Interaction between Fungal Communities, Soil Properties, and the Survival of Invading E. coli O157:H7 in Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sample Collection and Characterization
2.2. Soil Inoculation and Enumeration
2.3. Soil DNA Extraction, Illumina MiSeq and Sequence Data Analysis
2.4. Survival Data Modeling
2.5. Statistical Analysis.
3. Results
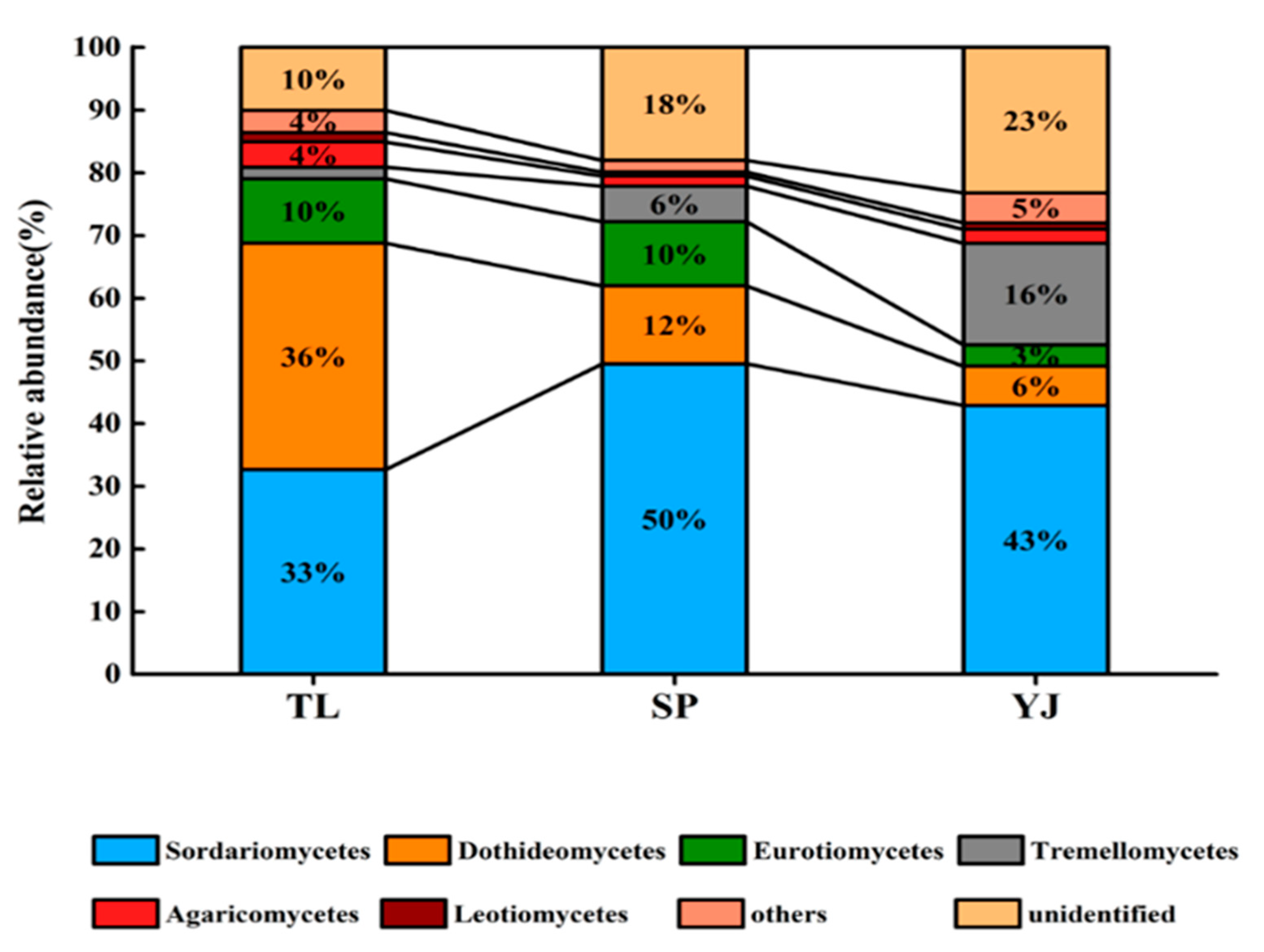
3.1. Fungal Community Composition and Structure
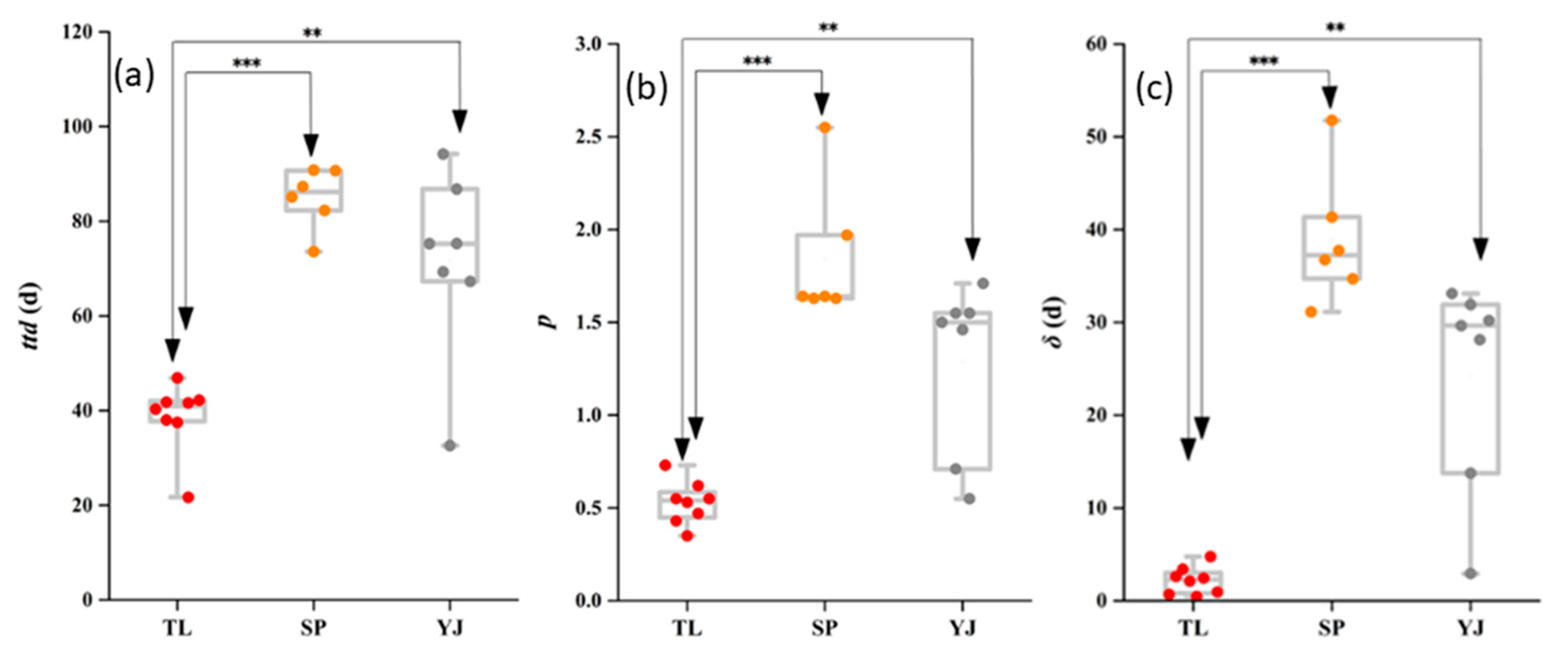
3.2. Comparison of Survival Profiles and Survival Parameters (ttd, p, and δ).
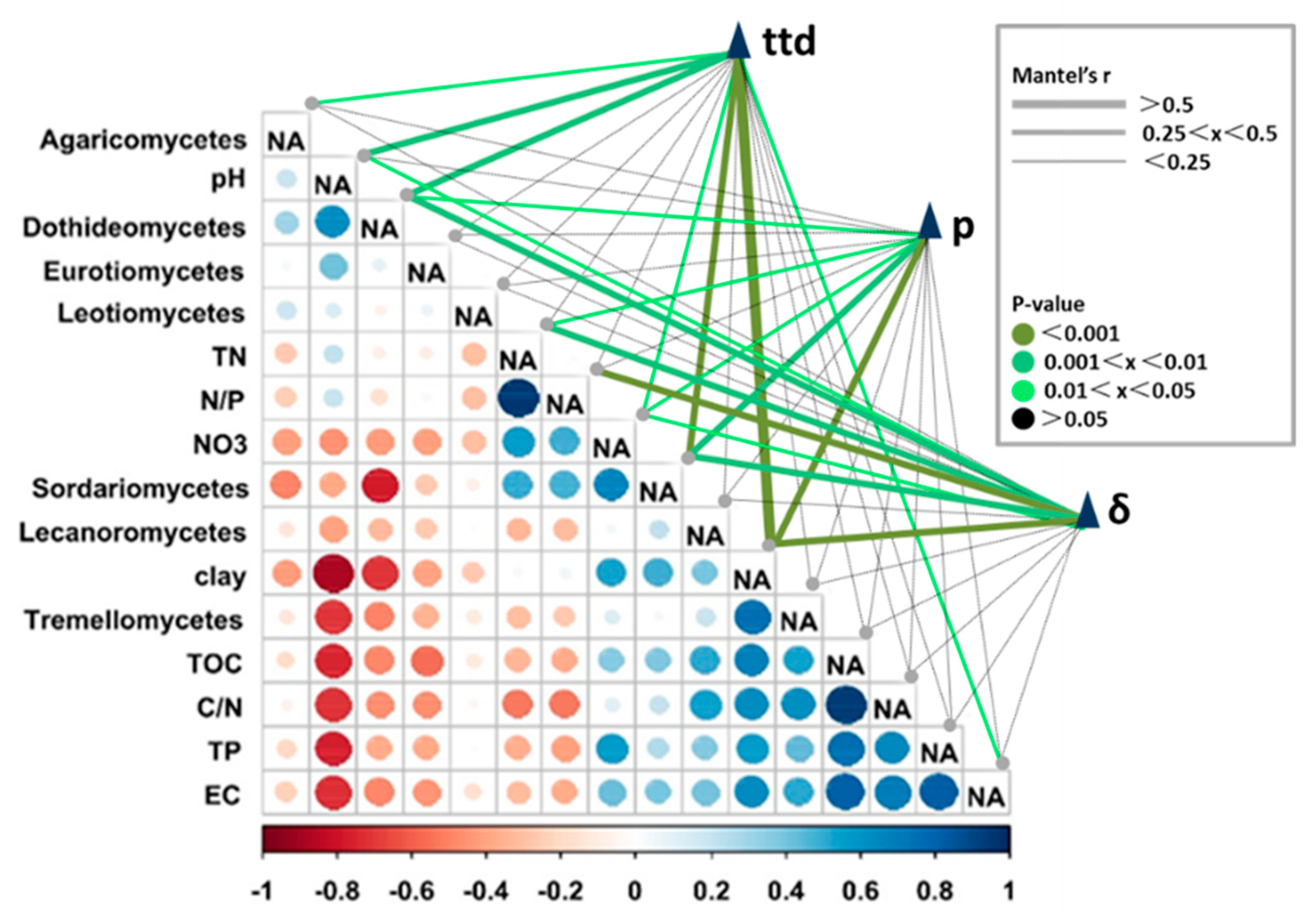
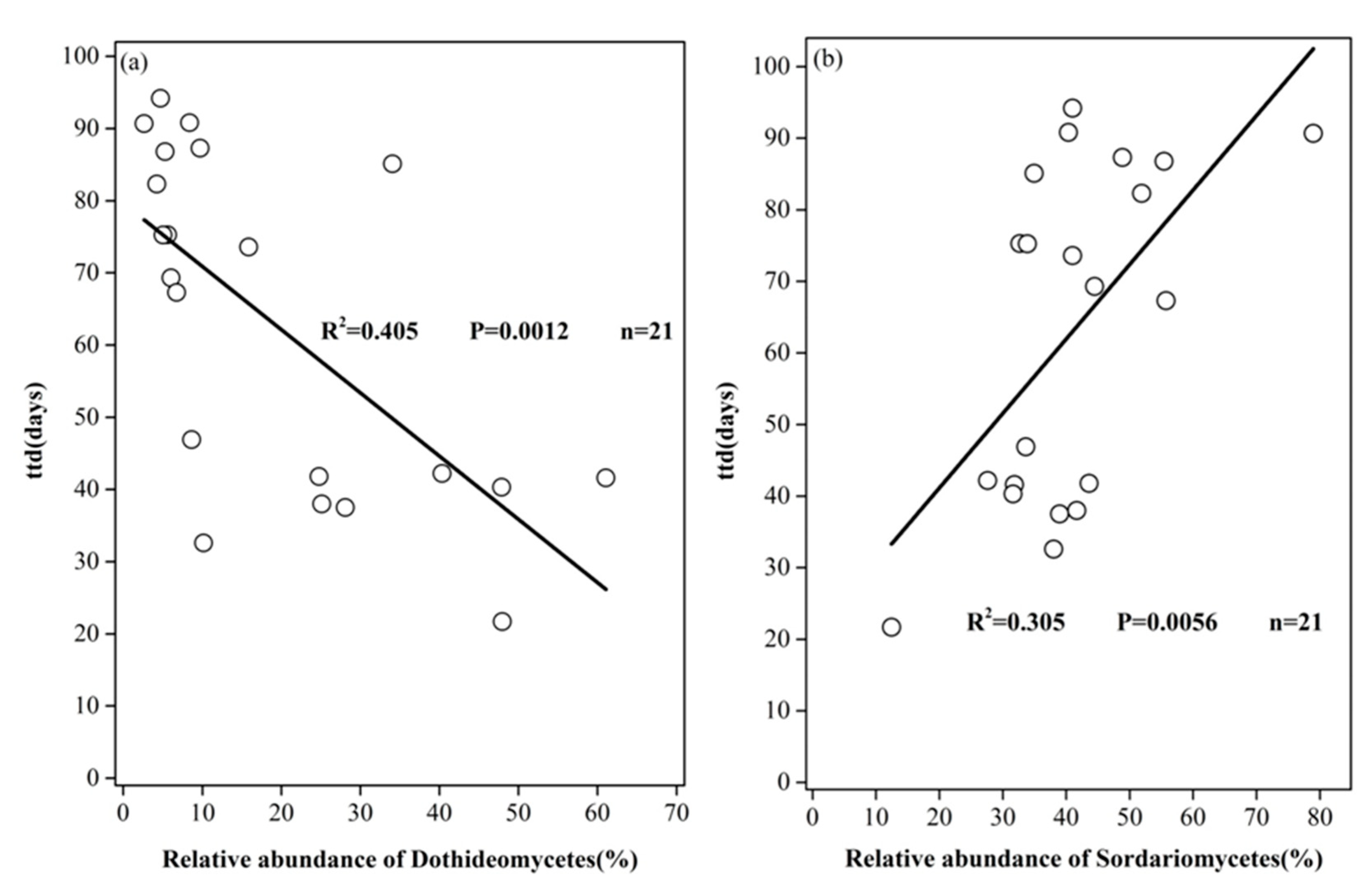
3.3. Pearson and Mantel Correlation between Soil Properties, Survival Parameters, and Relative Abundances of Selected Fungal Classes
3.4. Correlation Analysis between Fungal Community, Bacterial Community, and Survival Parameters
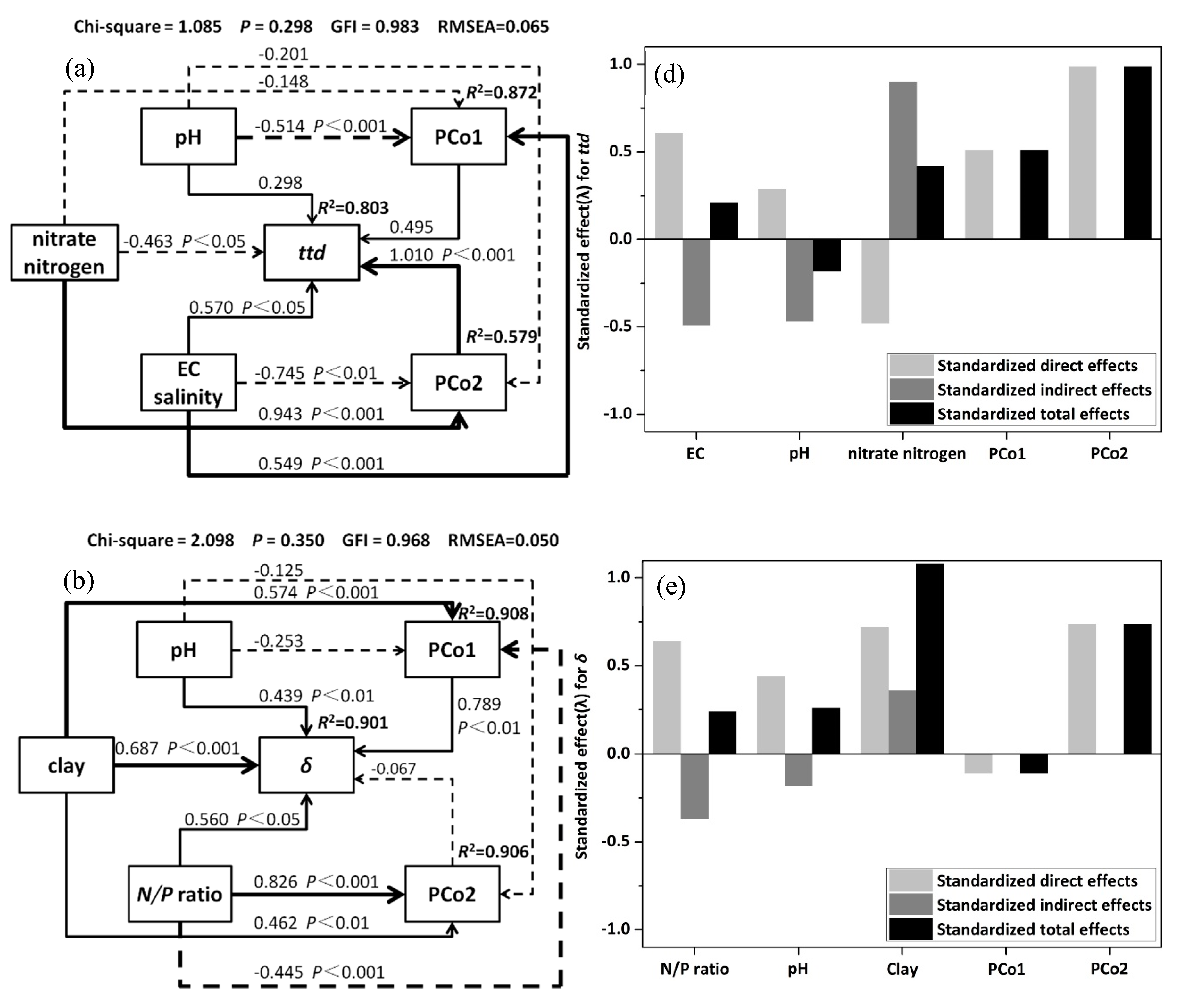
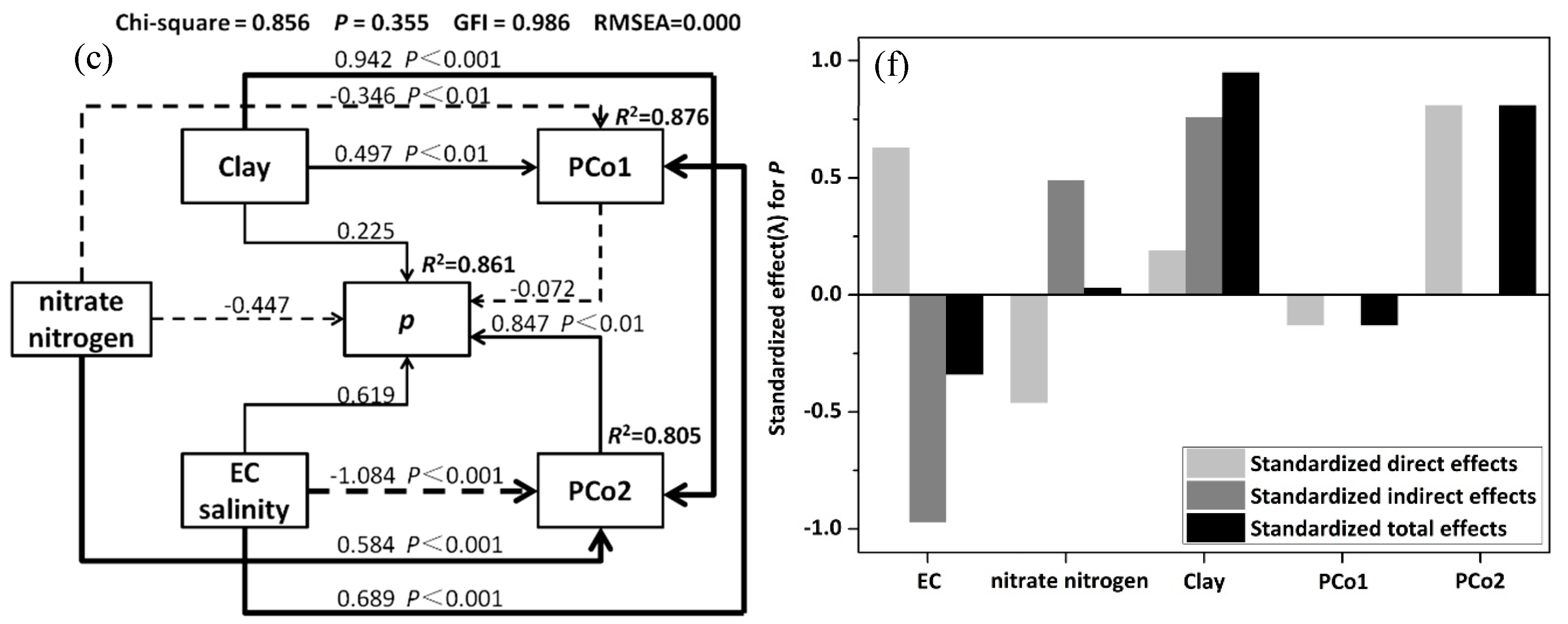
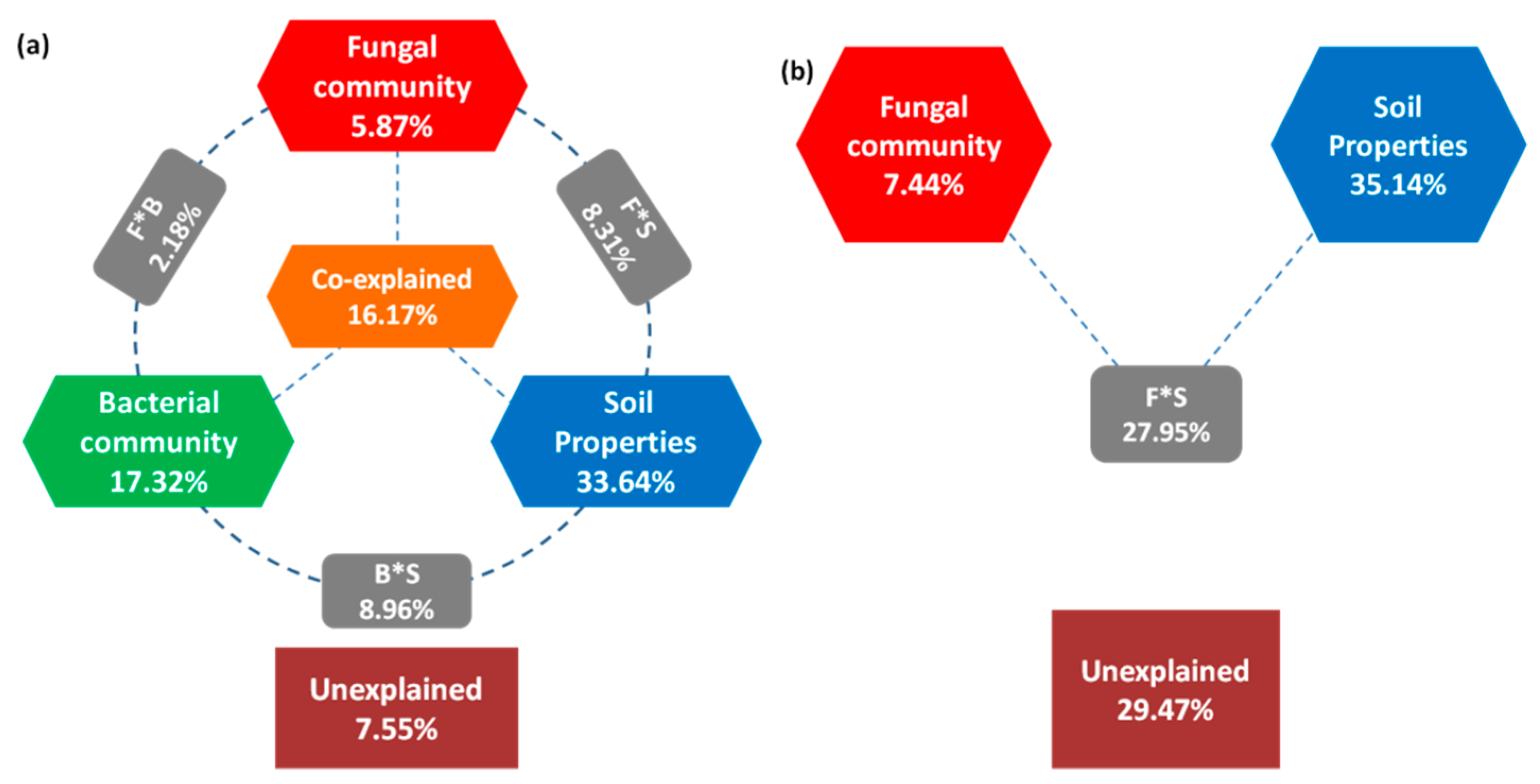
3.5. Structural Equation Model(SEM) and Variation Partition Analysis (VPA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, H.; Terajima, J.; Wada, A.; Inagaki, Y.; Itoh, K.I.; Tamura, K.; Watanabe, H. Molecular typing of enterohemorrhagic escherichia coli o157:H7 isolates in japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 1997, 35, 1675–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, M.P. Escherichia coli o157:H7 and its significance in foods. Int. J. Food Microbiol. 1991, 12, 289–301. [Google Scholar] [CrossRef]
- Besser, R.E.; Lett, S.M.; Weber, J.T.; Doyle, M.P.; Barrett, T.J.; Wells, J.G.; Griffin, P.M. An outbreak of diarrhea and hemolytic uremic syndrome from escherichia coli o157:H7 in fresh-pressed apple cider. JAMA 1993, 269, 2217–2220. [Google Scholar] [CrossRef] [PubMed]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-borne diseases-the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139 (Suppl. 1), S3–S15. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Watt, P.M.; Shouse, P.J.; Grieve, C.M. Fate of escherichia coli o157:H7 in irrigation water on soils and plants as validated by culture method and real-time pcr. Can. J. Microbiol. 2004, 50, 1007–1014. [Google Scholar] [CrossRef]
- Solomon, E.B.; Yaron, S.; Matthews, K.R. Transmission of escherichia coli o157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 2002, 68, 397–400. [Google Scholar] [CrossRef] [Green Version]
- Yamahara, K.M.; Layton, B.A.; Santoro, A.E.; Boehm, A.B. Beach sands along the california coast are diffuse sources of fecal bacteria to coastal waters. Environ. Sci. Technol. 2007, 41, 4515–4521. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Semenov, A.V.; Costa, R.; Trevors, J.T. Survival of escherichia coli in the environment: Fundamental and public health aspects. Isme J. 2011, 5, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Ongeng, D.; Geeraerd, A.H.; Springael, D.; Ryckeboer, J.; Muyanja, C.; Mauriello, G. Fate of escherichia coli o157:H7 and salmonella enterica in the manure-amended soil-plant ecosystem of fresh vegetable crops: A review. Crit. Rev. Microbiol. 2015, 41, 273–294. [Google Scholar] [CrossRef]
- Muniesa, M.; Jofre, J.; Garcia-Aljaro, C.; Blanch, A.R. Occurrence of escherichia coli o157: H7 and other enterohemorrhagic escherichia coli in the environment. Environ. Sci. Technol. 2006, 40, 7141–7149. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Ding, M.Y.; Han, Z.M.; Ma, J.C. Persistence of salmonella typhimurium in well waters from a rural area of changchun city, china. Int. J. Env. Res. Pub. Health 2018, 15, 1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fremaux, B.; Prigent-Combaret, C.; Delignette-Muller, M.L.; Mallen, B.; Dothal, M.; Gleizal, A.; Vernozy-Rozand, C. Persistence of shiga toxin-producing escherichia coli O26 in various manure-amended soil types. J. Appl. Microbiol. 2008, 104, 296–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Ibekwe, A.M.; Crowley, D.E.; Yang, C.-H. Persistence of escherichia coli o157:H7 in major leafy green producing soils. Environ. Sci. Technol. 2012, 46, 12154–12161. [Google Scholar] [CrossRef]
- NandaKafle, G.; Christie, A.A.; Vilain, S.; Brozel, V.S. Growth and extended survival of escherichia coli o157:H7 in soil organic matter. Front. Microbiol. 2018, 9, 762. [Google Scholar] [CrossRef] [Green Version]
- Cai, P.; Huang, Q.; Walker, S.L. Deposition and survival of escherichia coli o157:H7 on clay minerals in a parallel plate flow system. Environ. Sci. Technol. 2013, 47, 1896–1903. [Google Scholar] [CrossRef]
- Cai, P.; Liu, X.; Ji, D.D.; Yang, S.S.; Walker, S.L.; Wu, Y.C.; Gao, C.H.; Huang, Q.Y. Impact of soil clay minerals on growth, biofilm formation, and virulence gene expression of escherichia coli o157:H7. Environ. Pollut. 2018, 243, 953–960. [Google Scholar] [CrossRef]
- Yao, Z.; Yang, L.; Wang, H.; Wu, J.; Xu, J. Fate of escherichia coli o157: H7 in agricultural soils amended with different organic fertilizers. J. Hazard. Mater. 2015, 296, 30–36. [Google Scholar] [CrossRef]
- Semenov, A.V.; van Bruggen, A.H.C.; van Overbeek, L.; Termorshuizen, A.J.; Semenov, A.M. Influence of temperature fluctuations on escherichia coli o157: H7 and salmonella enterica serovar typhimurium in cow manure. FEMS Microbiol. Ecol. 2007, 60, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Semenov, A.V.; van Overbeek, L.; Termorshuizen, A.J.; van Bruggen, A.H.C. Influence of aerobic and anaerobic conditions on survival of escherichia coli o157:H7 and salmonella enterica serovar typhimurium in luria-bertani broth, farm-yard manure and slurry. J. Environ. Manag. 2011, 92, 780–787. [Google Scholar] [CrossRef]
- Franz, E.; Klerks, M.A.; De Vos, O.J.; Termorshuizen, A.J.; van Bruggen, A.H.C. Prevalence of shiga toxin-producing escherichia coli stx(1), stx(2), eaea, and rfbe genes and survival of e-coli o157: H7 in manure from organic and low-input conventional dairy farms. Appl. Environ. Microbiol. 2007, 73, 2180–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franz, E.; Semenov, A.V.; Termorshuizen, A.J.; de Vos, O.J.; Bokhorst, J.G.; van Bruggen, A.H.C. Manure-amended soil characteristics affecting the survival of e-coli o157: H7 in 36 dutch soils. Environ. Microbiol. 2008, 10, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, J. Role of pseudomonas aeruginosa biofilm in the initial adhesion, growth and detachment of escherichia coli in porous media. Environ. Sci. Technol. 2008, 42, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.J.; Wang, H.Z.; Brookes, P.C.; Salles, J.F.; Xu, J.M. Soil ph and microbial diversity constrain the survival of e-coli in soil. Soil Biol. Biochem. 2019, 128, 139–149. [Google Scholar] [CrossRef]
- Yao, Z.; Wang, H.; Wu, L.; Wu, J.; Brookes, P.C.; Xu, J. Interaction between the microbial community and invading escherichia coli o157:H7 in soils from vegetable fields. Appl. Environ. Microbiol. 2014, 80, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Ibekwe, A.M.; Yang, C.-H.; Crowley, D.E. Influence of bacterial communities based on 454-pyrosequencing on the survival of escherichia coli o157:H7 in soils. FEMS Microbiol. Ecol. 2013, 84, 542–554. [Google Scholar] [CrossRef] [Green Version]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of escherichia coli o157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef] [Green Version]
- Barker, J.; Humphrey, T.J.; Brown, M.W.R. Survival of escherichia coli o157 in a soil protozoan: Implications for disease. Fems Microbiol. Lett. 1999, 173, 291–295. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Ma, J.; Crowley, D.E.; Yang, C.H.; Johnson, A.M.; Petrossian, T.C.; Lum, P.Y. Topological data analysis of escherichia coli o157:H7 and non-o157 survival in soils. Front. Cell. Infect. Microbiol. 2014, 4, 122. [Google Scholar] [CrossRef] [Green Version]
- Murphy, H.M.; Thomas, M.K.; Schmidt, P.J.; Medeiros, D.T.; McFadyen, S.; Pintar, K.D.M. Estimating the burden of acute gastrointestinal illness due to giardia, cryptosporidium, campylobacter, E. coli O157 and norovirus associated with private wells and small water systems in canada. Epidemiol. Infect. 2016, 144, 1355–1370. [Google Scholar] [CrossRef] [Green Version]
- Artz, R.R.; Killham, K. Survival of escherichia coli o157:H7 in private drinking water wells: Influences of protozoan grazing and elevated copper concentrations. FEMS Microbiol. Lett. 2002, 216, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Kirk, T.K.; Farrell, R.L. Enzymatic “combustion”: The microbial degradation of lignin. Annu. Rev. Microbiol. 1987, 41, 465–501. [Google Scholar] [CrossRef] [PubMed]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. MMBR 2015, 79, 243–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Diepen, L.T.A.; Hobbie, E.A.; Mohan, J.E. Fungi, ecosystems and global change. Fungal Ecol. 2014, 10, 1–2. [Google Scholar] [CrossRef]
- Larney, F.J.; Yanke, L.J.; Miller, J.J.; McAllister, T.A. Fate of coliform bacteria in composted beef cattle feedlot manure. J. Environ. Qual. 2003, 32, 1508–1515. [Google Scholar] [CrossRef]
- Berger, C.N.; Sodha, S.V.; Shaw, R.K.; Griffin, P.M.; Pink, D.; Hand, P.; Frankel, G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010, 12, 2385–2397. [Google Scholar] [CrossRef]
- Patterson, L.; Navarro-Gonzalez, N.; Jay-Russell, M.T.; Aminabadi, P.; Antaki-Zukoski, E.; Pires, A.F.A. Persistence of escherichia coli in the soil of an organic mixed crop-livestock farm that integrates sheep grazing within vegetable fields. Zoonoses Public Health 2018, 65, 887–896. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Ding, M.; Li, J.; Liu, X.; Li, H.; Zhang, R.; Ma, J. Exploring links between water quality and e. Coli o157:H7 survival potential in well waters from a rural area of southern changchun city, china. J. Water Health 2018, 16, 300–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Ibekwe, A.M.; Yi, X.; Wang, H.; Yamazaki, A.; Crowley, D.E.; Yang, C.-H. Persistence of escherichia coli o157:H7 and its mutants in soils. PLoS ONE 2011, 6, e23191. [Google Scholar] [CrossRef]
- Coroller, L.; Leguerinel, I.; Mettler, E.; Savy, N.; Mafart, P. General model, based on two mixed weibull distributions of bacterial resistance, for describing various shapes of inactivation curves. Appl. Environ. Microbiol. 2006, 72, 6493–6502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mafart, P.; Couvert, O.; Gaillard, S.; Leguerinel, I. On calculating sterility in thermal preservation methods: Application of the weibull frequency distribution model. Int. J. Food Microbiol. 2002, 72, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Nergui, S.; Han, Z.; Huang, G.; Li, H.; Zhang, R.; Zhu, L.; Liao, J. The variation of the soil bacterial and fungal community is linked to land use types in northeast china. Sustainability 2019, 11, 3286. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Deng, M.; Yuan, A.; Wang, J.; Li, H.; Ma, J. Vertical variation of a black soil’s properties in response to freeze-thaw cycles and its links to shift of microbial community structure. Sci. Total Environ. 2018, 625, 106–113. [Google Scholar] [CrossRef]
- Ma, J.; Ibekwe, A.M.; Yang, C.-H.; Crowley, D.E. Bacterial diversity and composition in major fresh produce growing soils affected by physiochemical properties and geographic locations. Sci. Total Environ. 2016, 563–564, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Villarreal Ruiz, L.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Fungal biogeography. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, T.; Wei, G.; Wu, L.; Wu, J.; Xu, J. Survival of escherichia coli o157:H7 in soils under different land use types. Environ. Sci. Pollut. Res. Int. 2014, 21, 518–524. [Google Scholar] [CrossRef]
- Grandy, A.S.; Strickland, M.S.; Lauber, C.L.; Bradford, M.A.; Fierer, N. The influence of microbial communities, management, and soil texture on soil organic matter chemistry. Geoderma 2009, 150, 278–286. [Google Scholar] [CrossRef]
- Lucas, S.T.; D’Angelo, E.M.; Williams, M.A. Improving soil structure by promoting fungal abundance with organic soil amendments. Appl. Soil Ecol. 2014, 75, 13–23. [Google Scholar] [CrossRef]
- Rath, K.M.; Maheshwari, A.; Rousk, J. Linking microbial community structure to trait distributions and functions using salinity as an environmental filter. Mbio 2019, 10, e01607-19. [Google Scholar] [CrossRef] [Green Version]
- Baumann, K.; Marschner, P. Effects of salinity on microbial tolerance to drying and rewetting. Biogeochemistry 2013, 112, 71–80. [Google Scholar] [CrossRef]
- Lanzen, A.; Epelde, L.; Garbisu, C.; Anza, M.; Martin-Sanchez, I.; Blanco, F.; Mijangos, I. The community structures of prokaryotes and fungi in mountain pasture soils are highly correlated and primarily influenced by ph. Front. Microbiol. 2015, 6, 1321. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.; Dias, R.; de Quadros, P.D.; Davis-Richardson, A.; Camargo, F.A.O.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil ph determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 2015, 69, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of dothideomycetes. Stud. Mycol. 2009, 64, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ruibal, C.; Gueidan, C.; Selbmann, L.; Gorbushina, A.A.; Crous, P.W.; Groenewald, J.Z.; Muggia, L.; Grube, M.; Isola, D.; Schoch, C.L.; et al. Phylogeny of rock-inhabiting fungi related to dothideomycetes. Stud. Mycol. 2009, 64, 123–133. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Huang, S.-K.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.; D’souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, S.; Soria, I.; Ayala, N.; Portillo-Lopez, A. Culturable halotolerant fungal isolates from southern california gulf sediments. Open Agric. 2017, 2, 292–299. [Google Scholar] [CrossRef] [Green Version]
- Metcalf, J.L.; Xu, Z.Z.; Weiss, S.; Lax, S.; Van Treuren, W.; Hyde, E.R.; Song, S.J.; Amir, A.; Larsen, P.; Sangwan, N.; et al. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 2016, 351, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Zhang, M.M.; Peng, M.; Sui, X.; Li, W.; Sun, G.Y. Variations in soil functional fungal community structure associated with pure and mixed plantations in typical temperate forests of china. Front. Microbiol. 2019, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Penalva, M.A.; Arst, H.N., Jr. Recent advances in the characterization of ambient ph regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 2004, 58, 425–451. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum acidobacteria. Appl. Environ. Microbiol. 2011, 77, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westphal, A.; Williams, M.L.; Baysal-Gurel, F.; LeJeune, J.T.; Gardener, B.B.M. General suppression of escherichia coli o157:H7 in sand-based dairy livestock bedding. Appl. Environ. Microbiol. 2011, 77, 2113–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Mark Ibekwe, A.; Crowley, D.E.; Yang, C.-H. Persistence of escherichia coli o157 and non-o157 strains in agricultural soils. Sci. Total Environ. 2014, 490, 822–829. [Google Scholar] [CrossRef]
- Cortés-Lorenzo, C.; González-Martínez, A.; Smidt, H.; González-López, J.; Rodelas, B. Influence of salinity on fungal communities in a submerged fixed bed bioreactor for wastewater treatment. Chem. Eng. J. 2016, 285, 562–572. [Google Scholar] [CrossRef]
- Sigala, J.; Stringam, B.; Unc, A. In vitro examination of the application of saline concentrate to septic tank wastewater. Sustain. Water Resour. Manag. 2017, 3, 157–162. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.-J.; Banerjee, S.; White, J.F.; Zhou, N.; Zhao, Z.-Y.; Zhang, K.; Hu, M.-F.; Kingsley, K.; Tian, C.-Y. Not by salinity alone: How environmental factors shape fungal communities in saline soils. Soil Sci. Soc. Am. J. 2019, 83, 1387–1398. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, X.; Zhou, B.; Zhao, B.; Ma, M.; Guan, D.; Li, J.; Chen, S.; Cao, F.; Shen, D.; et al. Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast china. Soil Biol. Biochem. 2016, 95, 135–143. [Google Scholar] [CrossRef]
- Laughlin, R.J.; Stevens, R.J. Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 2002, 66, 1540–1548. [Google Scholar] [CrossRef]
- Shoun, H.; Kim, D.H.; Uchiyama, H.; Sugiyama, J. Denitrification by fungi. FEMS Microbiol. Lett. 1992, 73, 277–281. [Google Scholar] [CrossRef]
- Mothapo, N.; Chen, H.; Cubeta, M.A.; Grossman, J.M.; Fuller, F.; Shi, W. Phylogenetic, taxonomic and functional diversity of fungal denitrifiers and associated n2o production efficacy. Soil Biol. Biochem. 2015, 83, 160–175. [Google Scholar] [CrossRef]
- Kool, D.M.; Dolfing, J.; Wrage, N.; Van Groenigen, J.W. Nitrifier denitrification as a distinct and significant source of nitrous oxide from soil. Soil Biol. Biochem. 2011, 43, 174–178. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Yeoh, Y.K.; Kasinadhuni, N.R.; Lonhienne, T.G.; Robinson, N.; Hugenholtz, P.; Ragan, M.A.; Schmidt, S. Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci. Rep. 2015, 5, 8678. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Distance Method | MRPP | Adonis | Anosim | ||||
|---|---|---|---|---|---|---|---|
| δ | p | F | p | R | p | ||
| site | Bray | 0.6373 | 0.001 | 6.5737 | 0.001 | 0.8816 | 0.001 |
| Jaccard | 0.7697 | 0.001 | 4.2375 | 0.001 | 0.8816 | 0.001 | |
| Horn | 0.5839 | 0.001 | 8.0129 | 0.001 | 0.7586 | 0.001 | |
| Euclidean | 8357 | 0.001 | 4.1364 | 0.001 | 0.3711 | 0.001 | |
| Primary Matrix | Secondary Matrix | Correlation Methods | ||||||
|---|---|---|---|---|---|---|---|---|
| Mantel Test | Multiple Regression on Distance Matrices (MRM) | |||||||
| Pearson’s Product-Moment | Spearman’s Rank Correlation Rho | |||||||
| fungal community | survival parameters | r | p | r | p | R2 | F | p |
| 0.387 | 0.002 | 0.3857 | 0.003 | 0.1498 | 36.6372 | 0.002 | ||
| bacterial community | r | p | r | p | R2 | F | p | |
| 0.5174 | 0.001 | 0.4399 | 0.001 | 0.2677 | 72.0259 | 0.001 | ||
| Survival Parameter | Bacterial/FungalGroups | Controlling for | Mantel | Partial Mantel | ||
|---|---|---|---|---|---|---|
| r | p | r | p | |||
| ttd | Acidobacteria | other bacteria and fungi | 0.1922 | 0.024 | 0.0836 | 0.097 |
| Actinobacteria | 0.1694 | 0.055 | 0.0171 | 0.552 | ||
| Chloroflexi | 0.1963 | 0.015 | 0.2010 | 0.026 | ||
| Firmicutes | 0.2886 | 0.003 | 0.1976 | 0.024 | ||
| Gemmatimonadetes | 0.0430 | 0.265 | 0.0120 | 0.364 | ||
| Delta-Proteobacteria | 0.3316 | 0.004 | 0.3272 | 0.006 | ||
| Gamma-Proteobacteria | 0.3027 | 0.002 | 0.1844 | 0.024 | ||
| Dothideomycetes | 0.3435 | 0.003 | 0.2991 | 0.003 | ||
| Sordariomycetes | 0.2104 | 0.022 | 0.1308 | 0.076 | ||
| p | Acidobacteria | 0.2499 | 0.016 | 0.1668 | 0.033 | |
| Actinobacteria | 0.1793 | 0.055 | 0.0234 | 0.331 | ||
| Chloroflexi | 0.1351 | 0.058 | 0.1358 | 0.083 | ||
| Firmicutes | 0.1495 | 0.096 | 0.0523 | 0.224 | ||
| Gemmatimonadetes | 0.3775 | 0.017 | 0.3672 | 0.028 | ||
| Delta-Proteobacteria | 0.2164 | 0.011 | 0.2064 | 0.018 | ||
| Gamma-Proteobacteria | 0.3912 | 0.001 | 0.3123 | 0.002 | ||
| Dothideomycetes | 0.1922 | 0.048 | 0.1410 | 0.072 | ||
| Sordariomycetes | 0.4177 | 0.004 | 0.1922 | 0.050 | ||
| δ | Acidobacteria | 0.2358 | 0.008 | 0.1512 | 0.039 | |
| Actinobacteria | 0.1990 | 0.047 | 0.0499 | 0.185 | ||
| Chloroflexi | 0.1593 | 0.036 | 0.1608 | 0.038 | ||
| Firmicutes | 0.1754 | 0.042 | 0.0829 | 0.118 | ||
| Gemmatimonadetes | 0.2732 | 0.015 | 0.2590 | 0.031 | ||
| Delta-Proteobacteria | 0.2819 | 0.008 | 0.2746 | 0.008 | ||
| Gamma-Proteobacteria | 0.3841 | 0.002 | 0.3045 | 0.005 | ||
| Dothideomycetes | 0.2497 | 0.009 | 0.2037 | 0.014 | ||
| Sordariomycetes | 0.3335 | 0.001 | 0.2825 | 0.008 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, G.; Liao, J.; Han, Z.; Li, J.; Zhu, L.; Lyu, G.; Lu, L.; Xie, Y.; Ma, J. Interaction between Fungal Communities, Soil Properties, and the Survival of Invading E. coli O157:H7 in Soils. Int. J. Environ. Res. Public Health 2020, 17, 3516. https://doi.org/10.3390/ijerph17103516
Huang G, Liao J, Han Z, Li J, Zhu L, Lyu G, Lu L, Xie Y, Ma J. Interaction between Fungal Communities, Soil Properties, and the Survival of Invading E. coli O157:H7 in Soils. International Journal of Environmental Research and Public Health. 2020; 17(10):3516. https://doi.org/10.3390/ijerph17103516
Chicago/Turabian StyleHuang, Guannan, Jiafen Liao, Ziming Han, Jiahang Li, Liyue Zhu, Guangze Lyu, Lu Lu, Yuang Xie, and Jincai Ma. 2020. "Interaction between Fungal Communities, Soil Properties, and the Survival of Invading E. coli O157:H7 in Soils" International Journal of Environmental Research and Public Health 17, no. 10: 3516. https://doi.org/10.3390/ijerph17103516





