Data with Class Imbalance. Big Data Res. 2018, 13, 65–75. [Google Scholar] [CrossRef]
- Kharrazi, H. The Value of Unstructured Electronic Health Record Data in Geriatric Syndrome Case Identification. J. Am. Geriatr. Soc. 2018, 66, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Arenas, M.; Maturana, F.; Riveros, C.; Vrgoˇc, D. A framework for annotating CSV-like data. Proc. VLDB Endow. 2016, 9, 876–887. [Google Scholar] [CrossRef]
- Pooled Resource Open-Access ALS Clinical Trials Database. Available online: https://nctu.partners.org/proact/data/index (accessed on 27 February 2019).
- Feder, S.L. Data quality in electronic health records research: Quality domains and assessment methods. Western J. Nurs. Res. 2018, 40, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Reimer, M.A.M.E.; Andrew, P. Data quality assessment framework to assess electronic medical record data for use in research. Int. J. Med. Inform. 2016, 90, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-Y.; Cheng, C.-W.; Kaddi, C.D.; Venugopalan, J.; Hoffman, R.; Wang, M.D. Omic and electronic health record big data analytics for precision medicine. IEEE Trans. Biomed. Eng. 2017, 64, 263–273. [Google Scholar] [PubMed]
- Weiskopf, N.G.; Weng, C. Methods and dimensions of electronic health record data quality assessment: Enabling reuse for clinical research. J. Am. Med. Inform. Assoc. 2013, 20, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Tennison, J.; Kellogg, G.; Herman, I. Model for tabular data and metadata on the web, W3C Working Draft 8. Available online: https://www.w3.org/TR/tabular-data-model/ (accessed on 27 February 2019).
- Amaral, P.; Pinto, S.; de Carvalho, M.; Tom’as, P.; Madeira, S.C. Predicting the need for noninvasive ventilation in patients with amyotrophic lateral sclerosis. In Proceedings of the ACM SIGKDD Workshop on Health Informatics (HI-KDD 2012), Beijing, China, 12 August 2012. [Google Scholar]
- Carreiro, A.V.; Amaral, P.M.; Pinto, S.; Tom’as, P.; de Carvalho, M.; Madeira, S.C. Prognostic models based on patient snapshots and time windows: Predicting disease progression to assisted ventilation in amyotrophic lateral sclerosis. J. Biomed. Inform. 2015, 58, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.; Pereira, R.; Ferreira, J.; Vasconcelos, J.B.; Melo, F.; Velez, I. Predictive analysis in healthcare: Emergency wait time prediction. Adv. Intell. Syst. Comput. 2019, 806, 138–145. [Google Scholar]






| Model | Methods | Application | Sector | Ref | Year |
|---|---|---|---|---|---|
| Hybrid Model | NN and GA | Heart disease risk prediction | Cardiology | [17] | 2013 |
| Hybrid Model | K-means and clustering technologies | EMR data processing | Management | [24] | 2014 |
| Time Series Mining | Time series mining | Risk prediction of heart disease | Cardiology | [18] | 2014 |
| Association Rule Mining | Association rule mining | Diabetes risk prediction | Endocrinology | [21] | 2015 |
| Classification | Dynamic classification and hierarchical model | Re-hospitalization risk prediction | Management | [13] | 2015 |
| Classification | Multi-classifier method | Noncommunicable disease prediction | Epidemiology | [14] | 2015 |
| Classification | Support Vector Machine | Intensive Care units (ICU) risk prediction | Cardiology | [15] | 2015 |
| Classification | Decision Trees | Decision Support Systems | Management | [16] | 2015 |
| Clustering | Dynamic feature selection | Quality of medical service | Quality of Service | [23] | 2015 |
| Regression Algorithm | Ordinal regression framework | Suicide risk prediction | Psychiatry | [20,21] | 2015 |
| Time Series Mining | Time series mining | Risk prediction of colorectal cancer | Oncology | [19] | 2015 |
| Regression Algorithm | Multivariate logistic regression | Analysis of Cardiac Surgical Bed Demand | Management | [25] | 2016 |
| NLP Methods | Methods | Application | Reference | Year |
|---|---|---|---|---|
| Stemming, stop words, numbers and dates removed | Logistic Regression | Detection of Adverse Childhood Experiences | [28] | 2015 |
| Stop words, lowercase, (IDF) Inverse Document Frequency | Support Vector Machine (SVM) | Identification of fall-related injuries | [29] | 2015 |
| Self-Developed Algorithm | Coronary artery disease risk assessment | [30] | 2015 | |
| Tokenization, Term Frequency-Inverse Document Frequency (TF-IDF), Removal of Stopwords | Forward Neural Network with Back Propagation | Automatic diagnosis prediction | [27] | 2016 |
| Extracting | [6] | 2018 |
| Methods | Application | Sector | Ref | Year |
|---|---|---|---|---|
| Topic Modeling (LDA) | Identify Related Patient Safety | Events Security | [31] | 2015 |
| Bayesian Belief Networks and DT | Diagnosing Early Stages of Dementia Psychiatry | Psychiatry | [32] | 2018 |
| Logistic Regression | Predict Hospital Readmissions | Cardiology | [33] | 2018 |
| Clustering | Geriatric Syndrome Detection | Geriatric | [34] | 2018 |
| Characteristics | Description/Value | Comments |
|---|---|---|
| Data Source | Hospital Santa Maria, Lisbon, Portugal | |
| Data Format | Microsoft Excel Spreadsheet (xls) | Binary Format |
| Number of Sheets | 25 | Atrophy, Blood, Cramps, DeltoidEMG, Demographics, Diagnostic EMG, EPO-VGF, Fascic, Medication, MRC, NIV, NPO, PEG, Phrenic EMG, Reals, Reflexes, RFT, SCMEMG, SNIP, Spasticity, Treadmill |
| Static Information | Demographics (1 worksheet) | 71 field of static information about each patient |
| Temporal Information | 24 worksheets | |
| Number of Patients | 495 |
| Case | Date | Feature A | Feature B | Date | Feature A | Feature B | Date |
|---|---|---|---|---|---|---|---|
| 1 | 28 June 2010 | not done | 2.5 | 10 June 2011 | 10 | ||
| 2 | 5 November-2010 | 50 | N | 28 February 2011 | 2 | 3.5 | 12 June 2011 |
| 3 | 10 November 2006 | 12,3 | 12.5 | 11 December 2006 | 3 | 2,3 |
| Name | Birthday | Gender (1-Male; 2-Female) | Height (m) | Weight before 1st Symptoms (kg) |
|---|---|---|---|---|
| 1 | 6 June 1951 | 1 | 1.69 | 73 |
| 2 | 30 June 1951 | 1 | 1.80 | 58 |
| 3 | 3 January 2010 | 1 | ||
| 4 | 1 August 1927 | 1 | 1.65 | |
| 5 | 27 August 1941 | 1 | 1.57 | |
| 6 | 5 December 1940 | 2 | 1.58 | 57 |
| Name | Date | ALS-FRS | ALS-FRS-R | ALS-FRSb | R | NIV |
|---|---|---|---|---|---|---|
| 1 | 28 June 2010 | 37 | 44 | 12 | 10 | 0 |
| 1 | 6 October 2010 | 37 | 44 | 12 | 10 | 0 |
| 1 | 5 January011 | 37 | 44 | 12 | 10 | 1 |
| 1 | 5 May 2011 | 37 | 45 | 12 | 12 | 1 |
| 1 | 6 July 2011 | 38 | 46 | 12 | 12 | 1 |
| 10 | 19 May 2003 | 29 | 37 | 12 | 12 | 0 |
| 10 | 4 August 2003 | 30 | 38 | 12 | 12 | 0 |
| 10 | 3 November 2003 | 24 | 32 | 12 | 11 | 0 |
| Name | Date | %VC | %FVC | %MIP | %MEP | %P0.1 | PO2 | PCO2 | Weight | NIV |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 17 May 2010 | 85 | 86 | 29 | 52 | 64 | 84 | 43 | 73 | 0 |
| 10 | 12 May 2003 | 95 | 98 | 89 | 1,13 | 1,13 | 89 | 39 | 79 | 1 |
| 101 | 09 December 2004 | 1,06 | 1,1 | 69 | 1,21 | 61 | 91 | 35 | 1 | |
| 102 | 25 August 2004 | 38 | 40 | 23 | 25 | 30 | 70 | 63 | 73 | 0 |
| 104 | 04 August 2005 | 32 | 32 | 19 | 36 | 3,1 | 88 | 36 | 46 | 0 |
| Name | Date | SpO2mean | SpO2min | SpO2 < 9 0% | Disp 4% | Disp h < 4% | Disp3 | Disp h < 3% | Pattern | NIV |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 July 2010 | 95.56 | 92 | 0 | 5 | 0.96 | 10 | 1.92 | 1 | 1 |
| 1 | 4 April 2011 | 96.33 | 90 | 0 | 29 | 3.61 | 44 | 5.48 | 3 | 1 |
| 1 | 7 January 2003 | 94.25 | 89 | 0.06 | 3 | 0 | ||||
| 10 | 12 February 2003 | 94 | 85 | 0.71 | 23 | 2.57 | 51 | 5.69 | 3 | 0 |
| 10 | 30 January 2004 | 94.94 | 85 | 0.35 | 29 | 2.42 | 56 | 4.67 | 3 | 1 |
| Name | Date | PhrenMeanLat | PhrenMeanAmpl | PhrenMeanArea | NIV |
|---|---|---|---|---|---|
| 1 | 7 July2010 | 8 | 1 | 3 | 1 |
| 10 | 2 December 2003 | 9 | 0 | 2 | 0 |
| 100 | 1 January 2004 | 8 | 1 | 3 | 0 |
| 100 | 16 July 2004 | 7 | 1 | 3 | 1 |
| Name | Date | ALS-FRS | ALS-FRS-R | ALS-Frsb | R | Name_1 | Gender | BMI | MND | Age At Onset |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 28 June 2010 00:00:00.000 | 37 | 44 | 12 | 10 | 1 | 1 | 25.6 | 2 | 59 |
| 1 | 6 July 2011 00:00:00.000 | 38 | 46 | 12 | 12 | 1 | 1 | 25.6 | 2 | 59 |
| 10 | 10 May 2003 00:00:00.000 | 29 | 37 | 12 | 12 | 10 | 1 | 26.6 | 2 | 73 |
| 10 | 4 August 2003 00:00:00.000 | 30 | 38 | 12 | 12 | 10 | 1 | 26.6 | 2 | 73 |
| 10 | 3 November 200300:00:00.000 | 24 | 32 | 12 | 11 | 10 | 1 | 26.6 | 2 | 73 |
| Name | El Escorial Reviewed Criteria | Onset Form | Envolved Segment—1st Symptoms | Evolution Pattern | Name |
|---|---|---|---|---|---|
| 1 | pro | 1 | LL | RLL- LLL | 1 |
| 1 | pro | 1 | LL | RLL- LLL | 1 |
| 10 | pos | 1 | UL | LUL-LLL-RUL-RLL-R | 10 |
| 10 | pos | 1 | UL | LUL-LLL-RUL-RLL-R | 10 |
| 10 | pos | 1 | UL | LUL-LLL-RUL-RLL-R | 10 |
| Name | 1st Symptoms - 1st Visit | Alive\_Death\_Disappeared | Date of Death\_Disappearance | NIV |
|---|---|---|---|---|
| 1 | 18.1 | 0 | 0 | |
| 1 | 18.1 | 0 | 0 | |
| 10 | 28.3 | 1 | 20 December 2004 | 0 |
| 10 | 28.3 | 1 | 20 December 2004 | 0 |
| 10 | 28.3 | 1 | 20 December 2004 | 1 |
| Input File Names | Input Entries | Output Entries | Input Fields | Output Fields | Type Erros | Format Errors | Value Errors | Total Errors |
|---|---|---|---|---|---|---|---|---|
| demographics.csv | 616 | 481 | 71 | 13 | 2 | 15 | 5 | 22 |
| als-frs.csv | 502 | 780 | 243 | 6 | 122 | 25 | 4113 | 4260 |
| Input File | Input Entries | Output Entries | Regex Exclusions | Threshold Exclusions | Missing Value Exclusions | Total Exclusions |
|---|---|---|---|---|---|---|
| demographics.csv | 481 | 458 | 6 | 12 | 5 | 23 |
| als-frs.csv | 780 | 725 | 6 | 25 | 24 | 55 |
| Output File | Input Entries | Output Entries | Missing Key Exclusions |
|---|---|---|---|
| demographics.csv | 458 | 458 | 0 |
| als-frs\_with\_niv.csv | 725 | 725 | 24 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helgheim, B.I.; Maia, R.; Ferreira, J.C.; Martins, A.L. Merging Data Diversity of Clinical Medical Records to Improve Effectiveness. Int. J. Environ. Res. Public Health 2019, 16, 769. https://doi.org/10.3390/ijerph16050769
Helgheim BI, Maia R, Ferreira JC, Martins AL. Merging Data Diversity of Clinical Medical Records to Improve Effectiveness. International Journal of Environmental Research and Public Health. 2019; 16(5):769. https://doi.org/10.3390/ijerph16050769
Chicago/Turabian StyleHelgheim, Berit I., Rui Maia, Joao C. Ferreira, and Ana Lucia Martins. 2019. "Merging Data Diversity of Clinical Medical Records to Improve Effectiveness" International Journal of Environmental Research and Public Health 16, no. 5: 769. https://doi.org/10.3390/ijerph16050769
APA StyleHelgheim, B. I., Maia, R., Ferreira, J. C., & Martins, A. L. (2019). Merging Data Diversity of Clinical Medical Records to Improve Effectiveness. International Journal of Environmental Research and Public Health, 16(5), 769. https://doi.org/10.3390/ijerph16050769






