Flooding Irrigation Weakens the Molecular Ecological Network Complexity of Soil Microbes during the Process of Dryland-to-Paddy Conversion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Soil Sample Collection and Analysis
2.3. DNA Extraction, PCR Amplification, and Illumina Miseq Sequencing
2.4. Analysis of Soil Microbial Molecular Ecological Network
2.5. Data Statistic and Analysis
3. Results
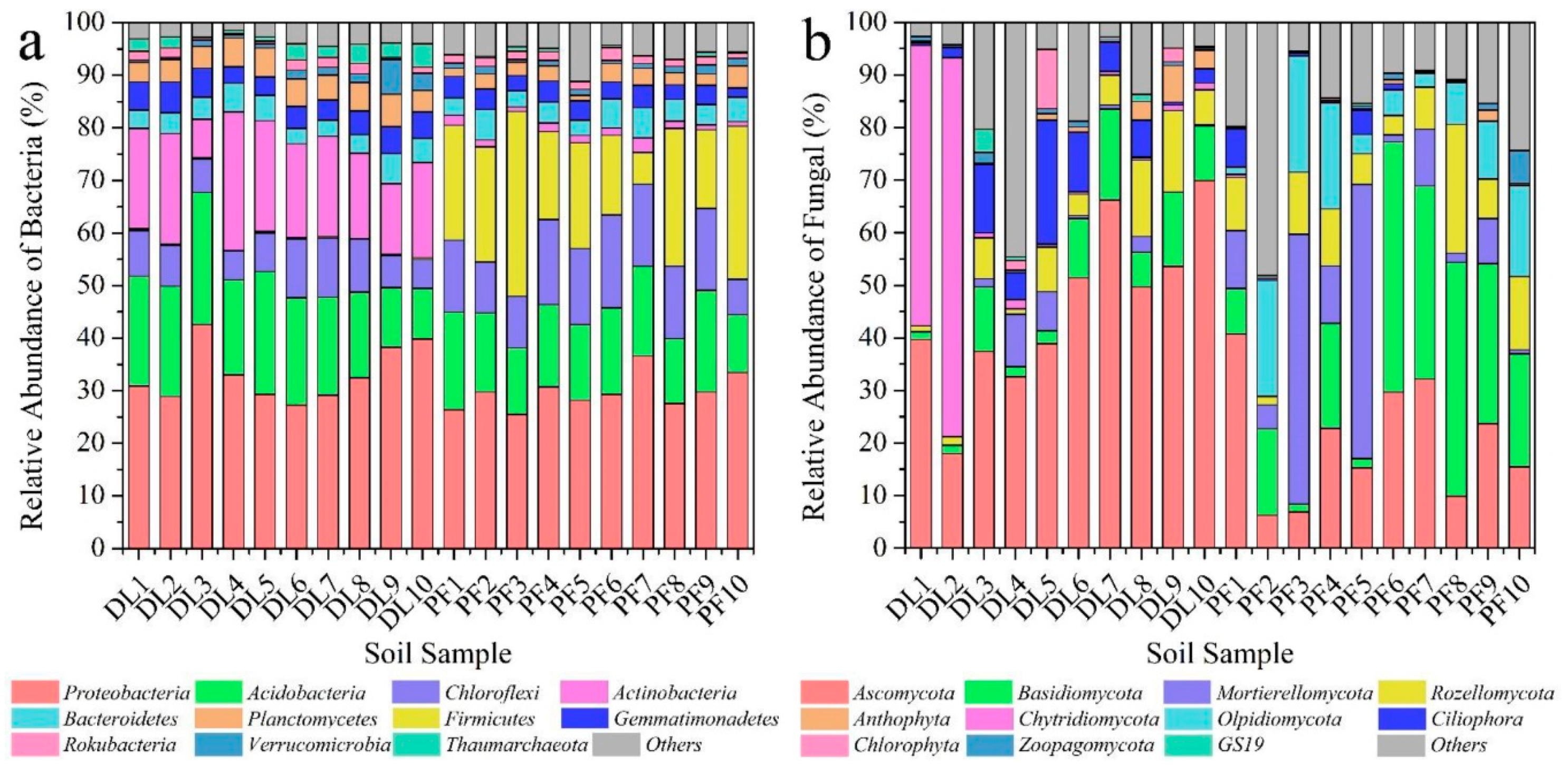
3.1. Effects of Dryland-to-Paddy Conversion on Soil Microbial Community Structure
3.1.1. Impact on Soil Microbial Community Diversity
3.1.2. Impact on the Soil Microbial Community Composition
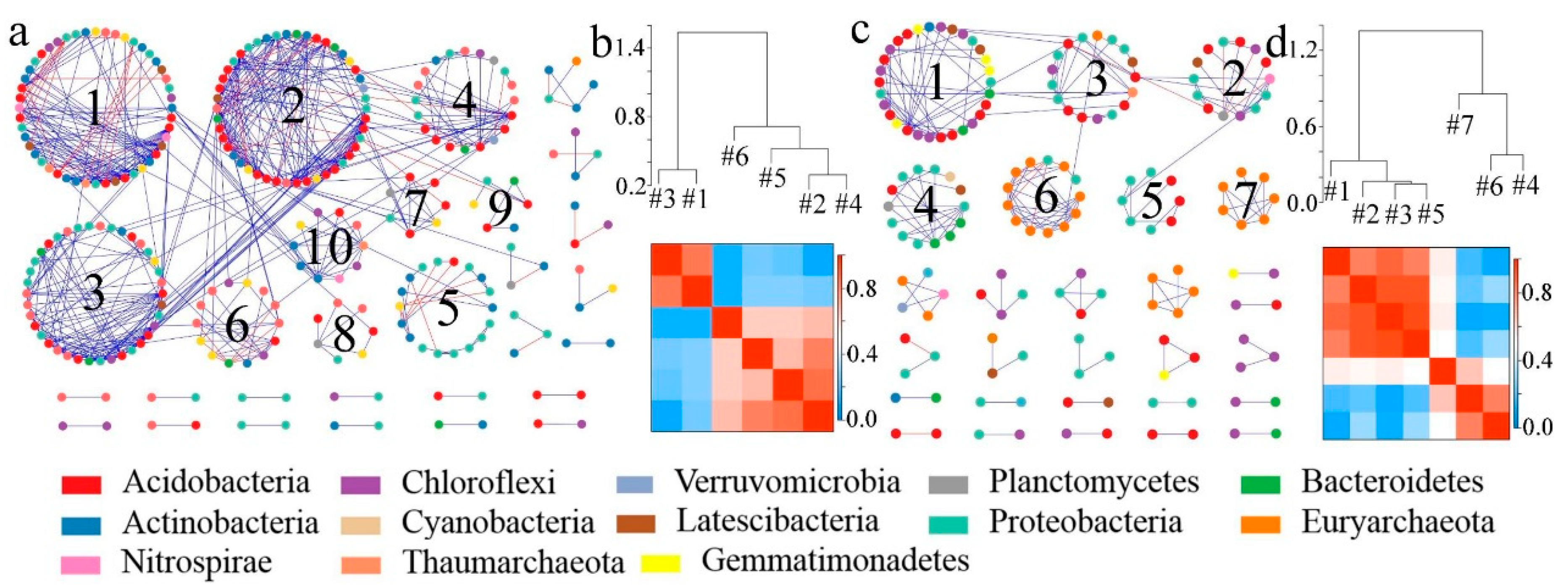
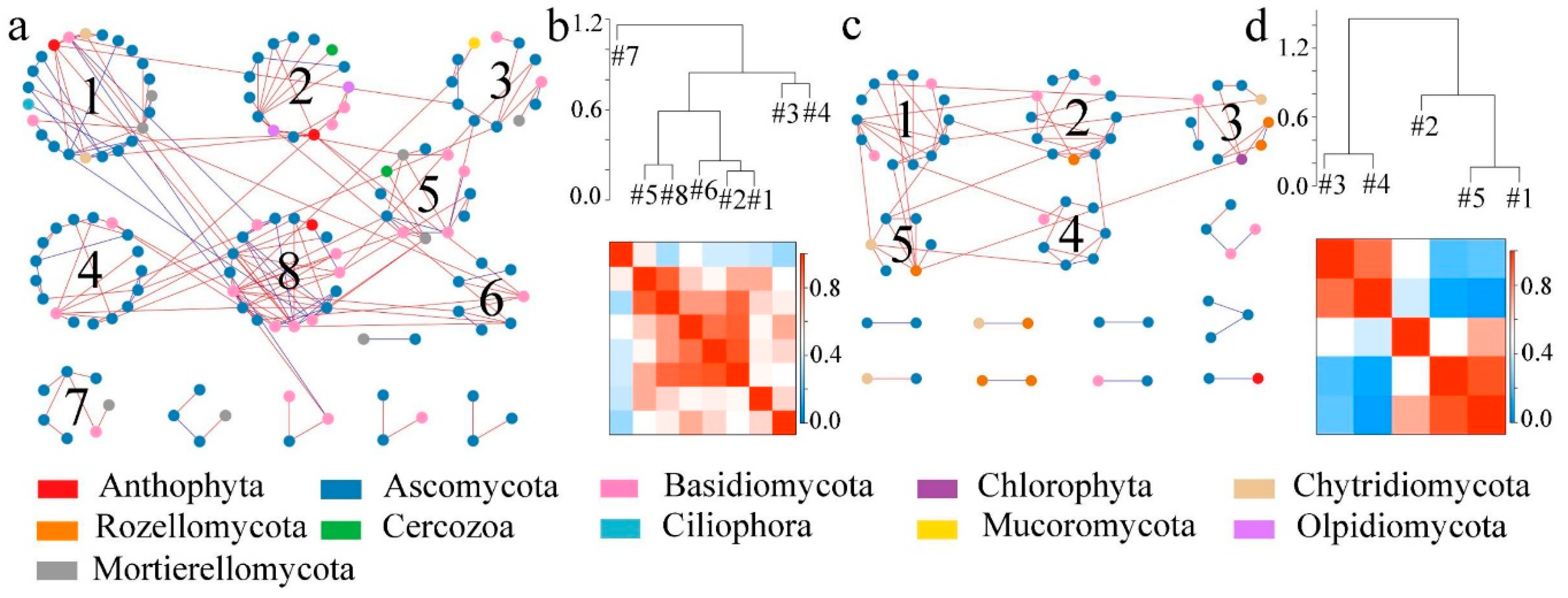
3.2. Effects of Dryland-to-Paddy Conversion on the Structure of Soil Microbial Molecular Ecological Networks
3.2.1. Changes in OTU Topological Structures of Molecular Ecological Networks Generated During the Process of Dryland-to-Paddy Conversion
3.2.2. Construction and Evaluation of Molecular Ecological Networks Generated during the Process of Dryland-to-Paddy Conversion
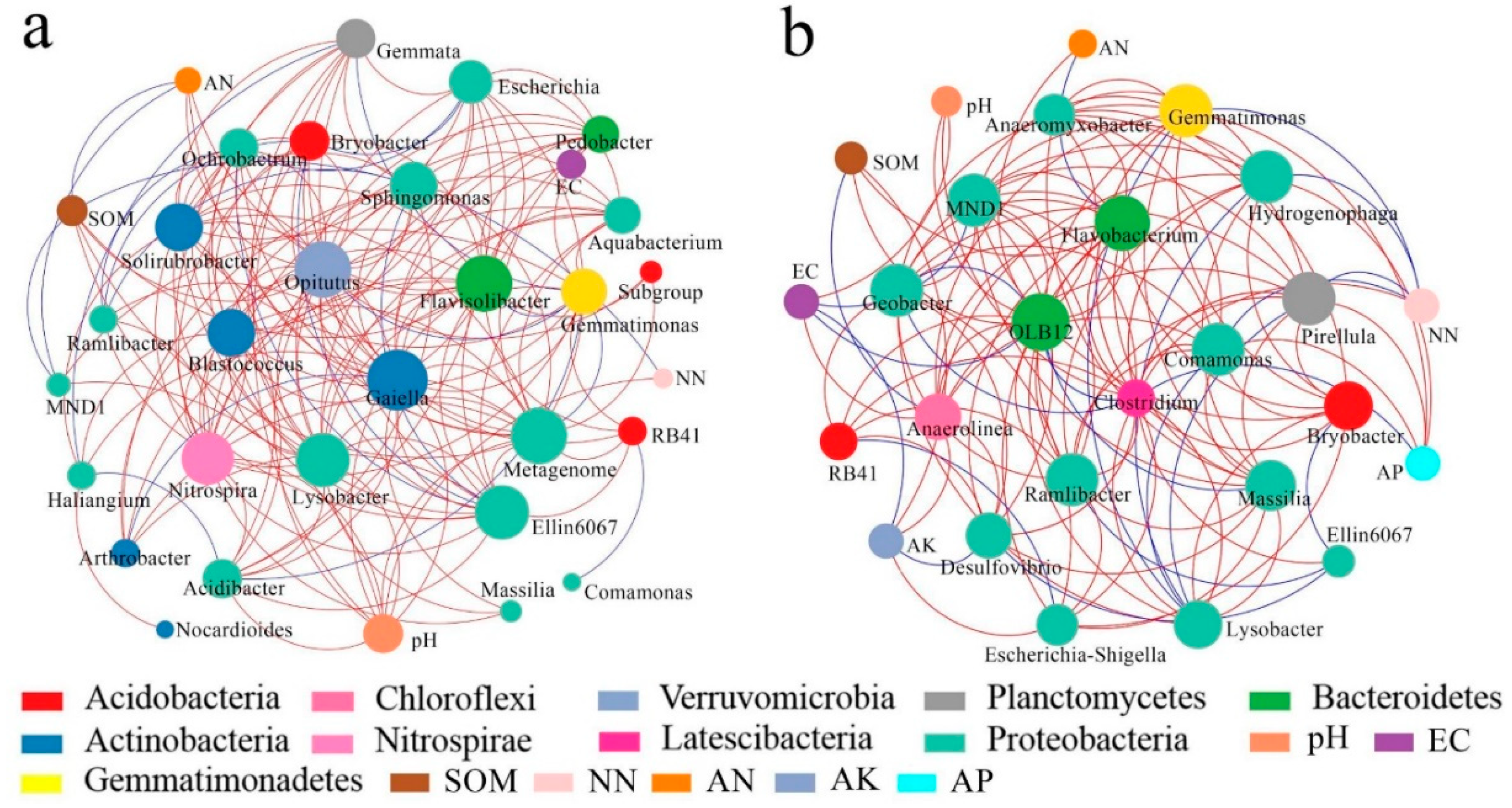
3.3. Mechanism of Interactions Between Soil Microbial Community and Soil Environmental Factors
3.3.1. Relationships between OTU Levels of Soil Microorganisms and Environmental Factors
3.3.2. Interactions between Soil Microbial Species and Environmental Factor Network
4. Discussion
4.1. Changes in Microbial Community Structure After Dryland-to-Paddy Conversion
4.2. Changes in the Structure of Microbial Molecular Ecological Networks After Dryland-to-Paddy Conversion
5. Conclusions
- (1)
- The abundance and diversity of soil bacteria and fungi decreased in a short time after the conversion process; the most dominant soil bacterial species in dry land and paddy field were Proteobacteria, and the most dominant fungal species in dry land and paddy field were Ascomycota and Basidiomycota, respectively. The abundance values of Actinobacteria, Firmicutes, and Olpidiomycota were greatly changed after the conversion, and these species can serve as indicator species for the evolution of soil microbial community diversity in the conversion process.
- (2)
- The dryland-to-paddy conversion had a great influence on the structure of microbial molecular ecological networks. Compared to dry land, the microbial interaction network modular structure and interspecific relationships of paddy field were simpler. The number of nodes, number of connecting lines, average connectivity and clustering coefficient of the soil microbial network of paddy field were lower. When the environment changed, the disturbance of environmental factors would affect the whole ecological network after a period of time, and the instability of network structure would increase. In the future, the dynamic variations of microbial network in conversion processes from dry land to paddy field should be continually investigated.
- (3)
- The soil microbial community structure was significantly related to environmental factors. The changes in pH, EC, OM and AK caused by the conversion were the main reasons for the change of soil microbial community structure (p < 0.05). The soil microbial community structure was closely related to the dominant microorganism species, and the soil environmental factors were the critical factors limiting the adaptive development of microorganisms.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crowther, T.W.; Todd-Brown, K.E.; Rowe, C.W.; Wieder, W.R.; Carey, J.C.; Machmuller, M.B.; Snoek, B.; Fang, S.; Zhou, G.; Allison, S.D. Quantifying global soil carbon losses in response to warming. Nature 2016, 540, 104. [Google Scholar] [CrossRef] [PubMed]
- Lyu, M.; Xie, J.; Ukonmaanaho, L.; Jiang, M.; Li, Y.; Chen, Y.; Yang, Z.; Zhou, Y.; Lin, W.; Yang, Y. Land use change exerts a strong impact on deep soil C stabilization in subtropical forests. J. Soils Sediments 2017, 17, 2305–2317. [Google Scholar] [CrossRef]
- Wang, S.; Liu, J.; Yu, G.; Pan, Y.; Chen, Q.; Li, K.; Li, J. Effects of land use change on the storage of soil organic carbon: A case study of the Qianyanzhou Forest Experimental Station in China. Clim. Chang. 2004, 67, 247–255. [Google Scholar] [CrossRef]
- Kueppers, L.M.; Snyder, M.A.; Sloan, L.C. Irrigation cooling effect: Regional climate forcing by land-use change. Geophys. Res. Lett. 2007, 34. [Google Scholar] [CrossRef] [Green Version]
- Houghton, R.; Hackler, J. Sources and sinks of carbon from land-use change in China. Glob. Biogeochem. Cycles 2003, 17. [Google Scholar] [CrossRef]
- Akala, V.; Lal, R. Potential of mine land reclamation for soil organic carbon sequestration in Ohio. Land Degrad. Dev. 2015, 11, 289–297. [Google Scholar] [CrossRef]
- Arneth, A.; Sitch, S.; Pongratz, J.; Stocker, B.D.; Ciais, P.; Poulter, B.; Bayer, A.D.; Bondeau, A.; Calle, L.; Chini, L.P. Historical carbon dioxide emissions caused by land-use changes are possibly larger than assumed. Nat. Geosci. 2017, 10, 79–84. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, E.; Baldock, J.; Xing, H. Potential soil organic carbon stock and its uncertainty under various cropping systems in Australian cropland. Soil Res. 2014, 52, 463–475. [Google Scholar] [CrossRef]
- Wu, H.; Guo, Z.; Gao, Q.; Peng, C. Distribution of soil inorganic carbon storage and its changes due to agricultural land use activity in China. Agric. Ecosyst. Environ. 2009, 129, 413–421. [Google Scholar] [CrossRef]
- Lai, L.; Huang, X.; Yang, H.; Chuai, X.; Zhang, M.; Zhong, T.; Chen, Z.; Chen, Y.; Wang, X.; Thompson, J.R. Carbon emissions from land-use change and management in China between 1990 and 2010. Sci. Adv. 2016, 2, e1601063. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Xiao, X.; Zhang, G.; Menarguez, M.; Choi, C.; Qin, Y.; Luo, P.; Zhang, Y.; Moore, B. Northward expansion of paddy rice in northeastern Asia during 2000–2014. Geophys. Res. Lett. 2016, 43, 3754–3761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Xia, F.; Bao, H.X. Strategic planning framework for land consolidation in China: A top-level design based on SWOT analysis. Habitat Int. 2015, 48, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Shao, Y.; Zhang, Z.; Resler, L.M.; Campbell, J.B.; Chen, G.; Zhou, Y. The evaluation of land consolidation policy in improving agricultural productivity in China. Sci. Rep. 2017, 7, 2792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolis, K.; Hiironen, J.; Riekkinen, K.; Vitikainen, A. Forest land consolidation and its effect on climate. Land Use Policy 2017, 61, 536–542. [Google Scholar] [CrossRef]
- Kang, S.; Eltahir, E.A. North China Plain threatened by deadly heatwaves due to climate change and irrigation. Nat. Commun. 2018, 9, 2894. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Eldridge, D.J.; Hamonts, K.; Reich, P.B.; Singh, B.K. Experimentally testing the species-habitat size relationship on soil bacteria: A proof of concept. Soil Biol. Biochem. 2018, 123, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, S.; Li, R.; Zhang, J.; Liu, Y.; Lv, L.; Zhu, H.; Wu, W.; Li, W. Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 2017, 109, 145–155. [Google Scholar] [CrossRef]
- He, X.; Su, Y.; Liang, Y.; Chen, X.; Zhu, H.; Wang, K. Land reclamation and short-term cultivation change soil microbial communities and bacterial metabolic profiles. J. Sci. Food Agric. 2012, 92, 1103–1111. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, Y.; Li, C.; Wang, F.; Sui, Y.; Suvannang, N.; Zhou, J.; Sun, B. Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol. Biochem. 2016, 95, 250–261. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Tang, Y.; Shen, J.; Wang, C.; Chen, C.; Yang, J.; Liu, Y.; Chen, X.; Li, Y.; Hou, H. Abundance of transcripts of functional gene reflects the inverse relationship between CH4 and N2O emissions during mid-season drainage in acidic paddy soil. Biol. Fertil. Soils 2018, 54, 885–895. [Google Scholar] [CrossRef]
- Kreye, C.; Dittert, K.; Zheng, X.; Zhang, X.; Lin, S.; Tao, H.; Sattelmacher, B. Fluxes of methane and nitrous oxide in water-saving rice production in north China. Nutr. Cycl. Agroecosyst. 2007, 77, 293–304. [Google Scholar] [CrossRef]
- Suleiman, A.K.A.; Manoeli, L.; Boldo, J.T.; Pereira, M.G.; Roesch, L.F.W. Shifts in soil bacterial community after eight years of land-use change. Syst. Appl. Microbiol. 2013, 36, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, J.; Yang, Y.; Hou, H.; Liu, G.-J.; Chen, F. Short-Term Response of Soil Microbial Community to Field Conversion from Dryland to Paddy under the Land Consolidation Process in North China. Agriculture 2019, 9, 216. [Google Scholar] [CrossRef] [Green Version]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. Fems Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef] [Green Version]
- Zechmeister-Boltenstern, S.; Keiblinger, K.M.; Mooshammer, M.; Peñuelas, J.; Richter, A.; Sardans, J.; Wanek, W. The application of ecological stoichiometry to plant–microbial–soil organic matter transformations. Ecol. Monogr. 2015, 85, 133–155. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol. Biochem. 2015, 83, 29–39. [Google Scholar] [CrossRef]
- Hopkins, D.W.; Sparrow, A.D.; Gregorich, E.G.; Novis, P.; Elberling, B.; Greenfield, L.G. Redistributed lacustrine detritus as a spatial subsidy of biological resources for soils in an Antarctic dry valley. Geoderma 2008, 144, 86–92. [Google Scholar] [CrossRef]
- Welbaum, G.E.; Sturz, A.V.; Dong, Z.; Nowak, J. Managing soil microorganisms to improve productivity of agro-ecosystems. Crit. Rev. Plant Sci. 2004, 23, 175–193. [Google Scholar] [CrossRef]
- Luo, F.; Yang, Y.; Zhong, J.; Gao, H.; Khan, L.; Thompson, D.K.; Zhou, J. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinform. 2007, 8, 299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, F.; Zhong, J.; Yang, Y.; Scheuermann, R.H.; Zhou, J. Application of random matrix theory to biological networks. Phys. Lett. A 2006, 357, 420–423. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Xue, K.; Xie, J.; Deng, Y.; Wu, L.; Cheng, X.; Fei, S.; Deng, S.; He, Z.; Van Nostrand, J.D. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Chang. 2012, 2, 106. [Google Scholar] [CrossRef]
- Luo, Z.; Ma, J.; Chen, F.; Li, X.; Hou, H.; Zhang, S. Cracks Reinforce the Interactions among Soil Bacterial Communities in the Coal Mining Area of Loess Plateau, China. Int. J. Environ. Res. Public Health 2019, 16, 4892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Zhao, H.; Deng, Y.; Zhou, J.; Li, G.; Sun, B. Long-term oil contamination alters the molecular ecological networks of soil microbial functional genes. Front. Microbiol. 2016, 7, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X. Functional molecular ecological networks. MBio 2010, 1, e00169-10. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Rodriguez-R, L.M.; Johnston, E.R.; Wu, L.; Cheng, L.; Xue, K.; Tu, Q.; Deng, Y.; He, Z.; Shi, J.Z. Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl. Environ. Microbiol. 2014, 80, 1777–1786. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Zhang, P.; Qin, Y.; Tu, Q.; Yang, Y.; He, Z.; Schadt, C.W.; Zhou, J. Network succession reveals the importance of competition in response to emulsified vegetable oil amendment for uranium bioremediation. Environ. Microbiol. 2016, 18, 205–218. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Yang, Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2011, 2, 00122-11. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Yin, S.; Liu, X.; Zhang, W.; Gu, T.; Shen, Q.; Qiu, H. Fungal networks in yield-invigorating and-debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Sang, S.; Liu, S.; Jia, J. Spatial distribution of pH and organic matter in urban soils and its implications on site-specific land uses in Xuzhou, China. C. R. Biol. 2014, 337, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhang, W.; Ma, J.; Yang, Y.; Zhang, S.; Chen, R. Experimental study on the effects of underground CO2 leakage on soil microbial consortia. Int. J. Greenh. Gas Control 2017, 63, 241–248. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Hou, L.; Zhang, Z.; Zhang, J.; Cheng, J.; Wei, G.; Lin, Y. Soil microbial diversity during 30 years of grassland restoration on the Loess Plateau, China: Tight linkages with plant diversity. Land Degrad. Dev. 2019, 30, 1172–1182. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lal, R. Soil carbon sequestration impacts on global climate change and food security. Science 2004, 304, 1623–1627. [Google Scholar] [CrossRef] [Green Version]
- Brye, K.; Slaton, N.; Mozaffari, M.; Savin, M.C.; Norman, R.; Miller, D. Short-term effects of land leveling on soil chemical properties and their relationships with microbial biomass. Soil Sci. Soc. Am. J. 2004, 68, 924–934. [Google Scholar] [CrossRef]
- Van Dijk, T. Complications for traditional land consolidation in Central Europe. Geoforum 2007, 38, 505–511. [Google Scholar] [CrossRef]
- Wang, J.; Yan, S.; Guo, Y.; Li, J.; Sun, G. The effects of land consolidation on the ecological connectivity based on ecosystem service value: A case study of Da’an land consolidation project in Jilin province. J. Geogr. Sci. 2015, 25, 603–616. [Google Scholar] [CrossRef]
- Yu, G.; Feng, J.; Che, Y.; Lin, X.; Hu, L.; Yang, S. The identification and assessment of ecological risks for land consolidation based on the anticipation of ecosystem stabilization: A case study in Hubei Province, China. Land Use Policy 2010, 27, 293–303. [Google Scholar] [CrossRef]
- Parfitt, J.M.B.; Timm, L.C.; Reichardt, K.; Pinto, L.F.S.; Pauletto, E.A.; Castilhos, D.D. Chemical and biological attributes of a lowland soil affected by land leveling. Pesqui. Agropecuária Bras. 2013, 48, 1489–1497. [Google Scholar] [CrossRef] [Green Version]
- Brye, K.R.; Slaton, N.A.; Norman, R.J. Soil physical and biological properties as affected by land leveling in a clayey aquert. Soil Sci. Soc. Am. J. 2006, 70, 631–642. [Google Scholar] [CrossRef]
- Sharifi, A.; Gorji, M.; Asadi, H.; Pourbabaee, A.A. Land leveling and changes in soil properties in paddy fields of Guilan Province, Iran. Paddy Water Environ. 2014, 12, 139–145. [Google Scholar] [CrossRef]
- Crecente, R.; Alvarez, C.; Fra, U. Economic, social and environmental impact of land consolidation in Galicia. Land Use Policy 2002, 19, 135–147. [Google Scholar] [CrossRef]
- Zhang, B.; Yao, S.H.; Hu, F. Microbial biomass dynamics and soil wettability as affected by the intensity and frequency of wetting and drying during straw decomposition. Eur. J. Soil Sci. 2007, 58, 1482–1492. [Google Scholar] [CrossRef]
- Edwards, J.; Santos-Medellín, C.; Nguyen, B.; Kilmer, J.; Liechty, Z.; Veliz, E.; Ni, J.; Phillips, G.; Sundaresan, V. Soil domestication by rice cultivation results in plant-soil feedback through shifts in soil microbiota. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Drenovsky, R.; Vo, D.; Graham, K.; Scow, K. Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microb. Ecol. 2004, 48, 424–430. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Roth, P.J.; Lehndorff, E.; Cao, Z.H.; Zhuang, S.; Bannert, A.; Wissing, L.; Schloter, M.; Kögel-Knabner, I.; Amelung, W. Accumulation of nitrogen and microbial residues during 2000 years of rice paddy and non-paddy soil development in the Yangtze River Delta, C hina. Glob. Chang. Biol. 2011, 17, 3405–3417. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, Y.; Su, Y.; Chen, B.; Fu, B.; Marschner, P. Effects of soil moisture and plant interactions on the soil microbial community structure. Eur. J. Soil Biol. 2007, 43, 31–38. [Google Scholar] [CrossRef]
- Brockett, B.F.; Prescott, C.E.; Grayston, S.J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 2012, 44, 9–20. [Google Scholar] [CrossRef]
- Legrand, F.; Picot, A.; Cobo-Díaz, J.F.; Carof, M.; Chen, W.; Le Floch, G. Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 2018, 132, 135–145. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Sanderlin, J.S.; Reeves, J.H.; Jenkins, M.B.; Endale, D.M.; Coleman, D.C.; Whitman, W.B. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol. Biochem. 2008, 40, 2843–2853. [Google Scholar] [CrossRef]
- Kragelund, C.; Caterina, L.; Borger, A.; Thelen, K.; Eikelboom, D.; Tandoi, V.; Kong, Y.; Van Der Waarde, J.; Krooneman, J.; Rossetti, S. Identity, abundance and ecophysiology of filamentous Chloroflexi species present in activated sludge treatment plants. FEMS Eicrobiol. Ecol. 2007, 59, 671–682. [Google Scholar] [CrossRef] [Green Version]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS ONE 2013, 8, e66146. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Yang, B.; Wang, H.; Yan, Q.; Cao, Y.; He, X. Changes in composition and diversity of fungal communities along Quercus mongolica forests developments in Northeast China. Appl. Soil Ecol. 2016, 100, 162–171. [Google Scholar] [CrossRef]
- Sterkenburg, E.; Bahr, A.; Durling, M.B.; Clemmensen, K.E.; Lindahl, B.D. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol. 2015, 207, 1145–1158. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Liu, J.; Wei, D.; Zhu, P.; Cui, X.A.; Zhou, B.; Chen, X.; Jin, J.; Liu, X.; Wang, G. Effects of over 30-year of different fertilization regimes on fungal community compositions in the black soils of northeast China. Agric. Ecosyst. Environ. 2017, 248, 113–122. [Google Scholar] [CrossRef]
- Yan, X.; Zhou, H.; Zhu, Q.; Wang, X.; Zhang, Y.; Yu, X.; Peng, X. Carbon sequestration efficiency in paddy soil and upland soil under long-term fertilization in southern China. Soil Tillage Res. 2013, 130, 42–51. [Google Scholar] [CrossRef]
- Unger, I.M.; Goyne, K.W.; Kremer, R.J.; Kennedy, A.C. Microbial community diversity in agroforestry and grass vegetative filter strips. Agrofor. Syst. 2013, 87, 395–402. [Google Scholar] [CrossRef]
- Wang, M.; Chen, S.; Zheng, H.; Li, S.; Chen, L.; Wang, D. The responses of cadmium phytotoxicity in rice and the microbial community in contaminated paddy soils for the application of different long-term N fertilizers. Chemosphere 2020, 238, 124700. [Google Scholar] [CrossRef] [PubMed]
- Hannula, S.E.; De Boer, W.; Van Veen, J. A 3-year study reveals that plant growth stage, season and field site affect soil fungal communities while cultivar and GM-trait have minor effects. PLoS ONE 2012, 7, e33819. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef] [Green Version]






| Parameters | Bacteria | Fungi | ||
|---|---|---|---|---|
| DL | PF | DL | PF | |
| Similarity threshold | 0.88 | 0.88 | 0.86 | 0.86 |
| Nodes | 261 | 160 | 126 | 70 |
| Links | 637 | 223 | 186 | 83 |
| Average degree (AvgK) | 4.881 | 2.788 | 2.952 | 2.371 |
| Average clustering coefficient (AvgCC) | 0.291 | 0.309 | 0.145 | 0.135 |
| Average path distance (AvgPD) | 6.36 | 5.699 | 4.362 | 4.006 |
| Density | 0.019 | 0.018 | 0.024 | 0.034 |
| Connectivity | 0.692 | 0.275 | 0.724 | 0.494 |
| Number of modules | 29 | 28 | 13 | 14 |
| Modular index | 0.694 | 0.822 | 0.659 | 0.678 |
| R2 | 0.814 | 0.848 | 0.856 | 0.859 |
| Soil Properties | Bacteria | Fungi | ||
|---|---|---|---|---|
| DL | PF | DL | PF | |
| pH | 0.438 * | 0.439 | 0.262 * | 0.189 |
| EC (mS·cm−3) | 0.350 * | 0.597 | 0.260 * | 0.234 |
| OM (g·kg−1) | 0.287 * | 0.330 * | −0.168 | −0.098 |
| NN (mg·kg−1) | 0.254 | 0.129 | −0.059 | −0.474 |
| AN (mg·kg−1) | 0.308 | 0.097 | 0.028 | 0.156 |
| AP (mg·kg−1) | 0.138 | −0.054 | −0.117 | 0.045 |
| AK (mg·kg−1) | −0.196 | 0.157 | −0.083 | 0.397 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhang, Q.; Ma, J.; Yang, Y.; Wang, Y.; Fu, C. Flooding Irrigation Weakens the Molecular Ecological Network Complexity of Soil Microbes during the Process of Dryland-to-Paddy Conversion. Int. J. Environ. Res. Public Health 2020, 17, 561. https://doi.org/10.3390/ijerph17020561
Li X, Zhang Q, Ma J, Yang Y, Wang Y, Fu C. Flooding Irrigation Weakens the Molecular Ecological Network Complexity of Soil Microbes during the Process of Dryland-to-Paddy Conversion. International Journal of Environmental Research and Public Health. 2020; 17(2):561. https://doi.org/10.3390/ijerph17020561
Chicago/Turabian StyleLi, Xiaoxiao, Qi Zhang, Jing Ma, Yongjun Yang, Yifei Wang, and Chen Fu. 2020. "Flooding Irrigation Weakens the Molecular Ecological Network Complexity of Soil Microbes during the Process of Dryland-to-Paddy Conversion" International Journal of Environmental Research and Public Health 17, no. 2: 561. https://doi.org/10.3390/ijerph17020561






