Neonatal Lead (Pb) Exposure and DNA Methylation Profiles in Dried Bloodspots
Abstract
:1. Introduction
2. Materials and Methods
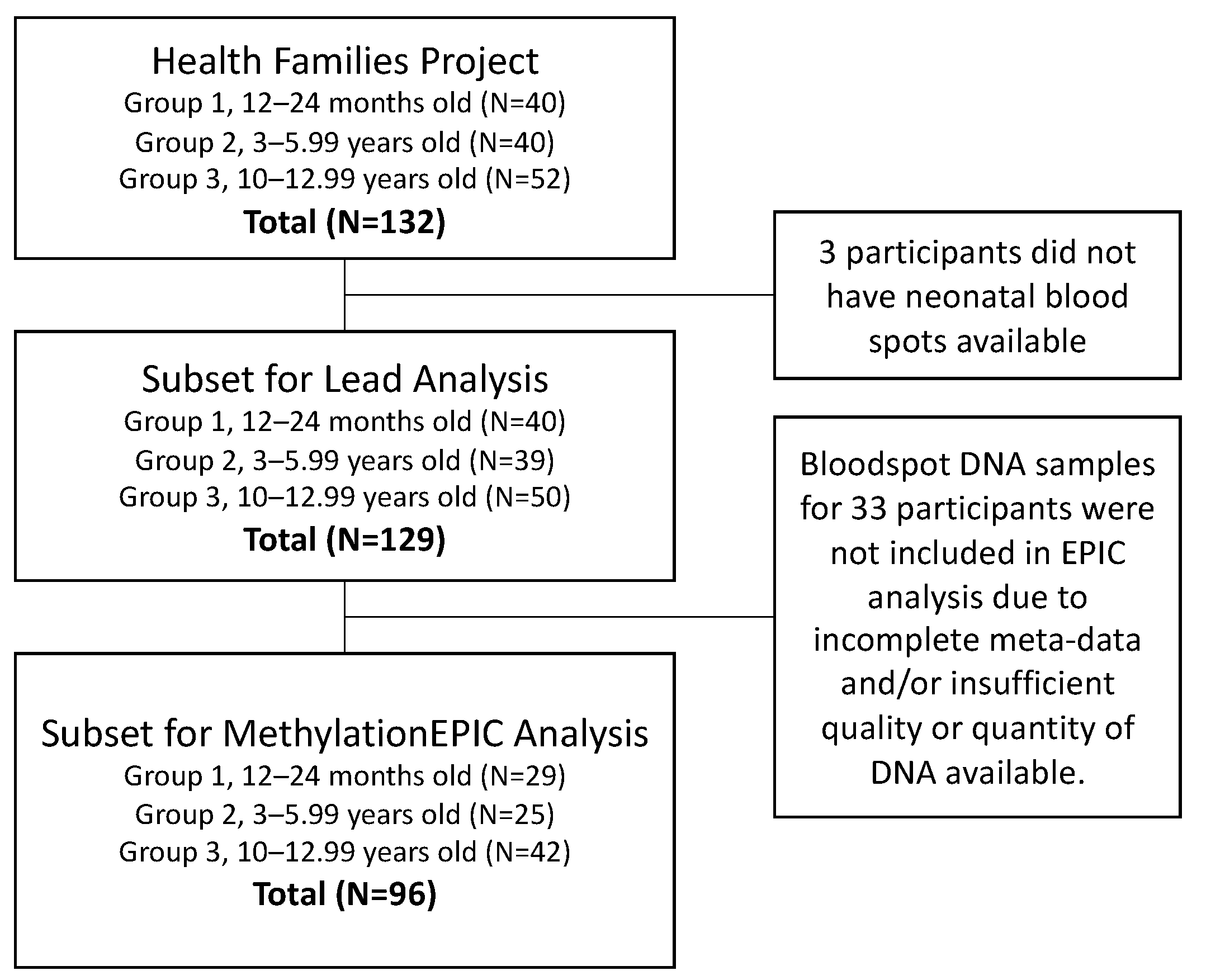
2.1. Healthy Families Project
2.2. Neonatal Bloodspots
2.3. Bloodspot Pb Assessment
2.4. DNA Isolation
2.5. MethylationEPIC Analysis
2.6. Statistical Methods
3. Results
3.1. Cohort Characteristics
3.2. Sample Characteristics
3.3. Pb Assessment in Dried Bloodspots
3.4. CpG Site Loci and Regions Differentially Methylated by Pb
3.5. Biological Pathways Enriched for Differential Methylation by Pb
3.6. Differential Variability by Pb Exposure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- ATSDR Toxicological Profile: Lead. Available online: https://www.atsdr.cdc.gov/toxprofiles/TP.asp?id=96&tid=22 (accessed on 20 April 2020).
- Cox, D.C.; Dewalt, G.; O’Haver, R.; Salatino, B. American Healthy Homes Survey Lead and Arsenic Findings; Department of Housing and Urban Development: Washington, DC, USA, 2011.
- Hanna-Attisha, M.; LaChance, J.; Sadler, R.C.; Champney Schnepp, A. Elevated Blood Lead Levels in Children Associated with the Flint Drinking Water Crisis: A Spatial Analysis of Risk and Public Health Response. Am. J. Public Health 2015, 106, 283–290. [Google Scholar] [CrossRef]
- Allaire, M.; Wu, H.; Lall, U. National trends in drinking water quality violations. Proc. Natl. Acad. Sci. USA 2018, 115, 2078–2083. [Google Scholar] [CrossRef] [Green Version]
- Pantic, I.; Tamayo-Ortiz, M.; Rosa-Parra, A.; Bautista-Arredondo, L.; Wright, R.O.; Peterson, K.E.; Schnaas, L.; Rothenberg, S.J.; Hu, H.; Téllez-Rojo, M.M. Children’s Blood Lead Concentrations from 1988 to 2015 in Mexico City: The Contribution of Lead in Air and Traditional Lead-Glazed Ceramics. Int. J. Environ. Res. Public Health 2018, 15, 2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kordas, K.; Ravenscroft, J.; Cao, Y.; McLean, E.V. Lead Exposure in Low and Middle-Income Countries: Perspectives and Lessons on Patterns, Injustices, Economics, and Politics. Int. J. Environ. Res. Public Health 2018, 15, 2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blood Lead Levels in Children|Lead|CDC. Available online: https://www.cdc.gov/nceh/lead/prevention/blood-lead-levels.htm (accessed on 18 June 2020).
- Senut, M.-C.; Cingolani, P.; Sen, A.; Kruger, A.; Shaik, A.; Hirsch, H.; Suhr, S.T.; Ruden, D. Epigenetics of early-life lead exposure and effects on brain development. Epigenomics 2012, 4, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Sharif, J.; Shinkai, Y.; Koseki, H. Is there a role for endogenous retroviruses to mediate long-term adaptive phenotypic response upon environmental inputs? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20110340. [Google Scholar] [CrossRef] [Green Version]
- Swope, C.B.; Hernández, D. Housing as a determinant of health equity: A conceptual model. Soc. Sci. Med. 2019, 243, 112571. [Google Scholar] [CrossRef]
- Loison, L. Lamarckism and epigenetic inheritance: A clarification. Biol. Philos. 2018, 33, 29. [Google Scholar] [CrossRef]
- Feinberg, A.P. The Key Role of Epigenetics in Human Disease Prevention and Mitigation | NEJM. N. Engl. J. Med. 2018, 378, 1323–1334. [Google Scholar] [CrossRef]
- Tzika, E.; Dreker, T.; Imhof, A. Epigenetics and Metabolism in Health and Disease. Front. Genet. 2018, 9, 361. [Google Scholar] [CrossRef] [Green Version]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic plasticity and the hallmarks of cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Jiao, B.; Shen, L. The Epigenetics of Alzheimer’s Disease: Factors and Therapeutic Implications. Front. Genet. 2018, 9, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, D.J.P. The developmental origins of adult disease. J. Am. Coll. Nutr. 2004, 23, 588S–595S. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faulk, C.; Barks, A.; Liu, K.; Goodrich, J.M.; Dolinoy, D.C. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 2013, 5, 487–500. [Google Scholar] [CrossRef] [Green Version]
- Leasure, J.L.; Giddabasappa, A.; Chaney, S.; Johnson, J.E.; Pothakos, K.; Lau, Y.S.; Fox, D.A. Low-Level Human Equivalent Gestational Lead Exposure Produces Sex-Specific Motor and Coordination Abnormalities and Late-Onset Obesity in Year-Old Mice. Environ. Health Perspect. 2008, 116, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Peterson, K.E.; Montgomery, K.; Sánchez, B.N.; Zhang, Z.; Afeiche, M.C.; Cantonwine, D.E.; Ettinger, A.S.; Cantoral, A.; Schnaas, L.; et al. Early lead exposure and childhood adiposity in Mexico city. Int. J. Hyg. Environ. Health 2019, 222, 965–970. [Google Scholar] [CrossRef]
- Liu, Y.; Ettinger, A.S.; Téllez-Rojo, M.; Sánchez, B.N.; Zhang, Z.; Cantoral, A.; Hu, H.; Peterson, K.E. Prenatal Lead Exposure, Type 2 Diabetes, and Cardiometabolic Risk Factors in Mexican Children at Age 10–18 Years. J. Clin. Endocrinol. Metab. 2020, 105, 210–218. [Google Scholar] [CrossRef]
- Montrose, L.; Faulk, C.; Francis, J.; Dolinoy, D.C. Perinatal lead (Pb) exposure results in sex and tissue-dependent adult DNA methylation alterations in murine IAP transposons. Environ. Mol. Mutagen. 2017, 58, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, J.M.; Sánchez, B.N.; Dolinoy, D.C.; Zhang, Z.; Hernández-Ávila, M.; Hu, H.; Peterson, K.E.; Téllez-Rojo, M.M. Quality control and statistical modeling for environmental epigenetics: A study on in utero lead exposure and DNA methylation at birth. Epigenetics 2015, 10, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Rygiel, C.A.; Dolinoy, D.C.; Perng, W.; Jones, T.R.; Solano, M.; Hu, H.; Téllez-Rojo, M.M.; Peterson, K.E.; Goodrich, J.M. Trimester-Specific Associations of Prenatal Lead Exposure WITH Infant Cord Blood DNA Methylation at Birth. Epigenetis Insights 2020, 13. [Google Scholar] [CrossRef]
- Sen, A.; Heredia, N.; Senut, M.-C.; Hess, M.; Land, S.; Qu, W.; Hollacher, K.; Dereski, M.O.; Ruden, D.M. Early life lead exposure causes gender-specific changes in the DNA methylation profile of DNA extracted from dried blood spots. Epigenomics 2015, 7, 379–393. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Hivert, M.-F.; Cardenas, A.; Zhong, J.; Rifas-Shiman, S.L.; Agha, G.; Colicino, E.; Just, A.C.; Amarasiriwardena, C.; Lin, X.; et al. Exposure to Low Levels of Lead in Utero and Umbilical Cord Blood DNA Methylation in Project Viva: An Epigenome-Wide Association Study. Environ. Health Perspect. 2017, 125, 087019. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.H.; Goldenberg, A.; Anderson, R.; Rothwell, E.; Botkin, J. State laws regarding the retention and use of residual newborn screening blood samples. Pediatrics 2011, 127, 703–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botkin, J.R.; Goldenberg, A.J.; Rothwell, E.; Anderson, R.A.; Lewis, M.H. Retention and Research Use of Residual Newborn Screening Bloodspots. Pediatrics 2013, 131, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, M.E.; Metternick-Jones, S.C.; Lister, K.J. International differences in the evaluation of conditions for newborn bloodspot screening: A review of scientific literature and policy documents. Eur. J. Hum. Genet. 2017, 25, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Acharya, Y.; Norton, E.C.; Lumeng, J.C. The Effect of Financial Compensation on Willingness to Supply a Child’s Blood Sample: A Randomized Controlled Trial. Eval. Health Prof. 2017, 40, 359–371. [Google Scholar] [CrossRef]
- Kochmanski, J.; Goodrich, J.M.; Peterson, K.E.; Lumeng, J.C.; Dolinoy, D.C. Neonatal bloodspot DNA methylation patterns are associated with childhood weight status in the Healthy Families Project. Pediatr. Res. 2019, 85, 848–855. [Google Scholar] [CrossRef]
- Basu, N.; Eng, J.W.L.; Perkins, M.; Santa-Rios, A.; Martincevic, G.; Carlson, K.; Neitzel, R.L. Development and application of a novel method to characterize methylmercury exposure in newborns using dried blood spots. Environ. Res. 2017, 159, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lee, M.S.J. Dried Blood Spots: Applications and Techniques; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Harkema, J.R.; Wagner, J.G.; Kaminski, N.E.; Morishita, M.; Keeler, G.J.; McDonald, J.D.; Barrett, E.G. HEI Health Review Committee Effects of concentrated ambient particles and diesel engine exhaust on allergic airway disease in Brown Norway rats. Res. Rep. Health Eff. Inst. 2009, 145, 5. [Google Scholar]
- CDC Laboratory Procedure Manual. Blood Metals Panel 3 (BMP3) ICP-DRC-MS. Method No: DLS 3016.8-04. 2012. Available online: https://www.cdc.gov/Nchs/Data/Nhanes/Nhanes_13_14/PbCd_H_MET.pdf (accessed on 15 July 2020).
- Fortin, J.-P.; Triche, T.J.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Langie, S.A.S.; De Boever, P.; Taylor, J.A.; Niu, L. RELIC: A novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genom. 2017, 18, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakulski, K.M.; Feinberg, J.I.; Andrews, S.V.; Yang, J.; Brown, S.; L McKenney, S.; Witter, F.; Walston, J.; Feinberg, A.P.; Fallin, M.D. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 2016, 11, 354–362. [Google Scholar] [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teschendorff, A.E.; Menon, U.; Gentry-Maharaj, A.; Ramus, S.J.; Gayther, S.A.; Apostolidou, S.; Jones, A.; Lechner, M.; Beck, S.; Jacobs, I.J.; et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS ONE 2009, 4, e8274. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Mansell, G.; Gorrie-Stone, T.J.; Bao, Y.; Kumari, M.; Schalkwyk, L.S.; Mill, J.; Hannon, E. Guidance for DNA methylation studies: Statistical insights from the Illumina EPIC array. BMC Genom. 2019, 20, 366. [Google Scholar] [CrossRef] [Green Version]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; V Lord, R.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Phipson, B.; Maksimovic, J.; Oshlack, A. missMethyl: An R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 2016, 32, 286–288. [Google Scholar] [CrossRef]
- Kochmanski, J.; Montrose, L.; Goodrich, J.M.; Dolinoy, D.C. Environmental Deflection: The Impact of Toxicant Exposures on the Aging Epigenome. Toxicol. Sci. 2017, 156, 325–335. [Google Scholar] [CrossRef] [Green Version]
- Phipson, B.; Oshlack, A. DiffVar: A new method for detecting differential variability with application to methylation in cancer and aging. Genome Biol. 2014, 15, 465. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Fourth National Report on Human Exposure to Environmental Chemicals Update. 2019. Available online: https://www.cdc.gov/exposurereport/pdf/FourthReport_UpdatedTables_Volume1_Jan2019-508.pdf (accessed on 15 July 2020).
- Goodrich, J.M.; Dolinoy, D.C.; Sánchez, B.N.; Zhang, Z.; Meeker, J.D.; Mercado-Garcia, A.; Solano-González, M.; Hu, H.; Téllez-Rojo, M.M.; Peterson, K.E. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolescence. Environ. Epigenetics 2016, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, A.; Cingolani, P.; Senut, M.-C.; Land, S.; Mercado-Garcia, A.; Tellez-Rojo, M.M.; Baccarelli, A.A.; Wright, R.O.; Ruden, D.M. Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics 2015, 10, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Montes-Castro, N.; Alvarado-Cruz, I.; Torres-Sánchez, L.; García-Aguiar, I.; Barrera-Hernández, A.; Escamilla-Núñez, C.; Del Razo, L.M.; Quintanilla-Vega, B. Prenatal exposure to metals modified DNA methylation and the expression of antioxidant- and DNA defense-related genes in newborns in an urban area. J. Trace Elem. Med. Biol. 2019, 55, 110–120. [Google Scholar] [CrossRef]
- Dou, J.F.; Farooqui, Z.; Faulk, C.D.; Barks, A.K.; Jones, T.; Dolinoy, D.C.; Bakulski, K.M. Perinatal Lead (Pb) Exposure and Cortical Neuron-Specific DNA Methylation in Male Mice. Genes 2019, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Dosunmu, R.; Alashwal, H.; Zawia, N.H. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech. Ageing Dev. 2012, 133, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Singh, G.; Singh, V.; Wang, Z.-X.; Voisin, G.; Lefebvre, F.; Navenot, J.-M.; Evans, B.; Verma, M.; Anderson, D.W.; Schneider, J.S. Effects of Developmental Lead Exposure on the Hippocampal Methylome: Influences of Sex and Timing and Level of Exposure. Toxicol. Lett. 2018, 290, 63–72. [Google Scholar] [CrossRef]
- Young, J.I.; Sivasankaran, S.K.; Wang, L.; Ali, A.; Mehta, A.; Davis, D.A.; Dykxhoorn, D.M.; Petito, C.K.; Beecham, G.W.; Martin, E.R.; et al. Genome-wide Brain DNA methylation analysis suggests epigenetic reprogramming in Parkinson disease. Neurol. Genet. 2019, 5, e342. [Google Scholar] [CrossRef] [Green Version]
- Sheftel, A.D.; Stehling, O.; Pierik, A.J.; Elsässer, H.-P.; Mühlenhoff, U.; Webert, H.; Hobler, A.; Hannemann, F.; Bernhardt, R.; Lill, R. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 11775–11780. [Google Scholar] [CrossRef] [Green Version]
- Rouault, T.A.; Tong, W.H. Iron–sulfur cluster biogenesis and human disease. Trends Genet. 2008, 24, 398–407. [Google Scholar] [CrossRef] [Green Version]
- Hoevenaar-Blom, M.P.; Wielders, J.P.; Groeneveld, H.; de Leeuw, E.; Schmits, R.J.; Pepermans, C.; Pasker-de Jong, P.C.; Hogeman, P.H. Prevalence and determinants of vitamin D deficiency in infants and toddlers in the Netherlands: A pilot study. Ann. Clin. Biochem. 2019, 56, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Kannan, K.; Wu, Q.; Bell, E.M.; Druschel, C.M.; Caggana, M.; Aldous, K.M. Analysis of polyfluoroalkyl substances and bisphenol A in dried blood spots by liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 4127–4138. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.M.; Yeung, E.H.; Ma, W.; Kannan, K.; Sundaram, R.; Smarr, M.M.; Buck Louis, G.M. Concentrations of Endocrine Disrupting Chemicals in Newborn Blood Spots and Infant Outcomes in the Upstate KIDS Study. Environ. Int. 2018, 121, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Traglia, M.; Croen, L.A.; Jones, K.L.; Heuer, L.S.; Yolken, R.; Kharrazi, M.; DeLorenze, G.N.; Ashwood, P.; Van de Water, J.; Weiss, L.A. Cross-genetic determination of maternal and neonatal immune mediators during pregnancy. Genome Med. 2018, 10, 67. [Google Scholar] [CrossRef]
- Nelson, P.G.; Kuddo, T.; Song, E.Y.; Dambrosia, J.M.; Kohler, S.; Satyanarayana, G.; Vandunk, C.; Grether, J.K.; Nelson, K.B. Selected neurotrophins, neuropeptides, and cytokines: Developmental trajectory and concentrations in neonatal blood of children with autism or Down syndrome. Int. J. Dev. Neurosci. 2006, 24, 73–80. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.; Kim, H.-J.; Lee, J.J.; Choi, G.; Choi, S.; Kim, S.; Kim, S.Y.; Moon, H.-B.; Kim, S.; et al. Association between Several Persistent Organic Pollutants and Thyroid Hormone Levels in Cord Blood Serum and Bloodspot of the Newborn Infants of Korea. PLoS ONE 2015, 10, e0125213. [Google Scholar] [CrossRef] [Green Version]
- Shields, B.M.; Knight, B.; Shakespeare, L.; Babrah, J.; Powell, R.J.; Clark, P.M.; Hattersley, A.T. Determinants of insulin concentrations in healthy 1-week-old babies in the community: Applications of a bloodspot assay. Early Hum. Dev. 2006, 82, 143–148. [Google Scholar] [CrossRef]
- Gao, L.; Millstein, J.; Siegmund, K.D.; Dubeau, L.; Maguire, R.; Gilliland, F.D.; Murphy, S.K.; Hoyo, C.; Breton, C.V. Epigenetic regulation of AXL and risk of childhood asthma symptoms. Clin. Epigenetics 2017, 9, 121. [Google Scholar] [CrossRef]
- Gonseth, S.; Shaw, G.M.; Roy, R.; Segal, M.R.; Asrani, K.; Rine, J.; Wiemels, J.; Marini, N.J. Epigenomic profiling of newborns with isolated orofacial clefts reveals widespread DNA methylation changes and implicates metastable epiallele regions in disease risk. Epigenetics 2019, 14, 198–213. [Google Scholar] [CrossRef] [Green Version]
- Perkins, M.; Basu, N. Dried blood spots for estimating mercury exposure in birds. Environ. Pollut. 2018, 236, 236–246. [Google Scholar] [CrossRef]
- Santa-Rios, A.; Barst, B.D.; Basu, N. Mercury Speciation in Whole Blood and Dried Blood Spots from Capillary and Venous Sources. Anal. Chem. 2020, 92, 3605–3612. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Heredia, N.; Senut, M.-C.; Land, S.; Hollocher, K.; Lu, X.; Dereski, M.O.; Ruden, D.M. Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci. Rep. 2015, 5, 14466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, J.W.; Edhlund, B.L.; Johnson, J.; Rosebush, C.E.; Holmquist, Z.S.; Swan, S.H.; Nguyen, R.H.N. Assessing a New Method for Measuring Fetal Exposure to Mercury: Newborn Bloodspots. Int. J. Environ. Res. Public Health 2016, 13, 692. [Google Scholar] [CrossRef] [Green Version]
- Funk, W.E.; McGee, J.K.; Olshan, A.F.; Ghio, A.J. Quantification of arsenic, lead, mercury and cadmium in newborn dried blood spots. Biomarkers 2013, 18, 174–177. [Google Scholar] [CrossRef] [Green Version]
- El-Hajjar, D.F.; Swanson, K.H.; Landmark, J.D.; Stickle, D.F. Validation of use of annular once-punched filter paper bloodspot samples for repeat lead testing. Clin. Chim. Acta 2007, 377, 179–184. [Google Scholar] [CrossRef]
- Funk, W.E. Use of Dried Blood Spots for Estimating Children’s Exposures to Heavy Metals in Epidemiological Research. J. Environ. Anal. Toxicol. 2015, 7. [Google Scholar] [CrossRef] [Green Version]
- Walker, R.M.; MacGillivray, L.; McCafferty, S.; Wrobel, N.; Murphy, L.; Kerr, S.M.; Morris, S.W.; Campbell, A.; McIntosh, A.M.; Porteous, D.J.; et al. Assessment of dried blood spots for DNA methylation profiling. Wellcome Open Res 2019, 4. [Google Scholar] [CrossRef]
| Included in MethylationEPIC Analysis, n = 96 | All HF Families, n = 129 | |||
|---|---|---|---|---|
| Characteristics | Mean ± SD or N (%) | Range | Mean ± SD or N (%) | Range |
| Mothers | ||||
| BMI (kg/m2) | 29.3 ± 8.9 | 15.4–66.4 | 29.5 ± 8.8 | 15.3–66.4 |
| Obesity | ||||
| Not obese | 60 (63%) | 77 (60%) | ||
| Obese | 34 (35%) | 55 (39%) | ||
| Race | ||||
| Non-white | 22 (23%) | 29 (22%) | ||
| White | 74 (77%) | 99 (77%) | ||
| Children | ||||
| Male (%) | 52 (54%) | 69 (53%) | ||
| Recruitment Group | ||||
| Group 1 | 29 (30%) | 40 (31%) | ||
| Group 2 | 25 (26%) | 39 (30%) | ||
| Group 3 | 42 (44%) | 50 (39%) | ||
| Estimated Cord Blood Cell Type % | ||||
| Nucleated Red Blood Cells (nRBC) | 4.2 ± 2.0 | 0.5–12.8 | na | na |
| CD8+ T cells | 7.5 ± 3.4 | 2.3–23.6 | na | na |
| CD4+ T cells | 19.3 ± 6.5 | 6.6–43.5 | na | na |
| B cells | 6.1 ± 2.8 | 1.4–21.0 | na | na |
| Monocytes | 9.0 ± 2.3 | 4.1–17.9 | na | na |
| Granulocytes | 57.0 ± 10.4 | 17.5–77.1 | na | na |
| Natural Killer Cells | 0.01 ± 0.1 | 0–0.96 | na | na |
| Pb Exposure Assessment | ||||
| Blood spot Pb (µg/dL) | 0.78 ± 0.85 | <MDL–5.27 | 0.93 ± 0.80 | <MDL–5.27 |
| Pb Category | ||||
| Low (<median 0.55 µg/dL) | 48 (50%) | 69 (53%) | ||
| High (≥median 0.55 µg/dL) | 48 (50%) | 60 (47%) | ||
| Probe ID | Position | Gene Name | Relationship to CpG Island | Estimate (per ug/dL Pb) | SE of Estimate | Average % Methylation at the CpG Site | Raw p-Value | q-Value |
|---|---|---|---|---|---|---|---|---|
| cg03744954 | chr7:23637556 | CCDC126 | Island | 0.010 | 0.002 | 2.2% | 1.33 × 10−6 | 0.12 |
| cg11961702 | chr10:1130138 | WDR37 | Open Sea | −0.007 | 0.001 | 98.1% | 1.08 × 10−6 | 0.12 |
| cg09489281 | chr5:43604149 | NNT | S. Shore | 0.027 | 0.005 | 5.1% | 1.19 × 10−6 | 0.12 |
| cg06157837 | chr16:50130934 | HEATR3 | Open Sea | −0.021 | 0.004 | 92.7% | 1.08 × 10−6 | 0.12 |
| cg12819470 | chr7:18013541 | Open Sea | −0.022 | 0.004 | 94.1% | 1.34 × 10−6 | 0.12 | |
| cg00694932 | chr22:49029825 | FAM19A5 | Open Sea | −0.027 | 0.005 | 87.6% | 4.91 × 10−7 | 0.12 |
| cg12666827 | chr17:81043176 | METRNL | Island | −0.026 | 0.005 | 92.1% | 5.19 × 10−7 | 0.12 |
| cg17533118 | chr2:204664739 | Open Sea | −0.030 | 0.006 | 88.3% | 8.35 × 10−7 | 0.12 | |
| cg16393928 | chr1:3135836 | PRDM16 | Open Sea | −0.024 | 0.005 | 90.8% | 1.46 × 10−6 | 0.12 |
| cg27500206 | chr11:4600140 | C11orf40 | Open Sea | −0.031 | 0.006 | 90.8% | 7.93 × 10−7 | 0.12 |
| cg25968149 | chr3:174255312 | Open Sea | −0.020 | 0.004 | 92.9% | 1.80 × 10−6 | 0.12 | |
| cg20383654 | chr1:39490535 | NDUFS5 | Open Sea | −0.018 | 0.004 | 93.0% | 1.82 × 10−6 | 0.12 |
| cg05355328 | chr19:33096524 | ANKRD27 | Island | −0.022 | 0.004 | 94.2% | 1.98 × 10−6 | 0.12 |
| cg25104648 | chr18:18566146 | ROCK1 | Open Sea | −0.022 | 0.004 | 95.0% | 2.57 × 10−6 | 0.13 |
| cg08236836 | chr2:95613608 | Open Sea | −0.017 | 0.003 | 98.4% | 2.54 × 10−6 | 0.13 | |
| cg25198485 | chr4:58086247 | Open Sea | −0.021 | 0.004 | 92.2% | 2.46 × 10−6 | 0.13 | |
| cg23724374 | chr6:150973466 | PLEKHG1 | Open Sea | −0.019 | 0.004 | 89.7% | 3.37 × 10−6 | 0.15 |
| cg11877273 | chr1:29182535 | OPRD1 | Open Sea | −0.019 | 0.004 | 94.1% | 3.61 × 10−6 | 0.15 |
| cg06019448 | chr4:190417346 | Open Sea | −0.023 | 0.005 | 90.5% | 3.53 × 10−6 | 0.15 | |
| cg06051201 | chr1:53925306 | DMRTB1 | Island | −0.026 | 0.005 | 95.1% | 4.37 × 10−6 | 0.17 |
| cg09980056 | chr6:52366854 | TRAM2 | Open Sea | −0.025 | 0.005 | 95.0% | 4.60 × 10−6 | 0.17 |
| cg09413029 | chr10:97152467 | SORBS1 | Open Sea | −0.022 | 0.005 | 93.0% | 4.49 × 10−6 | 0.17 |
| cg14810301 | chr5:88169099 | MEF2C | Open Sea | −0.018 | 0.004 | 93.3% | 5.41 × 10−6 | 0.17 |
| cg15406566 | chr16:4606279 | C16orf96 | Open Sea | 0.011 | 0.002 | 87.4% | 5.55 × 10−6 | 0.17 |
| cg05026437 | chr3:99527476 | MIR548G | Open Sea | −0.021 | 0.004 | 92.5% | 5.44 × 10−6 | 0.17 |
| cg14645844 | chr8:123831620 | ZHX2 | Open Sea | -0.024 | 0.005 | 93.7% | 5.17 × 10−6 | 0.17 |
| cg10174926 | chr14:72818968 | RGS6 | Open Sea | −0.018 | 0.004 | 93.0% | 5.81 × 10−6 | 0.17 |
| cg07431286 | chr5:72526496 | Island | 0.003 | 0.001 | 3.1% | 6.04 × 10−6 | 0.17 | |
| cg27094173 | chr11:73371753 | PLEKHB1 | N. Shore | −0.008 | 0.002 | 92.9% | 7.49 × 10−6 | 0.20 |
| cg00394874 | chr6:30747276 | Open Sea | 0.048 | 0.010 | 25.4% | 7.85 × 10−6 | 0.20 | |
| cg06534673 | chr16:1210311 | CACNA1H | Island | −0.027 | 0.006 | 71.4% | 7.73 × 10−6 | 0.20 |
| cg07366047 | chr2:135921205 | RAB3GAP1 | Open Sea | −0.029 | 0.006 | 94.1% | 7.97 × 10−6 | 0.20 |
| Position | Genes Overlapping This Region | # of CpG Sites | Mean Difference in Proportion Methylated per 1 µg/dL Pb | Min. Smoothed FDR q-Value |
|---|---|---|---|---|
| chr19:33096524-33096688 | ANKRD27 | 3 | −0.006 | 3.15 × 10−6 |
| chr1:53925145-53925368 | snoU13, Y_RNA, SCARNA16, U1, SCARNA18, SCARNA24, SNORD112, SNORA62, SNORA63, SNORD46, DMRTB1, SNORA2, SNORD81, U3, SNORA51, SNORA25, SCARNA20, SNORA67, U6, SNORA70, SNORA77, SNORA26, U8, SCARNA11, SNORA31, SNORA42, SNORA40, SNORD64, ACA64, snoU109, SNORD60, SNORD116 | 4 | −0.020 | 2.12 × 10−6 |
| chr9:123605229-123605666 | PSMD5-AS1, PSMD5 | 8 | −0.015 | 5.42 × 10−7 |
| chr15:93616424-93617402 | RGMA | 17 | −0.014 | 5.60 × 10−14 |
| chr11:111637306-111637615 | 4 | 0.010 | 3.06 × 10−5 | |
| chr17:6796745-6797034 | ALOX12P2, AC027763.2 | 4 | 0.016 | 6.21 × 10−5 |
| Pathway Identifier | Description of Pathway | # of Genes in Pathway | # Differentially Methylated Genes | p-Value | q-Value | # Differentially Methylated Genes | p-Value | q-Value | |
|---|---|---|---|---|---|---|---|---|---|
| GO Analysis—Dichotomized Pb | GO—Continuous Pb | ||||||||
| GO: BP | GO:0000902 | cell morphogenesis | 990 | 390 | 1.48 × 10−6 | 0.025 | 385 | 2.65 × 10−5 | 0.139 |
| GO: BP | GO:0000904 | cell morphogenesis involved in differentiation | 709 | 292 | 2.24 × 10−6 | 0.025 | ns | ||
| GO: BP | GO:0022610 | biological adhesion | 1369 | 467 | 5.47 × 10−6 | 0.034 | 472 | 1.23 × 10−5 | 0.131 |
| GO: BP | GO:0007155 | cell adhesion | 1363 | 465 | 5.93 × 10−6 | 0.034 | 469 | 1.73 × 10−5 | 0.131 |
| GO: BP | GO:0032989 | cellular component morphogenesis | 1098 | 417 | 1.03 × 10−5 | 0.047 | 421 | 9.31 × 10−6 | 0.131 |
| GO: BP | GO:0032990 | cell part morphogenesis | 653 | 269 | 1.25 × 10−5 | 0.047 | ns | ||
| GO: BP | GO:0048812 | neuron projection morphogenesis | 617 | 256 | 1.83 × 10−5 | 0.058 | ns | ||
| GO: BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 631 | 261 | 2.10 × 10−5 | 0.058 | ns | ||
| GO: BP | GO:0048858 | cell projection morphogenesis | 635 | 262 | 2.30 × 10−5 | 0.058 | ns | ||
| GO: BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 555 | 234 | 3.05 × 10−5 | 0.069 | ns | ||
| GO: BP | GO:0099173 | postsynapse organization | 150 | 72 | 6.64 × 10−5 | 0.136 | ns | ||
| GO: BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 11 | 11 | 7.18 × 10−5 | 0.136 | ns | ||
| GO: CC | GO:0071944 | cell periphery | 5163 | 1465 | 9.25 × 10−5 | 0.162 | ns | ||
| GO: MF | GO:0047372 | acylglycerol lipase activity | 9 | ns | 9 | 3.05 × 10−5 | 0.139 | ||
| KEGG Analysis—Dichotomized Pb | KEGG—Continuous Pb | ||||||||
| KEGG | path:hsa04360 | Axon guidance | 174 | 86 | 4.76 × 10−5 | 0.016 | ns | ||
| KEGG | path:hsa04727 | GABAergic synapse | 84 | 43 | 9.06 × 10−5 | 0.153 | ns | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montrose, L.; Goodrich, J.M.; Morishita, M.; Kochmanski, J.; Klaver, Z.; Cavalcante, R.; Lumeng, J.C.; Peterson, K.E.; Dolinoy, D.C. Neonatal Lead (Pb) Exposure and DNA Methylation Profiles in Dried Bloodspots. Int. J. Environ. Res. Public Health 2020, 17, 6775. https://doi.org/10.3390/ijerph17186775
Montrose L, Goodrich JM, Morishita M, Kochmanski J, Klaver Z, Cavalcante R, Lumeng JC, Peterson KE, Dolinoy DC. Neonatal Lead (Pb) Exposure and DNA Methylation Profiles in Dried Bloodspots. International Journal of Environmental Research and Public Health. 2020; 17(18):6775. https://doi.org/10.3390/ijerph17186775
Chicago/Turabian StyleMontrose, Luke, Jaclyn M. Goodrich, Masako Morishita, Joseph Kochmanski, Zachary Klaver, Raymond Cavalcante, Julie C. Lumeng, Karen E. Peterson, and Dana C. Dolinoy. 2020. "Neonatal Lead (Pb) Exposure and DNA Methylation Profiles in Dried Bloodspots" International Journal of Environmental Research and Public Health 17, no. 18: 6775. https://doi.org/10.3390/ijerph17186775






