Evaluation of DNA Methylation Changes and Micronuclei in Workers Exposed to a Construction Environment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Group and Biological Sample Collection
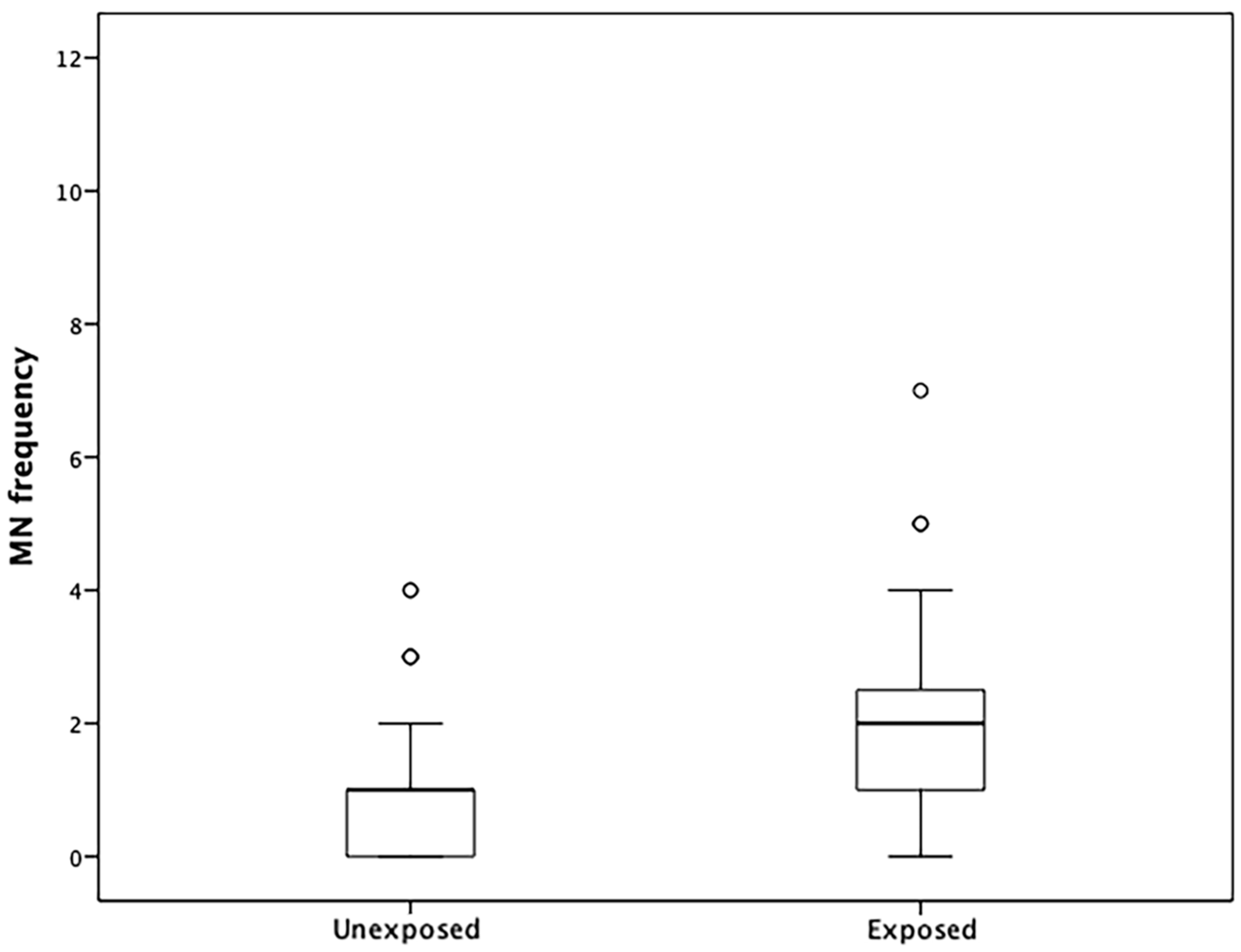
2.2. MNs in Lymphocytes
2.3. DNA Isolation, Bisulfite Conversion, and Pyrosequencing
2.4. Analysis of Trace Elements in Blood
2.5. Analysis of PM and Work Environment
2.6. Statistical Analysis
3. Results
3.1. General Characteristics
3.2. Analysis of Trace Elements in Blood
3.3. Analysis of PM2.5 Emitted in the Construction Environment
3.4. Tumor-Suppressor Genes and LINE-1 Methylation Levels by Exposure Group
3.5. Substances from the Work Environment and Their Effects on DNA Methylation
3.6. Confounding Factors and DNA Methylation Levels
3.7. MN Frequencies and Correlations with DNA Methylation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Consonni, D.; De Matteis, S.; Pesatori, A.C.; Bertazzi, P.A.; Olsson, A.C.; Kromhout, H.; Peters, S.; Vermeulen, R.C.; Pesch, B.; Bruning, T.; et al. Lung cancer risk among bricklayers in a pooled analysis of case-control studies. Int. J. Cancer 2015, 136, 360–371. [Google Scholar] [CrossRef]
- Donaldson, K.; Poland, C.A. Nanotoxicology: New insights into nanotubes. Nat. Nanotechnol. 2009, 4, 708–710. [Google Scholar] [CrossRef]
- Yang, M. A current global view of environmental and occupational cancers. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2011, 29, 223–249. [Google Scholar] [CrossRef]
- Lim, Y.H.; Kim, H.; Kim, J.H.; Bae, S.; Park, H.Y.; Hong, Y.C. Air pollution and symptoms of depression in elderly adults. Environ. Health Perspect. 2012, 120, 1023–1028. [Google Scholar] [CrossRef]
- Kampa, M.; Castanas, E. Human health effects of air pollution. Environ. Pollut. 2008, 151, 362–367. [Google Scholar] [CrossRef]
- Resende, F. Poluição Atmosférica por Emissão de Material Particulado: Avaliação e Controle nos Canteiros de obras de Edificios. Ph.D. Thesis, Escola Politécnica da Universidade de São Paulo, São Paulo, Brazil, 2007. [Google Scholar]
- Ghio, A.J.; Carraway, M.S.; Madden, M.C. Composition of air pollution particles and oxidative stress in cells, tissues, and living systems. J. Toxicol. Environ. Health B Crit. Rev. 2012, 15, 1–21. [Google Scholar] [CrossRef]
- Soberanes, S.; Gonzalez, A.; Urich, D.; Chiarella, S.E.; Radigan, K.A.; Osornio-Vargas, A.; Joseph, J.; Kalyanaraman, B.; Ridge, K.M.; Chandel, N.S.; et al. Particulate matter Air Pollution induces hypermethylation of the p16 promoter Via a mitochondrial ROS-JNK-DNMT1 pathway. Sci. Rep. 2012, 2, 275. [Google Scholar] [CrossRef]
- Hamra, G.B.; Guha, N.; Cohen, A.; Laden, F.; Raaschou-Nielsen, O.; Samet, J.M.; Vineis, P.; Forastiere, F.; Saldiva, P.; Yorifuji, T.; et al. Outdoor particulate matter exposure and lung cancer: A systematic review and meta-analysis. Environ. Health Perspect. 2014, 122, 906–911. [Google Scholar] [CrossRef]
- Zhao, J.; Gao, Z.; Tian, Z.; Xie, Y.; Xin, F.; Jiang, R.; Kan, H.; Song, W. The biological effects of individual-level PM(2.5) exposure on systemic immunity and inflammatory response in traffic policemen. Occup. Environ. Med. 2013, 70, 426–431. [Google Scholar] [CrossRef]
- Bolognesi, C.; Knasmueller, S.; Nersesyan, A.; Thomas, P.; Fenech, M. The HUMNxl scoring criteria for different cell types and nuclear anomalies in the buccal micronucleus cytome assay—An update and expanded photogallery. Mutat. Res. 2013, 753, 100–113. [Google Scholar] [CrossRef]
- Bonassi, S.; El-Zein, R.; Bolognesi, C.; Fenech, M. Micronuclei frequency in peripheral blood lymphocytes and cancer risk: Evidence from human studies. Mutagenesis 2011, 26, 93–100. [Google Scholar] [CrossRef]
- Duan, H.; He, Z.; Ma, J.; Zhang, B.; Sheng, Z.; Bin, P.; Cheng, J.; Niu, Y.; Dong, H.; Lin, H.; et al. Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon. Arch. Toxicol. 2013, 87, 2013–2022. [Google Scholar] [CrossRef]
- Herceg, Z. Epigenetics and cancer: Towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007, 22, 91–103. [Google Scholar] [CrossRef]
- Hou, L.; Zhang, X.; Tarantini, L.; Nordio, F.; Bonzini, M.; Angelici, L.; Marinelli, B.; Rizzo, G.; Cantone, L.; Apostoli, P.; et al. Ambient PM exposure and DNA methylation in tumor suppressor genes: A cross-sectional study. Part. Fibre Toxicol. 2011, 8, 25. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; He, Z.; Duan, H.; Gao, W.; Wang, H.; Yu, S.; Chen, W.; Zheng, Y. Associations between DNA methylation in DNA damage response-related genes and cytokinesis-block micronucleus cytome index in diesel engine exhaust-exposed workers. Arch. Toxicol. 2016, 90, 1997–2008. [Google Scholar] [CrossRef]
- Hossain, M.B.; Vahter, M.; Concha, G.; Broberg, K. Environmental arsenic exposure and DNA methylation of the tumor suppressor gene p16 and the DNA repair gene MLH1: Effect of arsenic metabolism and genotype. Metallomics 2012, 4, 1167–1175. [Google Scholar] [CrossRef]
- Wu, F.; Liu, J.; Qiu, Y.L.; Wang, W.; Zhu, S.M.; Sun, P.; Miao, W.B.; Li, Y.L.; Brandt-Rauf, P.W.; Xia, Z.L. Correlation of chromosome damage and promoter methylation status of the DNA repair genes MGMT and hMLH1 in Chinese vinyl chloride monomer (VCM)-exposed workers. Int. J. Occup. Med. Environ. Health 2013, 26, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Lorincz, A.T. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev. Mol. Diagn. 2014, 14, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Byun, H.M.; Zhong, J.; Motta, V.; Barupal, J.; Zheng, Y.; Dou, C.; Zhang, F.; McCracken, J.P.; Diaz, A.; et al. Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ. Mol. Mutagen. 2014, 55, 322–335. [Google Scholar] [CrossRef]
- Kato, T.; Suzuki, K.; Okada, S.; Kamiyama, H.; Maeda, T.; Saito, M.; Koizumi, K.; Miyaki, Y.; Konishi, F. Aberrant methylation of PSD disturbs Rac1-mediated immune responses governing neutrophil chemotaxis and apoptosis in ulcerative colitis-associated carcinogenesis. Int. J. Oncol. 2012, 40, 942–950. [Google Scholar] [CrossRef]
- Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007, 2, 1084–1104. [Google Scholar] [CrossRef] [Green Version]
- Silveira, H.C.; Schmidt-Carrijo, M.; Seidel, E.H.; Scapulatempo-Neto, C.; Longatto-Filho, A.; Carvalho, A.L.; Reis, R.M.; Saldiva, P.H. Emissions generated by sugarcane burning promote genotoxicity in rural workers: A case study in Barretos, Brazil. Environ. Health 2013, 12, 87. [Google Scholar] [CrossRef]
- Ramos, M.A.; Cury Fde, P.; Scapulatempo Neto, C.; Marques, M.M.; Silveira, H.C. Micronucleus evaluation of exfoliated buccal epithelial cells using liquid-based cytology preparation. Acta Cytol. 2014, 58, 582–588. [Google Scholar] [CrossRef]
- Vaissiere, T.; Hung, R.J.; Zaridze, D.; Moukeria, A.; Cuenin, C.; Fasolo, V.; Ferro, G.; Paliwal, A.; Hainaut, P.; Brennan, P.; et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009, 69, 243–252. [Google Scholar] [CrossRef]
- Batista, B.L.; Rodrigues, J.L.; Nunes, J.A.; Souza, V.C.; Barbosa, F., Jr. Exploiting dynamic reaction cell inductively coupled plasma mass spectrometry (DRC-ICP-MS) for sequential determination of trace elements in blood using a dilute-and-shoot procedure. Anal. Chim. Acta 2009, 639, 13–18. [Google Scholar] [CrossRef]
- Santos, A.G.; Regis, A.C.; da Rocha, G.O.; Bezerra Mde, A.; de Jesus, R.M.; de Andrade, J.B. A simple, comprehensive, and miniaturized solvent extraction method for determination of particulate-phase polycyclic aromatic compounds in air. J. Chromatogr. A 2016, 1435, 6–17. [Google Scholar] [CrossRef]
- Yassaa, N.; Meklati, B.Y.; Cecinato, A.; Marino, F. Organic aerosols in urban and waste landfill of Algiers metropolitan area: Occurrence and sources. Environ. Sci. Technol. 2001, 35, 306–311. [Google Scholar] [CrossRef]
- Ohura, T.; Amagai, T.; Fusaya, M.; Matsushita, H. Polycyclic aromatic hydrocarbons in indoor and outdoor environments and factors affecting their concentrations. Environ. Sci. Technol. 2004, 38, 77–83. [Google Scholar] [CrossRef]
- Rogge, W.F.; Hildemann, L.M.; Mazurek, M.A.; Cass, G.R.; Simoneit, B.R. Sources of fine organic aerosol. 2. Noncatalyst and catalyst-equipped automobiles and heavy-duty diesel trucks. Environ. Sci. Technol. 1993, 27, 636–651. [Google Scholar] [CrossRef]
- Yang, P.; Ma, J.; Zhang, B.; Duan, H.; He, Z.; Zeng, J.; Zeng, X.; Li, D.; Wang, Q.; Xiao, Y.; et al. CpG site-specific hypermethylation of p16INK4alpha in peripheral blood lymphocytes of PAH-exposed workers. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 182–190. [Google Scholar] [CrossRef]
- Ali, A.H.; Kondo, K.; Namura, T.; Senba, Y.; Takizawa, H.; Nakagawa, Y.; Toba, H.; Kenzaki, K.; Sakiyama, S.; Tangoku, A. Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol. Carcinog. 2011, 50, 89–99. [Google Scholar] [CrossRef]
- Ding, R.; Jin, Y.; Liu, X.; Zhu, Z.; Zhang, Y.; Wang, T.; Xu, Y. Characteristics of DNA methylation changes induced by traffic-related air pollution. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 796, 46–53. [Google Scholar] [CrossRef]
- Terry, M.B.; Delgado-Cruzata, L.; Vin-Raviv, N.; Wu, H.C.; Santella, R.M. DNA methylation in white blood cells: Association with risk factors in epidemiologic studies. Epigenetics 2011, 6, 828–837. [Google Scholar] [CrossRef]
- Rota, F.; Conti, A.; Campo, L.; Favero, C.; Cantone, L.; Motta, V.; Polledri, E.; Mercadante, R.; Dieci, G.; Bollati, V.; et al. Epigenetic and Transcriptional Modifications in Repetitive Elements in Petrol Station Workers Exposed to Benzene and MTBE. Int. J. Environ. Res. Public Health 2018, 15, 735. [Google Scholar] [CrossRef] [Green Version]
- Andreotti, G.; Karami, S.; Pfeiffer, R.M.; Hurwitz, L.; Liao, L.M.; Weinstein, S.J.; Albanes, D.; Virtamo, J.; Silverman, D.T.; Rothman, N.; et al. LINE1 methylation levels associated with increased bladder cancer risk in pre-diagnostic blood DNA among US (PLCO) and European (ATBC) cohort study participants. Epigenetics 2014, 9, 404–415. [Google Scholar] [CrossRef]
- Salas, L.A.; Villanueva, C.M.; Tajuddin, S.M.; Amaral, A.F.; Fernandez, A.F.; Moore, L.E.; Carrato, A.; Tardon, A.; Serra, C.; Garcia-Closas, R.; et al. LINE-1 methylation in granulocyte DNA and trihalomethane exposure is associated with bladder cancer risk. Epigenetics 2014, 9, 1532–1539. [Google Scholar] [CrossRef] [Green Version]
- Holland, N.; Bolognesi, C.; Kirsch-Volders, M.; Bonassi, S.; Zeiger, E.; Knasmueller, S.; Fenech, M. The micronucleus assay in human buccal cells as a tool for biomonitoring DNA damage: The HUMN project perspective on current status and knowledge gaps. Mutat. Res. 2008, 659, 93–108. [Google Scholar] [CrossRef]
- Hoyos-Giraldo, L.S.; Escobar-Hoyos, L.F.; Saavedra-Trujillo, D.; Reyes-Carvajal, I.; Munoz, A.; Londono-Velasco, E.; Tello, A.; Cajas-Salazar, N.; Ruiz, M.; Carvajal, S.; et al. Gene-specific promoter methylation is associated with micronuclei frequency in urothelial cells from individuals exposed to organic solvents and paints. J. Expo. Sci. Environ. Epidemiol. 2016, 26, 257–262. [Google Scholar] [CrossRef]
- Devoz, P.P.; Gomes, W.R.; De Araujo, M.L.; Ribeiro, D.L.; Pedron, T.; Greggi Antunes, L.M.; Batista, B.L.; Barbosa, F., Jr.; Barcelos, G.R.M. Lead (Pb) exposure induces disturbances in epigenetic status in workers exposed to this metal. J. Toxicol. Environ. Health A 2017, 80, 1098–1105. [Google Scholar] [CrossRef]
- Huang, H.; Ji, Y.; Zhang, J.; Su, Z.; Liu, M.; Tong, J.; Ge, C.; Chen, T.; Li, J. Aberrant DNA methylation in radon and/or cigarette smoke-induced malignant transformation in BEAS-2B human lung cell line. J. Toxicol. Environ. Health A 2017, 80, 1321–1330. [Google Scholar] [CrossRef]
- Miller, C.N.; Rayalam, S. The role of micronutrients in the response to ambient air pollutants: Potential mechanisms and suggestions for research design. J. Toxicol. Environ. Health B Crit. Rev. 2017, 20, 38–53. [Google Scholar] [CrossRef]
- de Castro Vasconcellos, P.; Sanchez-Ccoyllo, O.; Balducci, C.; Mabilia, R.; Cecinato, A. Occurrence and concentration levels of nitro-PAH in the air of three Brazilian cities experiencing different emission impacts. Water Air Soil Pollut. 2008, 190, 87–94. [Google Scholar] [CrossRef]
| Variable | Exposed (N = 59) | Unexposed (N = 49) | p Value | |
|---|---|---|---|---|
| Age | <35 | 25 (46.3) | 24 (49) | 0.60 a |
| ≥35 | 29 (53.7) | 25 (51) | ||
| Exposure time | Average (years) | 10 | - | - |
| Passive smoker | Yes | 37 (62.7) | 14 (31.1) | 0.001 a,* |
| No | 22 (37.3) | 31 (68.9) | ||
| Ex- smoker | Yes | 25 (47.2) | 7 (22.6) | 0.036 a,* |
| No | 28 (52.8) | 24(77.4) | ||
| Alcohol consumption | Yes | 36 (61) | 40 (83.3) | 0.01 a,* |
| No/Past | 23 (39) | 8 (16.7) | ||
| Ethnic group | White | 36 (63.2) | 28 (73.7) | 0.28 b |
| Black | 8 (14) | 2 (5.3) | ||
| Mixed | 13 (22.8) | 7 (18.4) | ||
| Asian | 0 | 1 (2.6) | ||
| Cp Genomic Sites | Unexposed Workers | Exposed Workers | p Value | FDR c Corrected p Values | ||||
|---|---|---|---|---|---|---|---|---|
| N | Mean ± SD | Median (IQR) | N | Mean ± SD | Median (IQR) | |||
| CDKN2A (%5mC) | ||||||||
| CpG site 1 | 44 | 2.85 ± 1.89 | 2.81 (0–5.75) | 57 | 3.48 ± 2.57 | 2.97 (0−10.06) | 0.48 b | 0.53 |
| CpG site 2 | 44 | 5.11 ± 2.81 | 5.39 (0–10.35) | 57 | 6.56 ± 3.03 | 6.82 (0−13.58) | 0.02 b,* | 0.04 * |
| CpG site 3 | 44 | 2.51 ± 2.31 | 2.50 (0–6.53) | 57 | 3.25 ± 2.57 | 2.81 (0−9.97) | 0.24 b | 0.31 |
| CpG site 4 | 44 | 4.42 ± 1.51 | 4.4 (0–7.58) | 57 | 5.02 ± 2.00 | 5.08 (0−10.01) | 0.11 b | 0.18 |
| CpG site 5 | 44 | 4.12 ± 2.76 | 4.66 (0–9.47) | 57 | 5.33 ± 2.86 | 5.3 (0−11.62) | 0.08 b | 0.14 |
| CpG site 6 | 44 | 1.93 ± 1.84 | 1.97 (0−5.14) | 57 | 2.47 ± 2.29 | 2.15 (0−8.00) | 0.40 b | 0.47 |
| CpG site 7 | 44 | 4.46 ± 2.35 | 4.67 (0−8.87) | 57 | 5.87 ± 2.37 | 5.68 (0−12.22) | 0.005 b,* | 0.02 * |
| All CpG sites combined | 44 | 3.63 ± 2.04 | 3.68 (0−7.47) | 57 | 4.79 ± 2.57 | 4.44 (0−10.73) | 0.05 b,* | |
| MLH1 (%5mC) | ||||||||
| CpG site 1 | 41 | 4.76 ± 1.54 | 4.35 (2.32−8.63) | 56 | 5.20 ± 1.48 | 5.26 (0−8.10) | 0.15 a | 0.22 |
| CpG site 2 | 41 | 3.69 ± 1.29 | 3.62 (0−5.85) | 56 | 3.70 ± 1.29 | 3.64 (0−6.42) | 0.76 b | 0.76 |
| CpG site 3 | 41 | 3.02 ± 1.80 | 3.27 (0−6.25) | 56 | 3.90 ± 1.27 | 4.03 (0−7.25) | 0.02 b,* | 0.04 * |
| CpG site 4 | 41 | 5.29 ± 2.02 | 5.13 (0−9.50) | 56 | 6.18 ± 1.57 | 6.26 (2.53−9.70) | 0.01 a,* | 0.03 * |
| CpG site 5 | 41 | 4.61 ± 1.87 | 4.10 (0−8.34) | 56 | 5.90 ± 1.50 | 5.92 (2.42−9.59) | 0.001 b,* | 0.01 * |
| CpG site 6 | 41 | 1.91 ± 2.01 | 2.06 (0−5.72) | 56 | 3.86 ± 1.47 | 3.95 (0−8.47) | 0.001 b,* | 0.01 * |
| All CpG sites combined | 41 | 4.22 ± 1.55 | 4.00 (1.74−7.49) | 56 | 5.04 ± 1.38 | 5.03 (1.61−9.76) | 0.007 a,* | |
| APC (%5mC) | ||||||||
| CpG site 1 | 47 | 8.78 ± 10.14 | 7.48 (0−68.05) | 58 | 8.08 ± 3.26 | 8.13 (2.59−21.49) | 0.65 b | 0.68 |
| CpG site 2 | 47 | 1.99 ± 2.72 | 0.00 (0−9.46) | 58 | 3.43 ± 2.21 | 3.57 (0−11.01) | 0.002 b,* | 0.01 * |
| CpG site 3 | 47 | 4.22 ± 2.20 | 4.00 (0−12.18) | 58 | 5.13 ± 1.66 | 5.03 (1.61−11.51) | 0.013 b,* | 0.03 * |
| All CpG sites combined | 47 | 5.47 ± 2.43 | 6.13 (1.29−11.54) | 58 | 6.43 ± 3.33 | 5.73 (1.17−15.48) | 0.29 b | |
| CpG Hot | 47 | 3.11 ± 2.10 | 2.61 (0−10.82) | 58 | 4.29 ± 1.83 | 4.29 (0.81−9.61) | <0.001 b,* | |
| LINE-1 (%5mC) | ||||||||
| CpG site 1 | 44 | 74.45 ± 1.36 | 74.72 (70.72−76.94) | 57 | 73.20 ± 5.08 | 73.83 (36.54−73.17) | 0.01 b,* | 0.03 * |
| CpG site 2 | 44 | 73.42 ± 7.30 | 68.87 (66.92−84.14) | 57 | 72.52 ± 7.78 | 67.84 (64.95−85.26) | 0.16 b | 0.22 |
| CpG site 3 | 44 | 63.13 ± 6.68 | 58.82 (55.87−73.13) | 57 | 62.00 ± 7.16 | 58.20 (49.44−74.35) | 0.08 b | 0.14 |
| CpG site 4 | 44 | 59.08 ± 5.04 | 62.30 (50.52−64.39) | 57 | 59.17 ± 4.38 | 61.24 (48.66−64.35) | 0.34 b | 0.42 |
| CpG site 5 | 44 | 66.18 ± 2.41 | 66.47 (57.53−72.28) | 57 | 67.26 ± 1.11 | 67.14 (63.51−70.59) | 0.005 b,* | 0.02 * |
| All CpG sites combined | 44 | 68.04 ± 1.55 | 68.63 (64.98−70.37) | 57 | 66.83 ± 2.33 | 65.93 (62.48−72.48) | 0.005 b,* | |
| Gene | Variable | Β | SD (β) | p Value |
|---|---|---|---|---|
| CDKN2A (n = 101) | Constant | 2.69 | 0.74 | <0.001 * |
| Group (case/control) | 1.14 | 0.61 | 0.06 | |
| Ex-smoker (yes/no) | 0.82 | 0.61 | 0.18 | |
| Passive smoker (yes/no) | 0.23 | 0.6 | 0.7 | |
| Drinking habit | 0.72 | 0.59 | 0.22 | |
| MLH1 (n = 97) | Constant | 2 | 0.55 | <0.001 * |
| Group (case/control) | 1.86 | 0.45 | <0.001 * | |
| Ex-smoker (yes/no) | 0.13 | 0.45 | 0.78 | |
| Passive smoker (yes/no) | −0.07 | 0.44 | 0.88 | |
| Drinking habit | 0.01 | 0.43 | 0.98 | |
| APC (n = 105) | Constant | 3.33 | 0.61 | <0.001 * |
| Group (case/control) | 1.05 | 0.5 | 0.03 * | |
| Ex-smoker (yes/no) | 0.44 | 0.51 | 0.39 | |
| Passive smoker (yes/no) | −0.44 | 0.49 | 0.37 | |
| Drinking habit | 0.16 | 0.49 | 0.74 | |
| LINE-1 (n = 101) | Constant | 67.22 | 0.64 | <0.001 * |
| Group (case/control) | −1.12 | 0.53 | 0.03 * | |
| Ex-smoker (yes/no) | −0.17 | 0.53 | 0.75 | |
| Passive smoker (yes/no) | 0.46 | 0.52 | 0.38 | |
| Drinking habit | 0.69 | 0.51 | 0.18 |
| Groups | Gene | N | B | IRR (CI95%) | p Value |
|---|---|---|---|---|---|
| Exposed | LINE-1 | 48 | −0.114 | 0.89 (0.8–0.99) | 0.039 |
| Unexposed | LINE-1 | 40 | 0.142 | 1.15 (0.93–1.43) | 0.196 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, I.R.; Ramos, M.C.A.S.; Arantes, L.M.R.B.; Lengert, A.V.H.; Oliveira, M.A.; Cury, F.P.; Martins Pereira, G.; Santos, A.G.; Barbosa, F., Jr.; Vasconcellos, P.C.; et al. Evaluation of DNA Methylation Changes and Micronuclei in Workers Exposed to a Construction Environment. Int. J. Environ. Res. Public Health 2019, 16, 902. https://doi.org/10.3390/ijerph16060902
Silva IR, Ramos MCAS, Arantes LMRB, Lengert AVH, Oliveira MA, Cury FP, Martins Pereira G, Santos AG, Barbosa F Jr., Vasconcellos PC, et al. Evaluation of DNA Methylation Changes and Micronuclei in Workers Exposed to a Construction Environment. International Journal of Environmental Research and Public Health. 2019; 16(6):902. https://doi.org/10.3390/ijerph16060902
Chicago/Turabian StyleSilva, Isana R., Manoela C. A. S. Ramos, Lídia M. R. B. Arantes, André V. H. Lengert, Marco A. Oliveira, Fernanda P. Cury, Guilherme Martins Pereira, Aldenor G. Santos, Fernando Barbosa, Jr., Pérola C. Vasconcellos, and et al. 2019. "Evaluation of DNA Methylation Changes and Micronuclei in Workers Exposed to a Construction Environment" International Journal of Environmental Research and Public Health 16, no. 6: 902. https://doi.org/10.3390/ijerph16060902
APA StyleSilva, I. R., Ramos, M. C. A. S., Arantes, L. M. R. B., Lengert, A. V. H., Oliveira, M. A., Cury, F. P., Martins Pereira, G., Santos, A. G., Barbosa, F., Jr., Vasconcellos, P. C., Cuenin, C., Herceg, Z., & Silveira, H. C. S. (2019). Evaluation of DNA Methylation Changes and Micronuclei in Workers Exposed to a Construction Environment. International Journal of Environmental Research and Public Health, 16(6), 902. https://doi.org/10.3390/ijerph16060902






