The Global Emergency of Novel Coronavirus (SARS-CoV-2): An Update of the Current Status and Forecasting
Abstract
1. Introduction
1.1. What Are Coronaviruses
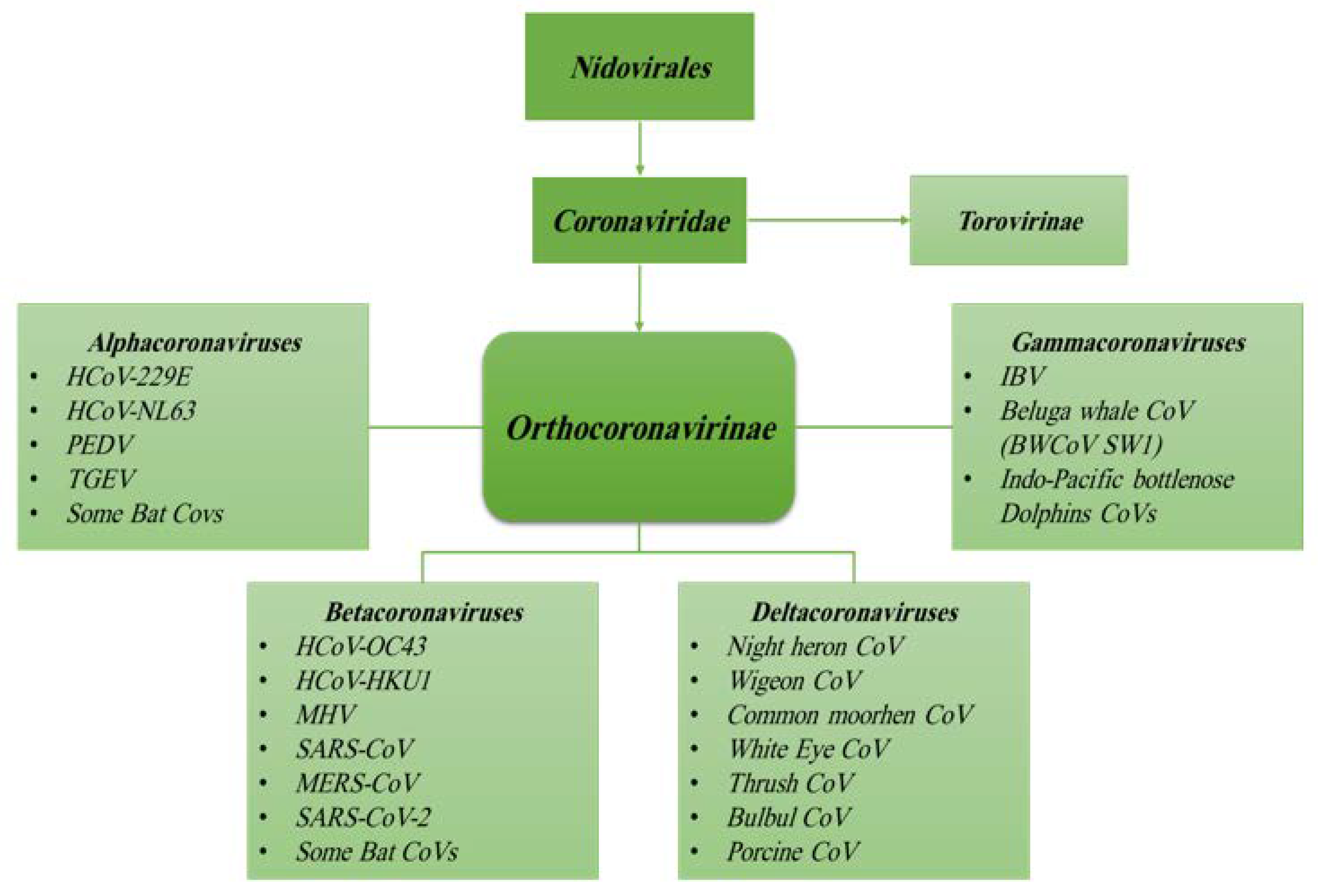
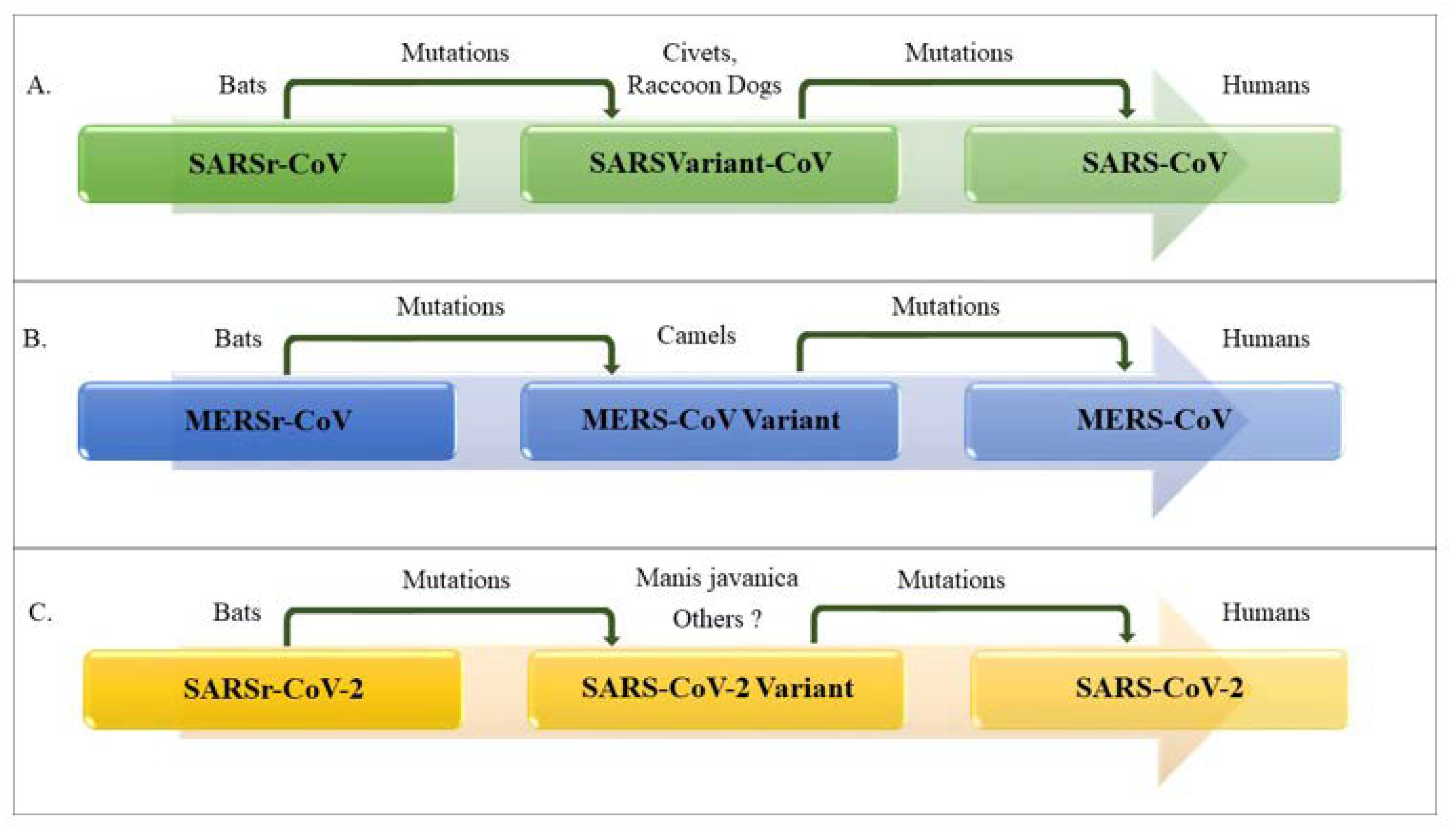
1.2. Origin, Family Member, Diversity and Taxonomy of Coronaviruses
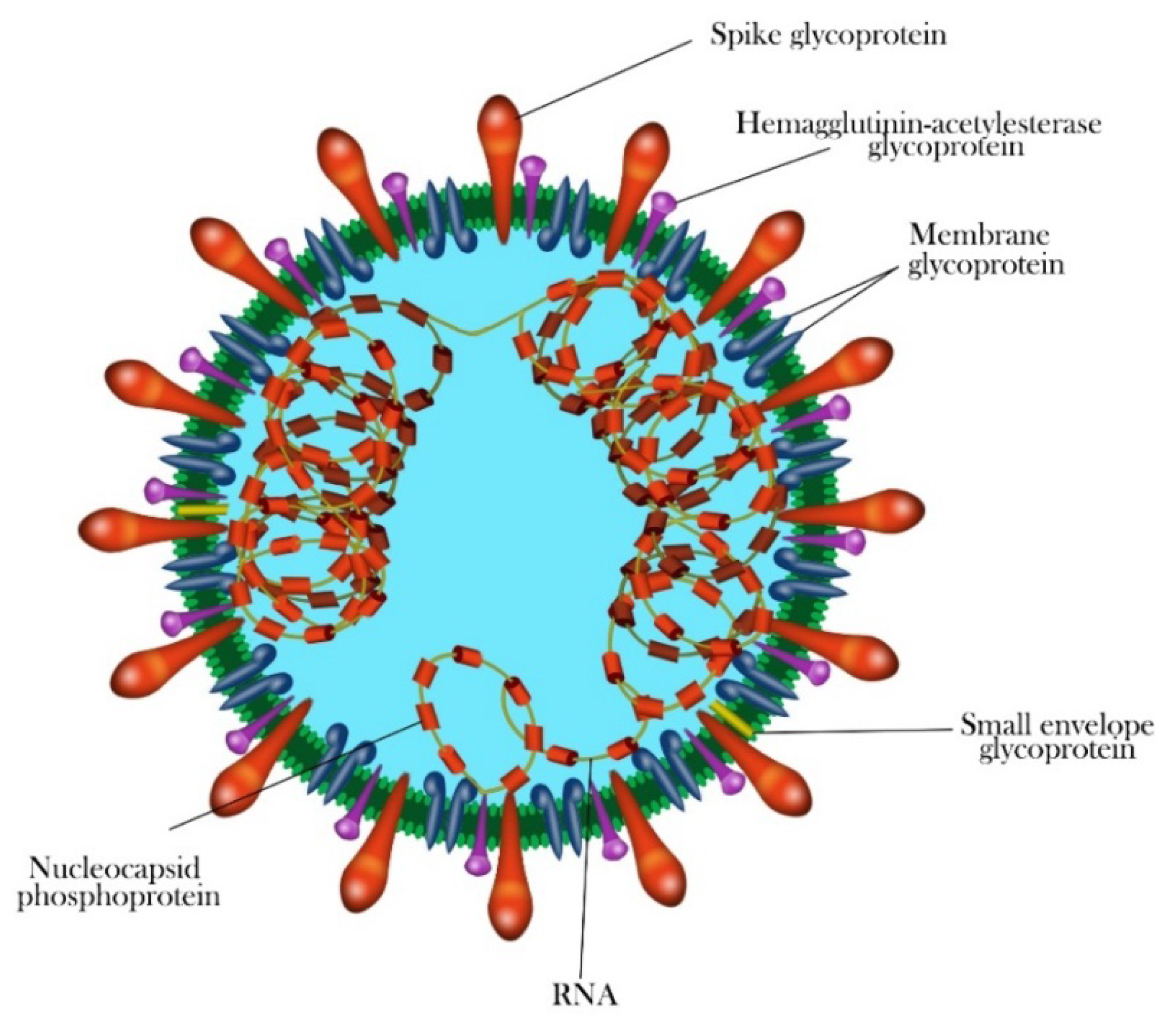
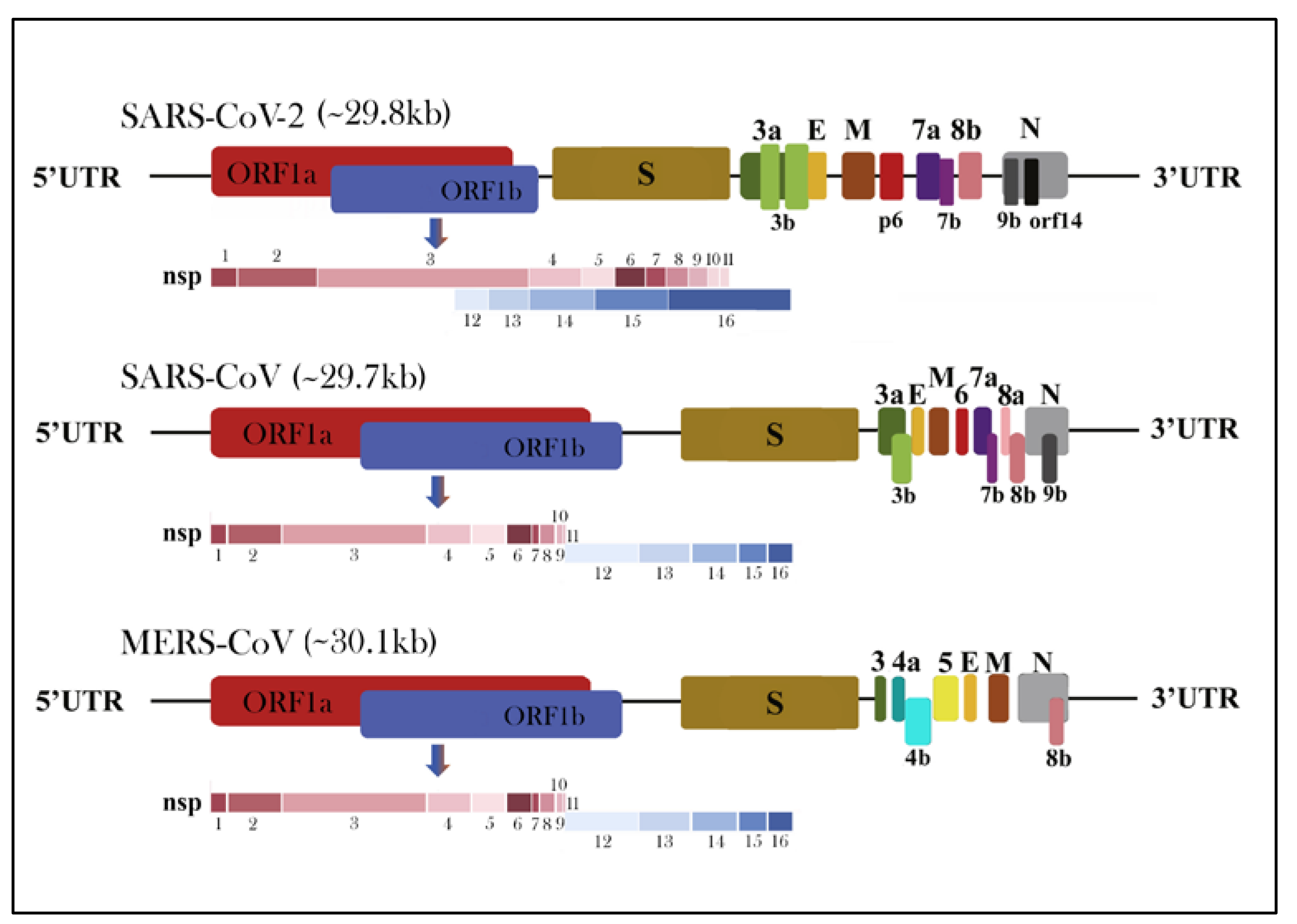
1.3. Morphology and Genomic Structure of HCoVs
1.4. Attachment, Entry and Life Cycle of HCoVs
2. Pathogenesis and Mode of Transmission
Human-to-Human Transmission and Viral Reservoir
3. Epidemiology
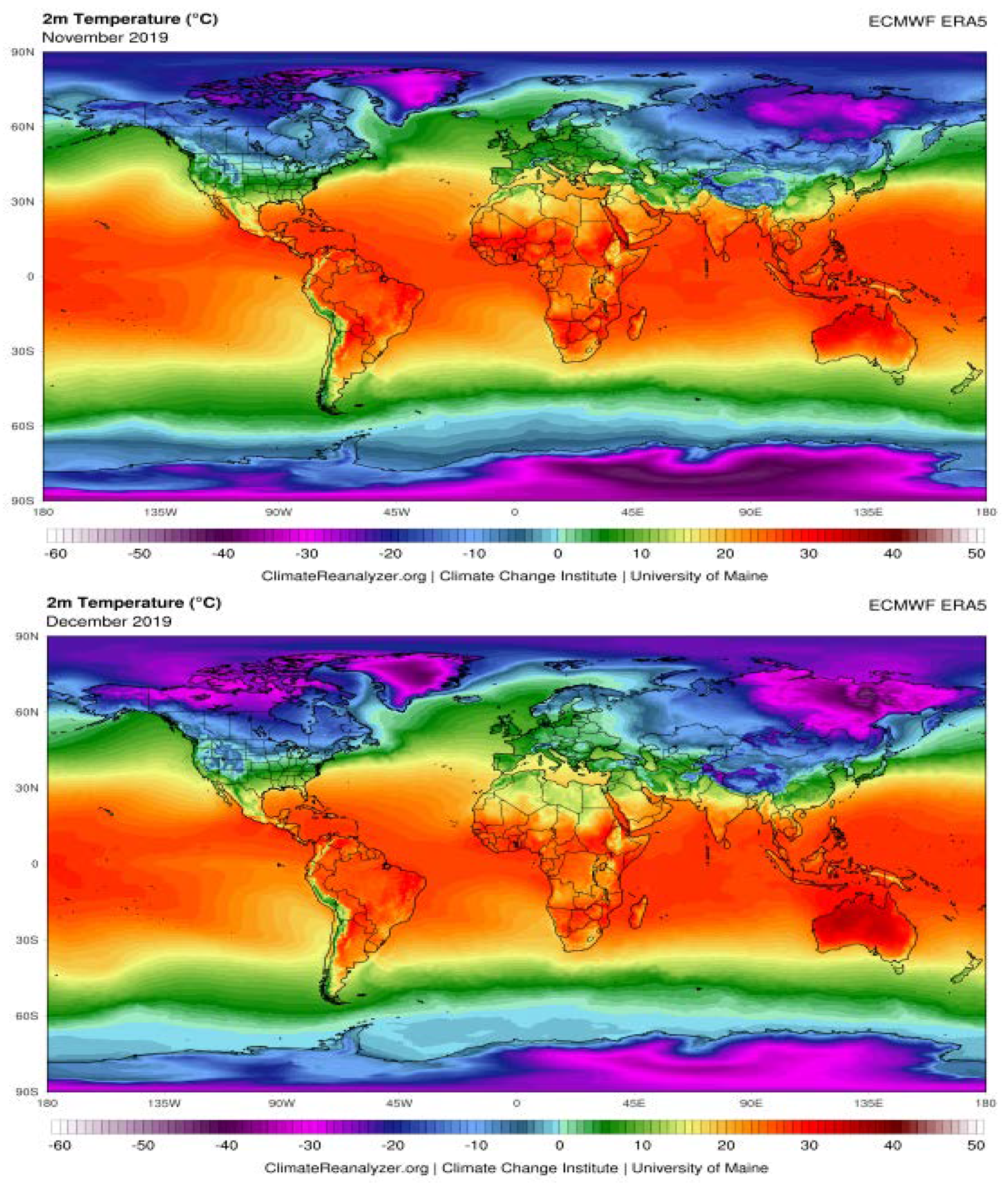
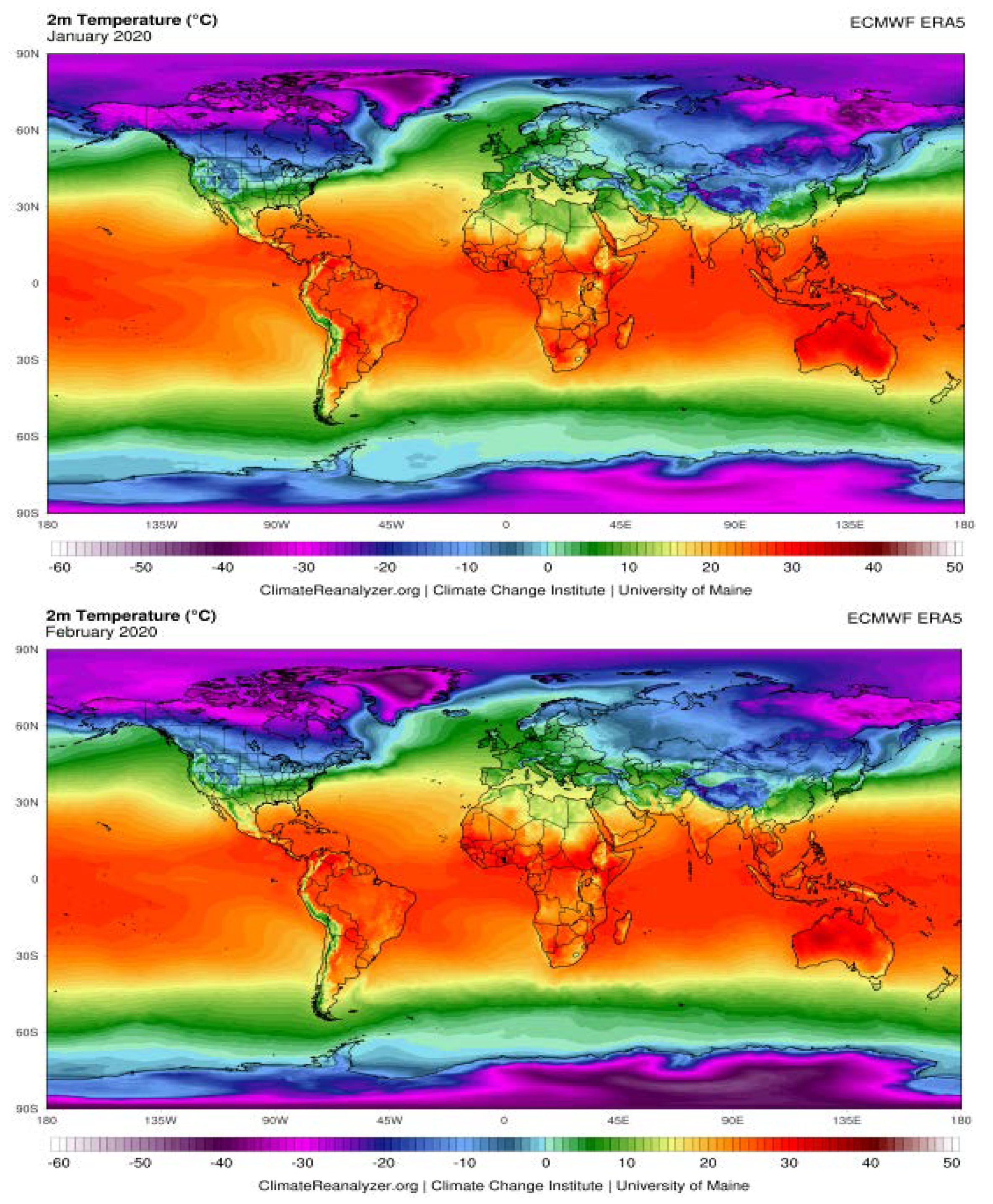
The Possible Effects of Weather Conditions on the Transmission of SARS-CoV-2
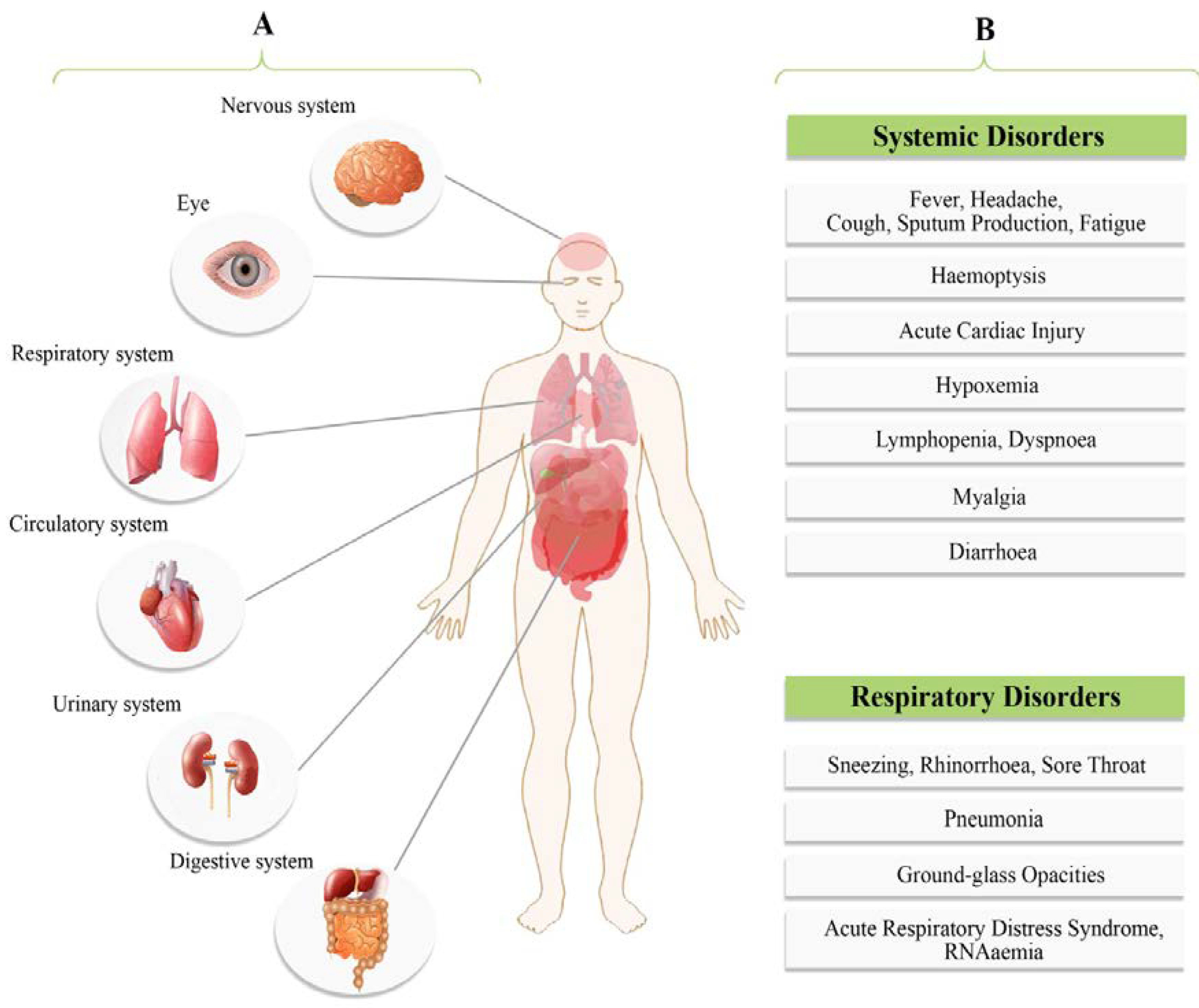
4. Clinical Presentations
5. Diagnosis
5.1. Nucleic Acid Detection Technology
5.2. CT scans and Serology Methods
6. SARS-CoV-2 Therapeutics Strategies
6.1. Inhibitors of SARS-CoV-2 Fusion/Entry
6.2. Inhibitors of SARS-CoV-2 Main Enzymes
6.3. Inhibitors of SARS-CoV-2 Replication
6.4. Modulators of SARS-CoV-2 Induced Inflammatory Response
6.5. Passive Immunization
6.6. Cell-Based Therapies
6.7. Vaccines
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chan, J.F.; Lau, S.K.; Woo, P.C. The emerging novel Middle East respiratory syndrome coronavirus: The “knowns” and “unknowns”. J. Formos. Med. Assoc. 2013, 112, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Hassan, S.A.; Sheikh, F.N.; Jamal, S.; Ezeh, J.K.; Akhtar, A. Coronavirus (COVID-19): A Review of Clinical Features, Diagnosis, and Treatment. Cureus 2020, 12, e7355. [Google Scholar] [CrossRef]
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965, 1, 1467–1470. [Google Scholar] [CrossRef]
- Woo, P.C.; Huang, Y.; Lau, S.K.; Yuen, K.Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef]
- Scott, F.W. Evaluation of risks and benefits associated with vaccination against coronavirus infections in cats. Adv. Vet. Med. 1999, 41, 347–358. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 38, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Stratton, C.W.; Tang, Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020, 92, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef]
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.M.; Elshabrawy, H.A. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020, 9, 186. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Drexler, J.F.; Gloza-Rausch, F.; Glende, J.; Corman, V.M.; Muth, D.; Goettsche, M.; Seebens, A.; Niedrig, M.; Pfefferle, S.; Yordanov, S.; et al. Genomic Characterization of Severe Acute Respiratory Syndrome-Related Coronavirus in European Bats and Classification of Coronaviruses Based on Partial RNA-Dependent RNA Polymerase Gene Sequences. J. Virol. 2010, 84, 11336–11349. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Peiris, J.S.; Lai, S.T.; Poon, L.L.; Guan, Y.; Yam, L.Y.; Lim, W.; Nicholls, J.; Yee, W.K.; Yan, W.W.; Cheung, M.T.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- Yin, Y.; Wunderink, R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018, 23, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Imai, N.; Dorigatti, I.; Cori, A.; Donnelly, C.; Riley, S.; Ferguson, N.M. Estimating the potential total number of novel Coronavirus cases in Wuhan City, China. Imp. Coll. Lond. 2020. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Miłek, J.; Blicharz-Domańska, K. Coronaviruses in Avian Species—Review with Focus on Epidemiology and Diagnosis in Wild Birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351.e2. [Google Scholar] [CrossRef]
- He, F.; Deng, Y.; Li, W. Coronavirus disease 2019: What we know? J. Med. Virol. 2020, 92, 719–725. [Google Scholar] [CrossRef]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 20. [Google Scholar] [CrossRef]
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Pyrc, K.; Berkhout, B.; van der Hoek, L. Identification of new human coronaviruses. Expert Rev. Anti-Infect. Ther. 2007, 5, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Fung, T.S.; Chong, K.K.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014, 109, 97–109. [Google Scholar] [CrossRef]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Báez-Santos, Y.M.; St John, S.E.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Nal, B.; Chan, C.; Kien, F.; Siu, L.; Tse, J.; Chu, K.; Kam, J.; Staropoli, I.; Crescenzo-Chaigne, B.; Escriou, N.; et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009, 83, 7221–7234. [Google Scholar] [CrossRef]
- Siu, Y.L.; Teoh, K.T.; Lo, J.; Chan, C.M.; Kien, F.; Escriou, N.; Tsao, S.W.; Nicholls, J.M.; Altmeyer, R.; Peiris, J.S.; et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008, 82, 11318–11330. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Marra, M.A.; Jones, S.J.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome sequence of the SARS-associated coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Bredenbeek, P.J.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.; Guan, Y.; Rozanov, M.; Spaan, W.J.; Gorbalenya, A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef]
- De Groot, R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. J. 2006, 23, 59–72. [Google Scholar] [CrossRef]
- Klausegger, A.; Strobl, B.; Regl, G.; Kaser, A.; Luytjes, W.; Vlasak, R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J. Virol. 1999, 73, 3737–3743. [Google Scholar] [CrossRef]
- Weiss, S.R.; Navas-Martin, S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Simmons, G.; Reeves, J.D.; Rennekamp, A.J.; Amberg, S.M.; Piefer, A.J.; Bates, P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 2004, 101, 4240–4245. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, J.; Du, L.; Yan, X.; Hu, G.; Zhou, Y.; Jiang, S. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: Revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine 2006, 24, 5498–5508. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72,314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Kindler, E.; Thiel, V.; Weber, F. Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response. Adv. Virus Res. 2016, 96, 219–243. [Google Scholar] [CrossRef]
- Sahin, A.R.; Erdogan, A.; Agaoglu, P.M.; Dineri, Y.; Cakirci, A.Y.; Senel, M.E.; Okyay, R.A.; Tasdogan, A.M. 2019 Novel Coronavirus (COVID-19) Outbreak: A Review of the Current Literature. EJMO 2020, 4, 1–7. [Google Scholar] [CrossRef]
- Gong, L.; Li, J.; Zhou, Q.; Xu, Z.; Chen, L.; Zhang, Y.; Xue, C.; Wen, Z.; Cao, Y. A New Bat-HKU2-like Coronavirus in Swine, China, 2017. Emerg. Infect. Dis. 2017, 23, 1607–1609. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tian, X.; Qin, P.; Wang, B.; Zhao, P.; Yang, Y.L.; Wang, L.; Wang, D.; Song, Y.; Zhang, X.; et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017, 211, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yan, M.; Yang, L.; Ding, B.; He, B.; Wang, Y.; Liu, X.; Liu, C.; Zhu, H.; You, B.; et al. SARS-associated coronavirus transmitted from human to pig. Emerg. Infect. Dis. 2005, 11, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Vergara-Alert, J.; van den Brand, J.M.; Widagdo, W.; Muñoz, M.; Raj, S.; Schipper, D.; Solanes, D.; Cordón, I.; Bensaid, A.; Haagmans, B.L.; et al. Livestock Susceptibility to Infection with Middle East Respiratory Syndrome Coronavirus. Emerg. Infect. Dis. 2017, 23, 232–240. [Google Scholar] [CrossRef]
- Huang, C.; Liu, W.J.; Xu, W.; Jin, T.; Zhao, Y.; Song, J.; Shi, Y.; Ji, W.; Jia, H.; Zhou, Y.; et al. A Bat-Derived Putative Cross-Family Recombinant Coronavirus with a Reovirus Gene. PLoS Pathog. 2016, 12, e1005883. [Google Scholar] [CrossRef]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Sui, J.; Kuhn, J.H.; Moore, M.J.; Luo, S.; Wong, S.K.; Huang, I.C.; Xu, K.; Vasilieva, N.; et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005, 24, 1634–1643. [Google Scholar] [CrossRef]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef]
- Ortega, J.T.; Serrano, M.L.; Pujol, F.H.; Rangel, H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020, 19. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Ciaglia, E.; Vecchione, C.; Puca, A.A. COVID-19 Infection and Circulating ACE2 Levels: Protective Role in Women and Children. Front. Pediatr. 2020, 8, 206. [Google Scholar] [CrossRef]
- Kim, H.; Paul, A. Contact Tracing: A game of big numbers in the time of COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: http://www.ncbi.nlm.nih.gov/pubmed/32150360 (accessed on 4 July 2020).
- Han, Y.; Yang, H. The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID-19): A Chinese perspective. J. Med. Virol. 2020, 10. [Google Scholar] [CrossRef]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef]
- Lu, C.W.; Liu, X.F.; Jia, Z.F. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet 2020, 395, e39. [Google Scholar] [CrossRef]
- Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine Association. An update on the epidemiological characteristics of novel coronavirus pneumonia (COVID-19). Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 139–144. [Google Scholar] [CrossRef]
- Chen, H.; Guo, J.; Wang, C.; Luo, F.; Yu, X.; Zhang, W.; Li, J.; Zhao, D.; Xu, D.; Gong, Q.; et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020, 395, 809–815. [Google Scholar] [CrossRef]
- Wei, M.; Yuan, J.; Liu, Y.; Fu, T.; Yu, X.; Zhang, Z.J. Novel Coronavirus Infection in Hospitalized Infants Under 1 Year of Age in China. JAMA 2020, 323, 1313–1314. [Google Scholar] [CrossRef]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Chan-Yeung, M.; Xu, R.H. SARS: Epidemiology. Respirology 2003, 8, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Gretebeck, L.M.; Subbarao, K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015, 13, 123–129. [Google Scholar] [CrossRef]
- Du, T.A. Outbreak of a novel coronavirus. Nat. Rev. Microbiol. 2020, 18, 123. [Google Scholar] [CrossRef]
- National Health Commission of the People’s Republic of China. Pneumonia Epidemic Situation of New Coronavirus Infection on 23 January 2020. Available online: http://www.nhc.gov.cn/yjb/s3578/202001/5d19a4f6d3154b9fae328918ed2e3c8a.shtml (accessed on 23 January 2020).
- Bogoch, I.I.; Watts, A.; Thomas-Bachli, A.; Huber, C.; Kraemer, M.U.G.; Khan, K. Pneumonia of unknown aetiology in Wuhan, China: Potential for international spread via commercial air travel. J. Travel Med. 2020, 27. [Google Scholar] [CrossRef]
- McCreary, E.K.; Pogue, J.M. Coronavirus Disease 2019 Treatment: A Review of Early and Emerging Options. Open Forum Infect. Dis. 2020, 7. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Shan, J. 2019 Novel coronavirus: Where we are and what we know. Infection 2020, 48, 155–163. [Google Scholar] [CrossRef]
- Lin, Q.; Chiu, A.P.; Zhao, S.; He, D. Modeling the spread of Middle East respiratory syndrome coronavirus in Saudi Arabia. Stat. Methods Med. Res. 2018, 27, 1968–1978. [Google Scholar] [CrossRef] [PubMed]
- Lipsitch, M.; Cohen, T.; Cooper, B.; Robins, J.M.; Ma, S.; James, L.; Gopalakrishna, G.; Chew, S.K.; Tan, C.C.; Samore, M.H.; et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 2003, 300, 1966–1970. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of Respiratory Viral Infections. Annu. Rev. Virol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Huang, R.; Zhuang, L.; Zhang, K. Assessment of China’s Offshore Wind Resources Based on the Integration of Multiple Satellite Data and Meteorological Data. Remote Sens. 2019, 11, 2680. [Google Scholar] [CrossRef]
- Wang, J.; Tang, K.; Feng, K.; Lin, X.; Lv, W.; Chen, K.; Wang, F. High Temperature and High Humidity Reduce the Transmission of COVID-19. SSRN 2020. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, A.; Gong, L.; Luo, L.; Guo, W.; Li, C.; Zheng, J.; Li, C.; Yang, B.; Zeng, J.; et al. Temperature significant change COVID-19 transmission in 429 cities. MedRxiv 2020. [Google Scholar] [CrossRef]
- Bannister-Tyrrell, M.; Meyer, A.; Faverjon, C.; Cameron, A. Preliminary evidence that higher temperatures are associated with lower incidence of COVID-19, for cases reported globally up to 29 February 2020. medRxiv 2020. [Google Scholar] [CrossRef]
- Wu, Y.; Jing, W.; Liu, J.; Ma, Q.; Yuan, J.; Wang, Y.; Du, M.; Liu, M. Effects of temperature and humidity on the daily new cases and new deaths of COVID-19 in 166 countries. Sci. Total Environ. 2020, 729, 139051. [Google Scholar] [CrossRef]
- Scafetta, N. Distribution of the SARS-CoV-2 Pandemic and Its Monthly Forecast Based on Seasonal Climate Patterns. Int. J. Environ. Res. Public Health 2020, 17, 3493. [Google Scholar] [CrossRef]
- Raines, K.S.; Doniach, S.; Bhanot, G. The transmission of SARS-CoV-2 is likely comodulated by temperature and by relative humidity. medRxiv 2020. [Google Scholar] [CrossRef]
- Majidi Nezhad, M.; Groppi, D.; Marzialetti, P.; Fusilli, L.; Laneve, G.; Cumo, F.; Astiasogarsia, D. Wind energy potential analysis using Sentinel-1 satellite: A review and a case study on Mediterranean islands. Renew. Sustain. Energy Rev. 2019, 109, 499–513. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Chen, Y.; Qin, Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.; Lau, E.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Kannan, S.; Shaik Syed Ali, P.; Sheeza, A.; Hemalatha, K. COVID-19 (Novel Coronavirus 2019) -recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2006–2011. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Wang, W.; Tang, J.; Wei, F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2020, 92, 441–447. [Google Scholar] [CrossRef]
- Gralinski, L.E.; Menachery, V.D. Return of the Coronavirus: 2019-nCoV. Viruses 2020, 12, 135. [Google Scholar] [CrossRef]
- Phan, L.T.; Nguyen, T.V.; Luong, Q.C.; Nguyen, T.V.; Nguyen, H.T.; Le, H.Q.; Nguyen, T.T.; Cao, T.M.; Pham, Q.D. Importation and Human-to-Human Transmission of a Novel Coronavirus in Vietnam. N. Engl. J. Med. 2020, 382, 872–874. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhou, Z.; Guo, D.; Li, C.; Fang, Z.; Chen, L.; Yang, R.; Li, X.; Zeng, W. Coronavirus disease 2019: Initial chest CT findings. Eur. Radiol. 2020, 30, 4398–4406. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Li, J.; Li, X.; Qi, X. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology 2020, 295, 18. [Google Scholar] [CrossRef] [PubMed]
- Salehi, S.; Abedi, A.; Balakrishnan, S.; Gholamrezanezhad, A. Coronavirus Disease 2019 (COVID-19): A Systematic Review of Imaging Findings in 919 Patients. AJR Am. J. Roentgenol. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Thomsen, T.; Sell, N.; Goldsmith, A.J. Abdominal and testicular pain: An atypical presentation of COVID-19. Am. J. Emerg. Med. 2020, 38, 1542.e1–1542.e3. [Google Scholar] [CrossRef] [PubMed]
- Tay, H.S.; Harwood, R. Atypical presentation of COVID-19 in a frail older person. Age Ageing 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, T. Diagnosis and clinical management of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection: An operatioal recommendation of Peking Union Medical College Hospital (V2.0). Emerg. Microbes Infect. 2020, 9, 582–585. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Zhang, W.; Du, R.H.; Li, B.; Zheng, X.S.; Yang, X.L.; Hu, B.; Wang, Y.Y.; Xiao, G.F.; Yan, B.; Shi, Z.L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Farfour, E.; Lesprit, P.; Visseaux, B.; Pascreau, T.; Jolly, E.; Houhou, N.; Mazaux, L.; Asso-Bonnet, M.; Vasse, M.; SARS-CoV-2 Foch Hospital Study Group. The Allplex 2019-nCoV (Seegene) assay: Which performances are for SARS-CoV-2 infection diagnosis? [published online ahead of print, 28 May 2020]. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 10. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, M.; Shen, C.; Wang, F.; Yuan, J.; Li, J.; Zhang, M.; Wang, Z.; Xing, L.; Wei, J.; et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. MedRxiv 2020. [Google Scholar] [CrossRef]
- Binnicker, M.J. Emergence of a Novel Coronavirus Disease (COVID-19) and the Importance of Diagnostic Testing: Why Partnership between Clinical Laboratories, Public Health Agencies, and Industry Is Essential to Control the Outbreak. Clin. Chem. 2020, 66, 664–666. [Google Scholar] [CrossRef]
- Merindol, N.; Pépin, G.; Marchand, C.; Rheault, M.; Peterson, C.; Poirier, A.; Houle, C.; Germain, H.; Danylo, A. SAS-CoV-2 detection by direct rRT-PCR without RNA extraction. J. Clin. Virol. 2020, 128, 104423. [Google Scholar] [CrossRef]
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020, 20, 425–434. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Q.; Xu, H.; Lu, H.; Liu, S.; Li, H. Imaging of coronavirus disease 2019: A Chinese expert consensus statement. Eur. J. Radiol. 2020, 127, 109008. [Google Scholar] [CrossRef]
- Pan, F.; Ye, T.; Sun, P.; Gui, S.; Liang, B.; Li, L.; Zheng, D.; Wang, J.; Hesketh, R.L.; Yang, L.; et al. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19). Radiology 2020, 295, 715–721. [Google Scholar] [CrossRef]
- Pan, Y.; Guan, H.; Zhou, S.; Wang, Y.; Li, Q.; Zhu, T.; Hu, Q.; Xia, L. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): A study of 63 patients in Wuhan, China. Eur. Radiol. 2020, 30, 3306–3309. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef]
- Chassagnon, G.; Vakalopoulou, M.; Paragios, N.; Revel, M.P. Artificial intelligence applications for thoracic imaging. Eur. J. Radiol. 2020, 123, 108774. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Iwano, S.; Naganawa, S. A review on the use of artificial intelligence for medical imaging of the lungs of patients with coronavirus disease 2019. Diagn. Interv. Radiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, C. Radiological role in the detection, diagnosis and monitoring for the coronavirus disease 2019 (COVID-19). Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4523–4528. [Google Scholar] [CrossRef]
- Li, X.; Zeng, W.; Li, X.; Chen, H.; Shi, L.; Li, X.; Xiang, H.; Cao, Y.; Chen, H.; Liu, C.; et al. CT imaging changes of corona virus disease 2019 (COVID-19): A multi-center study in Southwest China. J. Transl. Med. 2020, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Luo, Z.; Jia, Y.; Zhou, C.; He, J.; Lyu, J.; Shen, X. CT differential diagnosis of COVID-19 and non-COVID-19 in symptomatic suspects: A practical scoring method. BMC Pulm. Med. 2020, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 10. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 10. [Google Scholar] [CrossRef]
- Kohmer, N.; Westhaus, S.; Rühl, C.; Ciesek, S.; Rabenau, H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020, 129, 104480. [Google Scholar] [CrossRef]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Jiang, D.; Zhou, P.; Cheng, L.; Li, Y.; Ma, X.; Jin, T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020. [Google Scholar] [CrossRef]
- Diao, B.; Wen, K.; Chen, J.; Liu, Y.; Yuan, Z.; Han, C.; Chen, J.; Pan, Y.; Chen, L.; Dan, Y.; et al. Diagnosis of Acute Respiratory Syndrome Coronavirus 2 Infection by Detection of Nucleocapsid Protein. medRxiv 2020. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Meenakshisundaram, S.; Manickam, M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today 2020, 25, 668–688. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. This scientist hopes to test coronavirus drugs on animals in locked-down Wuhan. Nature 2020, 577, 607. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.S.; Sircar, S.; Bhat, S.; Sharun, K.; Dhama, K.; Dadar, M.; Tiwari, R.; Chaicumpa, W. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020, 40, 68–76. [Google Scholar] [CrossRef]
- Zaher, N.H.; Mostafa, M.I.; Altaher, A.Y. Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharm. 2020, 70, 145–159. [Google Scholar] [CrossRef]
- Li, H.; Zhou, Y.; Zhang, M.; Wang, H.; Zhao, Q.; Liu, J. Updated Approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Ren, J.L.; Zhang, A.H.; Wang, X.J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol. Res. 2020, 155, 104743. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Yang, Y.; Shi, W.; Xue, M.; Yang, W.; Song, Z.; Yao, C.; Yin, J.; Shi, D.; Zhang, Y.; et al. Complementary and alternative medicine is expected to make greater contribution in controlling the prevalence of influenza. Biosci. Trends 2013, 7, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.X.; Li, S.X.; Zheng, F.Y. Application of in Vitro Bionic Digestion and Biomembrane Extraction for Metal Speciation Analysis, Bioavailability and Risk Assessment in Lianhua Qingwen Capsule. Zhongguo Zhong Yao Za Zhi 2014, 39, 2330–2335. [Google Scholar]
- Wang, Z.; Chen, X.; Lu, Y.; Chen, F.; Zhang, W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci. Trends 2020, 1, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Kong, W.P.; Huang, Y.; Roberts, A.; Murphy, B.R.; Subbarao, K.; Nabel, G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 2004, 428, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; He, Y.; Liu, S. SARS vaccine development. Emerg. Infect. Dis. 2005, 11, 1016–1020. [Google Scholar] [CrossRef]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Widjaja, I.; Wang, C.; van Haperen, R.; Gutiérrez-Álvarez, J.; van Dieren, B.; Okba, N.; Raj, V.S.; Li, W.; Fernandez-Delgado, R.; Grosveld, F.; et al. Towards a solution to MERS: Protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg. Microbes Infect. 2019, 8, 516–530. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Kruse, R.L. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Research 2020, 9, 72. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Zheng, M.; Song, L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell. Mol. Immunol. 2020, 17, 536–538. [Google Scholar] [CrossRef]
- Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, J.; Li, S.; Sun, J.; Teng, Y.; Wu, M.; Li, J.; Li, Y.; Hu, N.; Wang, H.; et al. Epitope-Based Vaccine Target Screening against Highly Pathogenic MERS-CoV: An In Silico Approach Applied to Emerging Infectious Diseases. PLoS ONE 2015, 10, e0144475. [Google Scholar] [CrossRef]
- Xie, Q.; He, X.; Yang, F.; Liu, X.; Li, Y.; Liu, Y.; Yang, Z.; Yu, J.; Zhang, B.; Zhao, W. Analysis of the Genome Sequence and Prediction of B-Cell Epitopes of the Envelope Protein of Middle East Respiratory Syndrome-Coronavirus. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 1344–1350. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Inoue, Y.; Tanaka, N.; Tanaka, Y.; Inoue, S.; Morita, K.; Zhuang, M.; Hattori, T.; Sugamura, K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007, 81, 8722–8729. [Google Scholar] [CrossRef]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30–e31. [Google Scholar] [CrossRef]
- Neveu, G.; Ziv-Av, A.; Barouch-Bentov, R.; Berkerman, E.; Mulholland, J.; Einav, S. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J. Virol. 2015, 89, 4387–4404. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases? Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C.; et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Wang, N.; Li, H.; Shi, Y.; Guo, G.; Liu, K.; Zeng, H.; Zou, Q. Therapeutic Drugs Targeting 2019-nCoV Main Protease by High-Throughput Screening. BioRxiv 2020, 1. [Google Scholar] [CrossRef]
- Chen, Y.W.; Yiu, C.B.; Wong, K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research 2020, 9, 129. [Google Scholar] [CrossRef]
- Martinez, M.A. Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef]
- Zha, L.; Li, S.; Pan, L.; Tefsen, B.; Li, Y.; French, N.; Chen, L.; Yang, G.; Villanueva, E.V. Corticosteroid treatment of patients with coronavirus disease 2019 (COVID-19). Med. J. Aust. 2020, 212, 416–420. [Google Scholar] [CrossRef]
- Zhou, W.; Liu, Y.; Tian, D.; Wang, C.; Wang, S.; Cheng, J.; Hu, M.; Fang, M.; Gao, Y. Potential benefits of precise corticosteroids therapy for severe 2019-nCoV pneumonia. Signal Transduct. Target Ther. 2020, 5, 1–3. [Google Scholar] [CrossRef]
- Russell, C.D.; Millar, J.E.; Baillie, J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020, 395, 473–475. [Google Scholar] [CrossRef]
- Khan, F.; Fabbri, L.; Stewart, I.; Robinson, K.; Smyth, A.R.; Jenkins, G. A systematic review of Anakinra, Tocilizumab, Sarilumab and Siltuximab for coronavirus-related infections. medRxiv 2020. [Google Scholar] [CrossRef]
- Fu, Y.; Cheng, Y.; Wu, Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. [published online ahead of print, 3 March 2020]. Virol. Sin. 2020, 1–6. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Luo, P.; Liu, Y.; Qiu, L.; Liu, X.; Liu, D.; Li, J. Tocilizumab treatment in COVID-19: A single center experience. J. Med. Virol. 2020, 92, 814–818. [Google Scholar] [CrossRef]
- Michot, J.M.; Albiges, L.; Chaput, N.; Saada, V.; Pommeret, F.; Griscelli, F.; Balleyguier, C.; Besse, B.; Marabelle, A.; Netzer, F.; et al. Tocilizumab, an anti-IL-6 receptor antibody, to treat COVID-19-related respiratory failure: A case report. Ann. Oncol. 2020, 31, 961–964. [Google Scholar] [CrossRef]
- Zhang, X.; Song, K.; Tong, F.; Fei, M.; Guo, H.; Lu, Z.; Wang, J.; Zheng, C. First case of COVID-19 in a patient with multiple myeloma successfully treated with tocilizumab. Blood Adv. 2020, 4, 1307–1310. [Google Scholar] [CrossRef]
- Marano, G.; Vaglio, S.; Pupella, S.; Facco, G.; Catalano, L.; Liumbruno, G.M.; Grazzini, G. Convalescent plasma: New evidence for an old therapeutic tool? Blood Transfus. 2016, 14, 152–157. [Google Scholar] [CrossRef][Green Version]
- Chen, L.; Xiong, J.; Bao, L.; Shi, Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020, 20, 398–400. [Google Scholar] [CrossRef]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Al-Homoud, A.H.; Memish, Z.A. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med. Infect. Dis. 2020, 34, 101615. [Google Scholar] [CrossRef] [PubMed]
- Seesuay, W.; Jittavisutthikul, S.; Sae-Lim, N.; Sookrung, N.; Sakolvaree, Y.; Chaicumpa, W. Human transbodies that interfere with the functions of Ebola virus VP35 protein in genome replication and transcription and innate immune antagonism. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.; Cuenca, J.; Cruz, F.F.; Figueroa, F.E.; Rocco, P.; Weiss, D.J. Current status of cell-based therapies for respiratory virus infections: Applicability to COVID-19. Eur. Respir. J. 2020, 55, 2000858. [Google Scholar] [CrossRef] [PubMed]
- Connick, P.; Kolappan, M.; Crawley, C.; Webber, D.J.; Patani, R.; Michell, A.W.; Du, M.Q.; Luan, S.L.; Altmann, D.R.; Thompson, A.J.; et al. Autologous mesenchymal stem cells for the treatment of secondary progressive multiple sclerosis: An open-label phase 2a proof-of-concept study. Lancet Neurol. 2012, 11, 150–156. [Google Scholar] [CrossRef]
- Wilson, J.G.; Liu, K.D.; Zhuo, H.; Caballero, L.; McMillan, M.; Fang, X.; Cosgrove, K.; Vojnik, R.; Calfee, C.S.; Lee, J.W.; et al. Mesenchymal stem (stromal) cells for treatment of ARDS: A phase 1 clinical trial. Lancet Respir. Med. 2015, 3, 24–32. [Google Scholar] [CrossRef]
- Bari, E.; Ferrarotti, I.; Saracino, L.; Perteghella, S.; Torre, M.L.; Corsico, A.G. Mesenchymal Stromal Cell Secretome for Severe COVID-19 Infections: Premises for the Therapeutic Use. Cells 2020, 9, 924. [Google Scholar] [CrossRef]
- Hashmi, S.; Ahmed, M.; Murad, M.H.; Litzow, M.R.; Adams, R.H.; Ball, L.M.; Prasad, V.K.; Kebriaei, P.; Ringden, O. Survival after mesenchymal stromal cell therapy in steroid-refractory acute graft-versus-host disease: Systematic review and meta-analysis. Lancet Haematol. 2016, 3, e45–e52. [Google Scholar] [CrossRef]
- Xu, J. Therapeutic Applications of Mesenchymal Stem Cells for Systemic Lupus Erythematosus. Adv. Exp. Med. Biol. 2018, 1089, 73–85. [Google Scholar] [CrossRef]
- Galipeau, J.; Sensébé, L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell 2018, 22, 824–833. [Google Scholar] [CrossRef]
- Bernardo, M.E.; Fibbe, W.E. Mesenchymal stromal cells: Sensors and switchers of inflammation. Cell Stem Cell 2013, 13, 392–402. [Google Scholar] [CrossRef]
- Hopcraft, S.E.; Damania, B. Tumour viruses and innate immunity. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160267. [Google Scholar] [CrossRef]
- Li, W.; Ren, G.; Huang, Y.; Su, J.; Han, Y.; Li, J.; Chen, X.; Cao, K.; Chen, Q.; Shou, P.; et al. Mesenchymal stem cells: A double-edged sword in regulating immune responses. Cell Death Differ. 2012, 19, 1505–1513. [Google Scholar] [CrossRef]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2- Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020, 11, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, C.; Chen, L.; Tang, L.; Zhu, Y.; Xu, X.; Chen, L.; Gao, H.; Lu, X.; Yu, L.; et al. Clinical study of mesenchymal stem cell treating acute respiratory distress syndrome induced by epidemic Influenza A (H7N9) infection, a hint for COVID-19 treatment. Engineering 2020, 10. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, R.; Shi, D.; Liu, X.; Chen, Y.; Dou, X.; Zhu, X.; Lu, C.; Liang, W.; Liao, L.; et al. Mesenchymal stem cells induce mature dendritic cells into a novel Jagged-2-dependent regulatory dendritic cell population. Blood 2009, 113, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Wang, M.X.; Ang, I.; Tan, S.; Lewis, R.F.; Chen, J.I.; Gutierrez, R.A.; Gwee, S.; Chua, P.; Yang, Q.; et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J. Clin. Med. 2020, 9, 623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y. Potential interventions for novel coronavirus in China: A systematic review. J. Med. Virol. 2020, 92, 479–490. [Google Scholar] [CrossRef]
- CEPI. CEPI to Fund Three Programmes to Develop Vaccines against the Novel Coronavirus, nCoV-2019. 2020. Available online: https://cepi.net/news_cepi/cepi-to-fund-three-programmes-todevelop-vaccines-against-the-novel-coronavirus-ncov-2019/ (accessed on 15 February 2020).
- Kim, E.; Erdos, G.; Huang, S.; Kenniston, T.W.; Balmert, S.C.; Carey, C.D.; Raj, V.S.; Epperly, M.W.; Klimstra, W.B.; Haagmans, B.L.; et al. Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development. EBioMedicine 2020, 55, 102743. [Google Scholar] [CrossRef]
- Balmert, S.C.; Carey, C.D.; Falo, G.D.; Sethi, S.K.; Erdos, G.; Korkmaz, E.; Falo, L.D., Jr. Dissolving undercut microneedle arrays for multicomponent cutaneous vaccination. J. Control. Release 2020, 317, 336–346. [Google Scholar] [CrossRef]
- Zhao, X.; Birchall, J.C.; Coulman, S.A.; Tatovic, D.; Singh, R.K.; Wen, L.; Wong, F.S.; Dayan, C.M.; Hanna, S.J. Microneedle delivery of autoantigen for immunotherapy in type 1 diabetes. J. Control. Release 2016, 223, 178–187. [Google Scholar] [CrossRef]
- Kabashima, K.; Honda, T.; Ginhoux, F.; Egawa, G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019, 19, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.R.; Larregina, A.T. Professional antigen-presenting cells of the skin. Immunol. Res. 2006, 36, 127–136. [Google Scholar] [CrossRef]
- Morelli, A.E.; Rubin, J.P.; Erdos, G.; Tkacheva, O.A.; Mathers, A.R.; Zahorchak, A.F.; Thomson, A.W.; Falo, L.D., Jr.; Larregina, A.T. CD4+ T cell responses elicited by different subsets of human skin migratory dendritic cells. J. Immunol. 2005, 175, 7905–7915. [Google Scholar] [CrossRef]
- Sumpter, T.L.; Balmert, S.C.; Kaplan, D.H. Cutaneous immune responses mediated by dendritic cells and mast cells. JCI Insight 2019, 4, e123947. [Google Scholar] [CrossRef]
- Arya, J.; Prausnitz, M.R. Microneedle patches for vaccination in developing countries. J. Control. Release 2016, 240, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, J. Organization of the Skin Immune System and Compartmentalized Immune Responses in Infectious Diseases. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.A.; Wu, J.; Kaplan, D.H. Neuronal Regulation of Cutaneous Immunity. J. Immunol. 2020, 204, 264–270. [Google Scholar] [CrossRef]
- Dicks, M.D.; Spencer, A.J.; Edwards, N.J.; Wadell, G.; Bojang, K.; Gilbert, S.C.; Hill, A.V.; Cottingham, M.G. A novel chimpanzee adenovirus vector with low human seroprevalence: Improved systems for vector derivation and comparative immunogenicity. PLoS ONE 2012, 7, e40385. [Google Scholar] [CrossRef]
- Alharbi, N.K.; Padron-Regalado, E.; Thompson, C.P.; Kupke, A.; Wells, D.; Sloan, M.A.; Grehan, K.; Temperton, N.; Lambe, T.; Warimwe, G.; et al. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine 2017, 35, 3780–3788. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef]


| Replication Stage | Host Receptor | Virus | Function |
|---|---|---|---|
| Attachment and entry | Human angiotensin-converting enzyme 2 (ACE2) | SARS-CoV and most probably SARS-CoV-2 | Cellular receptor |
| Human dipeptidyl peptidase 4 (DPP4 or CD26) | MERS-CoV | Cellular receptor | |
| Cathepsin L | SARS-CoV | Cleave and activate S protein | |
| Furin | MERS-CoV | Cleave and activate S protein | |
| TMPRSS11D | SARS-CoV | Cleave and activate S protein | |
| IFITM | SARS-CoV, MERS-CoV | Restrict virus entry | |
| Genome replication and transcription | GSK3 | SARS-CoV | Phosphorylate N protein and facilitate viral replication |
| hnRNPA1 | SARS-CoV | Regulate viral RNA synthesis | |
| Translation of structural proteins | N-linked glycosylation enzymes | SARS-CoV | Modify S and M protein; N-linked glycosylation of the S protein facilitates lectin-mediated virion attachment and constitutes some neutralizing epitopes |
| ER chaperones | SARS-CoV | Proper folding and maturation of S protein |
| Virus | Receptor | Primary Host | Intermediate Host | Incubation Period | Number of Cases | Number of Deaths | Fatality | R0 |
|---|---|---|---|---|---|---|---|---|
| SARS-CoV | ACE2 | Bats | Civets and raccoon dogs | between 2 and 10 days and up to 14 days | 8098 | 916 | ~10% | 2–5 |
| MERS-CoV | DPP4 (CD26) | Bats | Camels | between 2 and 14 days | 2494 | 858 | ~35% | 2–5 |
| SARS-CoV-2 | ACE2 | Bats | Manis javanica, others? | Current estimates between 2 and 10 days and up to 14 days | 17,889,134 August 3rd, 2020 | Over 686,145 August 3rd, 2020 | ~3.8% August 3rd, 2020 | 2–6.47 |
| City | Average Humidity (%) | Temperature (°C) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nov. | Dec. | Jan. | Feb. | Mar. | Nov. | Dec. | Jan. | Feb. | Mar. | |
| Rome | 71 | 65 | 66 | 63 | 63 | 15 | 1 | 11 | 13 | 14 |
| Paris | 78 | 76 | 79 | 74 | 66 | 9 | 8 | 8 | 9 | 9 |
| Madrid | 70 | 67 | 68 | 63 | 61 | 10 | 10 | 9 | 13 | 12 |
| Milan | 77 | 74 | 69 | 58 | 62 | 11 | 8 | 7 | 11 | 11 |
| Lisbon | 75 | 74 | 77 | 76 | 71 | 15 | 14 | 12 | 15 | 15 |
| Rabat | 72 | 71 | 69 | 72 | 74 | 16 | 17 | 14 | 17 | 16 |
| Algiers | 64 | 62 | 61 | 61 | 66 | 16 | 17 | 15 | 18 | 16 |
| Tunis | 61 | 66 | 72 | 65 | 70 | 17 | 16 | 14 | 15 | 15 |
| Tripoli | 80 | 83 | 73 | 75 | 71 | 15 | 10 | 8 | 9 | 11 |
| Cairo | 45 | 52 | 55 | 53 | 46 | 25 | 18 | 16 | 18 | 22 |
| Clinical Types | Symptoms |
|---|---|
| Mild | In 81% of all confirmed COVID-19 cases. Dry cough, mild fever, sore throat, nasal congestion, muscle pain, headache and malaise. Absence of serious symptoms like dyspnea, also the absence of radiograph features. It may rapidly deteriorate into severe or critical cases, non-pneumonia or mild pneumonia. |
| Moderate | Dry cough, tachypnea and shortness of breath. |
| Severe | Acute respiratory distress syndrome (ARDS), severe pneumonia, severe dyspnea, sepsis or septic shock, tachypnea (respiratory frequency) ≥ 30/min, blood oxygen saturation (SpO2) ≤ 93%, partial pressure of arterial oxygen to fraction of inspired oxygen ratio (PaO2/FiO2) < 300, and/or lung infiltrates > 50% within 24 to 48 h. Fever can be absent or moderate. |
| Critical | In 5% of all confirmed COVID-19 cases. Respiratory failure, septic shock, RNAemia, cardiac injury and/or multiple organ dysfunction or failure. Case fatality rate is 49% (higher case fatality rate for patients with preexisting co-morbidities and lower-case fatality rate (0.9%) for patients without co-morbidities). Cardiovascular disease (10.5%), diabetes (7.3%), respiratory disease (6.5%), hypertension (6%) and oncological complications (5.6%). |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hozhabri, H.; Piceci Sparascio, F.; Sohrabi, H.; Mousavifar, L.; Roy, R.; Scribano, D.; De Luca, A.; Ambrosi, C.; Sarshar, M. The Global Emergency of Novel Coronavirus (SARS-CoV-2): An Update of the Current Status and Forecasting. Int. J. Environ. Res. Public Health 2020, 17, 5648. https://doi.org/10.3390/ijerph17165648
Hozhabri H, Piceci Sparascio F, Sohrabi H, Mousavifar L, Roy R, Scribano D, De Luca A, Ambrosi C, Sarshar M. The Global Emergency of Novel Coronavirus (SARS-CoV-2): An Update of the Current Status and Forecasting. International Journal of Environmental Research and Public Health. 2020; 17(16):5648. https://doi.org/10.3390/ijerph17165648
Chicago/Turabian StyleHozhabri, Hossein, Francesca Piceci Sparascio, Hamidreza Sohrabi, Leila Mousavifar, René Roy, Daniela Scribano, Alessandro De Luca, Cecilia Ambrosi, and Meysam Sarshar. 2020. "The Global Emergency of Novel Coronavirus (SARS-CoV-2): An Update of the Current Status and Forecasting" International Journal of Environmental Research and Public Health 17, no. 16: 5648. https://doi.org/10.3390/ijerph17165648
APA StyleHozhabri, H., Piceci Sparascio, F., Sohrabi, H., Mousavifar, L., Roy, R., Scribano, D., De Luca, A., Ambrosi, C., & Sarshar, M. (2020). The Global Emergency of Novel Coronavirus (SARS-CoV-2): An Update of the Current Status and Forecasting. International Journal of Environmental Research and Public Health, 17(16), 5648. https://doi.org/10.3390/ijerph17165648










