Antibiotic Resistance in Recreational Waters: State of the Science
Abstract
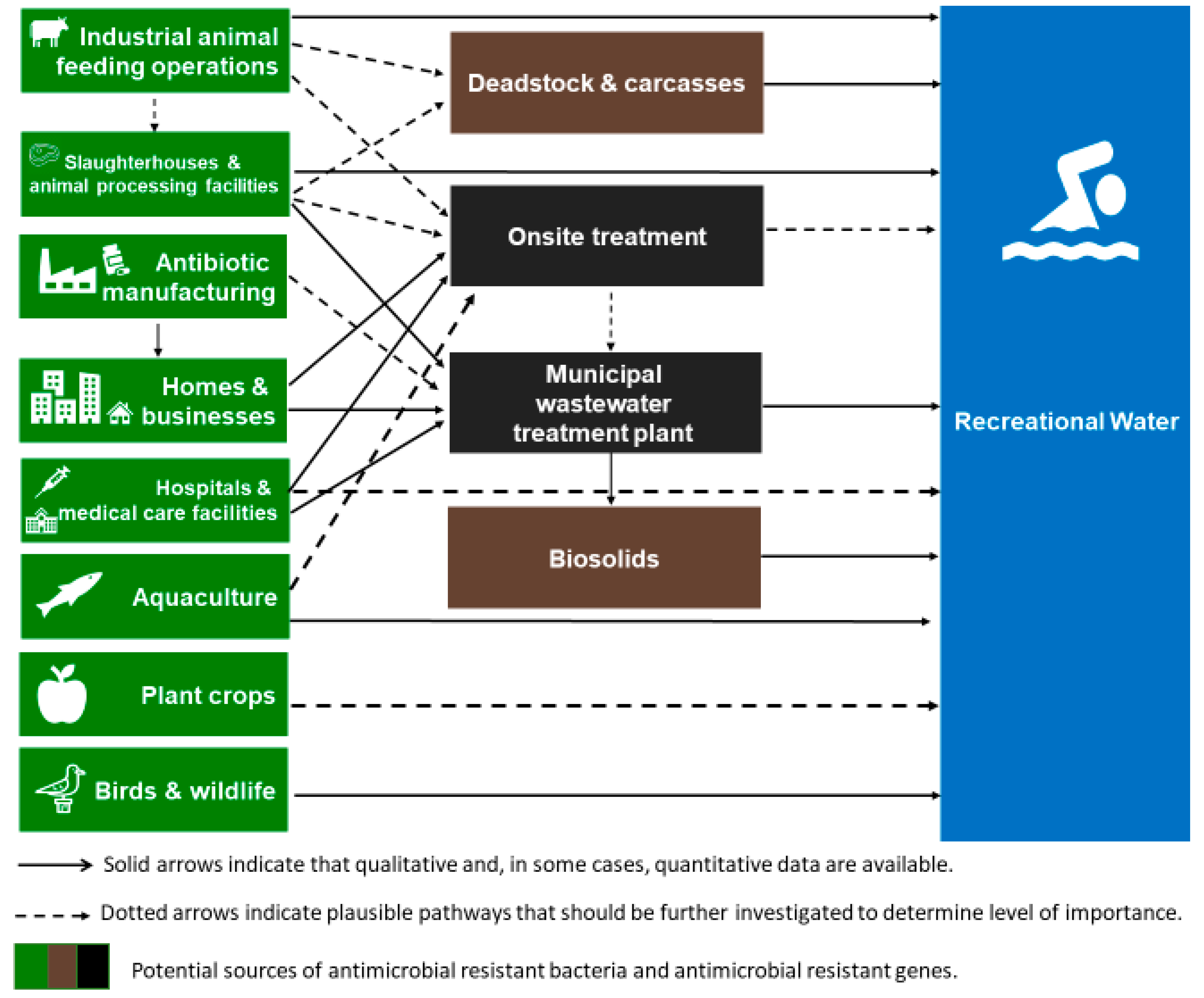
:1. Introduction
2. Mechanisms of Gene Persistence and Transfer in Surface Waters
3. Anthropogenic Sources of Antibiotic Resistance in Recreational Waters
3.1. Untreated Wastewater
Medical Waste
3.2. Treated Wastewater
Medical Waste
3.3. Biosolids
3.4. Agriculture and Aquaculture
3.5. Birds and Other Wildlife
4. Health Assessments
4.1. Health Endpoints and Pathogens of Concern
4.2. Epidemiological Studies
4.3. Exposure Estimates and Risk Assessments
5. Future Research Needs
5.1. Source Contribution
5.2. Fate and Transport—Across Treatment and in the Environment
5.3. Human Health Risks
5.4. Standardized Methods
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States; CDC: Atlanta, GA, USA, 2019.
- The Council of Canadian Academies. When Antibiotics Fail: The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada; The Council of Canadian Academies: Ottawa, ON, USA, 2019. [Google Scholar]
- World Health Organization (WHO). Ten Threats to Global Health in 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Antimicrobial Resistance; WHO: Geneva, Switzerland, 2019; Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 18 May 2020).
- Food and Agricultural Organization (FAO); World Health Organization (WHO). Joint FAO/WHO Expert Meeting in Collaboration with OIE on Foodborne Antimicrobial Resistance: Role of the Environment, Crops and Biocides; FAO and WHO: Rome, Italy, 2019. [Google Scholar]
- World Health Organization (WHO). Technical Brief on Water, Sanitation, Hygiene and Wastewater Management to Prevent Infections and Reduce the Spread of Antimicrobial Resistance; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Martinez, J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Williams-Nguyen, J.; Sallach, J.B.; Bartelt-Hunt, S.; Boxall, A.B.; Durso, L.M.; McLain, J.E.; Singer, R.S.; Snow, D.D.; Zilles, J.L. Antibiotics and antibiotic resistance in agroecosystems: State of the science. J. Environ. Qual. 2016, 45, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Quintela-Baluja, M.; Chan, C.W.; Ezzat Alnakip, M.; Abouelnaga, M.; Graham, D.W. Sanitation, water quality and antibiotic resistance dissemination. In The Battle against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs; Formatex Research Center: Badajoz, Spain, 2015; pp. 965–975. [Google Scholar]
- Pepper, I.L.; Brooks, J.P.; Gerba, C.P. Antibiotic resistant bacteria in municipal wastes: Is there reason for concern? Environ. Sci. Technol. 2018, 52, 3949–3959. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.F.; Weisberg, S.B.; Arnold, B.F.; Cao, Y.; Schiff, K.C.; Colford, J.M., Jr. Epidemiologic evaluation of multiple alternate microbial water quality monitoring indicators at three California beaches. Water Res. 2016, 94, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Thapaliya, D.; Hellwig, E.J.; Kadariya, J.; Grenier, D.; Jefferson, A.J.; Dalman, M.; Kennedy, K.; DiPerna, M.; Orihill, A.; Taha, M.; et al. Prevalence and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus on public recreational beaches in Northeast Ohio. Geohealth 2017, 1, 320–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, A.F.C.; Yin, X.L.; Zhang, T.; Hui, M.; Gaze, W.H. A coliform-targeted metagenomic method facilitating human exposure estimates to Escherichia coli-borne antibiotic resistance genes. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Hawkey, P.M.; Murray, A.K.; Ukoumunne, O.C.; Gaze, W.H. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: Environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ. Int. 2018, 114, 326–333. [Google Scholar] [CrossRef]
- Plano, L.R.; Shibata, T.; Garza, A.C.; Kish, J.; Fleisher, J.M.; Sinigalliano, C.D.; Gidley, M.L.; Withum, K.; Elmir, S.M.; Hower, S.; et al. Human-associated methicillin-resistant Staphylococcus aureus from a subtropical recreational marine beach. Microb. Ecol. 2013, 65, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.P.; Frank, K.M. Salmonella, Shigella, and Yersinia. Clin. Lab. Med. 2015, 35, 225–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogarty, L.R.; Haack, S.K.; Johnson, H.E.; Brennan, A.K.; Isaacs, N.M.; Spencer, C. Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) at ambient freshwater beaches. J. Water Health 2015, 13, 680–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokari, T.G.; Ibiebele, D.D.; Ottih, R.M. Antibiotic resistance among coliforms and Pseudomonas spp. from bodies of water around Port Harcourt, Nigeria. J. Appl. Bacteriol. 1988, 64, 355–359. [Google Scholar] [CrossRef]
- Ghosh, A.R.; Nair, G.B.; Naik, T.N.; Sarkar, S.K.; Mazumdar, R.; Pal, S.C.; Sen, D. Serovars of multi-antibiotic resistant Escherichia coli from the freshwater environs of Calcutta, India. Microbiol. Immunol. 1991, 35, 273–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Bürgmann, H. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [Green Version]
- Santiago-Rodriguez, T.M.; Rivera, J.I.; Coradin, M.; Toranzos, G.A. Antibiotic-resistance and virulence genes in Enterococcus isolated from tropical recreational waters. J. Water Health 2013, 11, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Colomer-Lluch, M.; Mora, A.; Lopez, C.; Mamani, R.; Dahbi, G.; Marzoa, J.; Herrera, A.; Viso, S.; Blanco, J.E.; Blanco, M.; et al. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J. Antimicrob. Chemother. 2013, 68, 758–765. [Google Scholar] [CrossRef] [Green Version]
- Alm, E.W.; Zimbler, D.; Callahan, E.; Plomaritis, E. Patterns and persistence of antibiotic resistance in faecal indicator bacteria from freshwater recreational beaches. J. Appl. Microbiol. 2014, 117, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.F.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Overbey, K.N.; Hatcher, S.M.; Stewart, J.R. Water quality and antibiotic resistance at beaches of the Galápagos Islands. Front. Environ. Sci. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Schijven, J.F.; Blaak, H.; Schets, F.M.; de Roda Husman, A.M. Fate of extended-spectrum beta-cactamase-producing Escherichia coli from faecal sources in surface water and probability of human exposure through swimming. Environ. Sci. Technol. 2015, 49, 11825–11833. [Google Scholar] [CrossRef]
- Kadykalo, S.; Thomas, J.; Parmley, E.J.; Pintar, K.; Fleury, M. Antimicrobial resistance of Salmonella and generic Escherichia coli isolated from surface water samples used for recreation and a source of drinking water in southwestern Ontario, Canada. Zoonoses Public Health 2020, 67, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2019, 50, 2016–2059. [Google Scholar] [CrossRef]
- Boczek, L.A.; Rice, E.W.; Johnston, B.; Johnson, J.R. Occurrence of antibiotic-resistant uropathogenic Escherichia coli clonal group A in wastewater effluents. Appl. Environ. Microbiol. 2007, 73, 4180–4184. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.; Cohen, T.; Grad, Y.H.; Hanage, W.P.; O’Brien, T.F.; Lipsitch, M. Origin and proliferation of multiple-drug resistance in bacterial pathogens. Microbiol. Mol. Biol. Rev. 2015, 79, 101–116. [Google Scholar] [CrossRef] [Green Version]
- Bueno, I.; Williams-Nguyen, J.; Hwang, H.; Sargeant, J.M.; Nault, A.J.; Singer, R.S. Systematic review: Impact of point sources on antibiotic-resistant bacteria in the natural environment. Zoonoses Public Health 2018, 65, e162–e184. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency (USEPA). Recreational Water Quality Criteria; EPA 820-F-12-058; United States Environmental Protection Agency, Office of Water: Washington, DC, USA, 2012.
- Health Canada. Guidelines for Canadian Recreational Water Quality, 3rd ed.; Health Canada: Ottawa, ON, USA, 2012; ISBN 978-1-100-20892-3. [Google Scholar]
- European Environment Agency (EEA). Bathing Water Quality; EEA: Copenhagen, Denmark, 2005. [Google Scholar]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Gantzer, C.; Maul, A.; Audic, J.M.; Schwartzbrod, L. Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl. Environ. Microbiol. 1998, 64, 4307–4312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debroas, D.; Siguret, C. Viruses as key reservoirs of antibiotic resistance genes in the environment. ISME J. 2019, 13, 2856–2867. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J. Antibiot. 2010, 63, 506–511. [Google Scholar] [CrossRef]
- Adu-Oppong, B.; Gasparrini, A.J.; Dantas, G. Genomic and functional techniques to mine the microbiome for novel antimicrobials and antimicrobial resistance genes. Ann. N. Y. Acad. Sci. 2017, 1388, 42–58. [Google Scholar] [CrossRef] [Green Version]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Cytryn, E. The soil resistome: The Anthropogenic, the nature and the unknown. Soil Biol. Biochem. 2013, 63, 18–23. [Google Scholar] [CrossRef]
- Diaz, K.S.; Rich, V.I.; McLain, J.E. Searching for antibiotic resistance genes in a pristine Arctic wetland. J. Contemp. Water Res. Educ. 2017, 160, 42–59. [Google Scholar] [CrossRef] [Green Version]
- Durso, L.M.; Wedin, D.A.; Gilley, J.E.; Miller, D.N.; Marx, D.B. Assessment of selected antibiotic resistances in ungrazed native Nebraska prairie soils. J. Environ. Qual. 2016, 45, 454–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.H.; Nonaka, L.; Tago, R.; Suzuki, S. Occurrence of two genotypes of tetracycline (TC) resistance gene tet(M) in the TC-resistant bacteria in marine sediments of Japan. Environ. Sci. Technol. 2008, 42, 5055–5061. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.L.S.; Ferreira, G.F.; de Freitas, G.J.C.; Ferreira, R.L.; Assiss, D.; Santos, D.; de Resende-Stoianoff, M.A. Screening of antifungal susceptibility in cave dwelling aspergilli and report of an amphotericin B-resistant Aspergillus flavus. Int. J. Speleol. 2017, 46, 369–378. [Google Scholar] [CrossRef] [Green Version]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- Kummerer, K. Antibiotics in the aquatic environment—A review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sandiford, S.K.; van Wezel, G.P. Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol. 2014, 41, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.R. Horizontal gene transfer. Evol. Med. Public Health 2015, 2015, 193–194. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Woegerbauer, M.; Bellanger, X.; Merlin, C. Cell-Free DNA: An underestimated source of antibiotic resistance gene dissemination at the interface between human activities and downstream environments in the context of wastewater reuse. Front. Microbiol. 2020, 11, 671. [Google Scholar] [CrossRef] [Green Version]
- Smalla, K.; Cook, K.; Djordjevic, S.P.; Klumper, U.; Gillings, M. Environmental dimensions of antibiotic resistance: Assessment of basic science gaps. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [Green Version]
- Joseph, S.M.; Battaglia, T.; Maritz, J.M.; Carlton, J.M.; Blaser, M.J. Longitudinal comparison of bacterial diversity and antibiotic resistance genes in New York City sewage. MSystems 2019, 4, e00327-19. [Google Scholar] [CrossRef] [Green Version]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef] [Green Version]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Baquero, F.; Coque, T.M. Widening the spaces of selection: Evolution along sublethal antimicrobial gradients. MBio 2014, 5, e02270. [Google Scholar] [CrossRef] [Green Version]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.E.; Agouri, S.R.; Tyrrell, J.M.; Walsh, T.R. Heavy metal resistance genes are associated with blaNDM-1- and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, A.W.; Power, A.; Hansen, M.G.; Brandt, K.K.; Piliposian, G.; Appleby, P.; O’Neill, P.A.; Jones, R.T.; Sierocinski, P.; Koskella, B.; et al. Heavy metal pollution and co-selection for antibiotic resistance: A microbial palaeontology approach. Environ. Int. 2019, 132, 105117. [Google Scholar] [CrossRef] [PubMed]
- Hiller, C.X.; Hubner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Almakki, A.; Jumas-Bilak, E.; Marchandin, H.; Licznar-Fajardo, P. Antibiotic resistance in urban runoff. Sci. Total Environ. 2019, 667, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Gordon, L.; Giraud, E.; Ganiere, J.P.; Armand, F.; Bouju-Albert, A.; de la Cotte, N.; Mangion, C.; Le Bris, H. Antimicrobial resistance survey in a river receiving effluents from freshwater fish farms. J. Appl. Microbiol. 2007, 102, 1167–1176. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Burgmann, H.; Sorum, H.; Norstrom, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Nzimande, S.B.; Mkize, N.G. Antimicrobial resistance and virulence signatures of Listeria and Aeromonas species recovered from treated wastewater effluent and receiving surface water in Durban, South Africa. BMC Microbiol. 2015, 15, 234. [Google Scholar] [CrossRef]
- Chaix, G.; Roger, F.; Berthe, T.; Lamy, B.; Jumas-Bilak, E.; Lafite, R.; Forget-Leray, J.; Petit, F. Distinct aeromonas populations in water column and associated with copepods from estuarine environment (Seine, France). Front. Microbiol. 2017, 8, 1259. [Google Scholar] [CrossRef] [Green Version]
- Ferreira da Silva, M.; Vaz-Moreira, I.; Gonzalez-Pajuelo, M.; Nunes, O.C.; Manaia, C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007, 60, 166–176. [Google Scholar] [CrossRef]
- Galvin, S.; Boyle, F.; Hickey, P.; Vellinga, A.; Morris, D.; Cormican, M. Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl. Environ. Microbiol. 2010, 76, 4772–4779. [Google Scholar] [CrossRef] [Green Version]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Książek, S.; Olańczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Soraas, A.; Sundsfjord, A.; Sandven, I.; Brunborg, C.; Jenum, P.A. Risk factors for community-acquired urinary tract infections caused by ESBL-producing enterobacteriaceae—A case-control study in a low prevalence country. PLoS ONE 2013, 8, e69581. [Google Scholar] [CrossRef] [Green Version]
- Hooban, B.; Joyce, A.; Fitzhenry, K.; Chique, C.; Morris, D. The role of the natural aquatic environment in the dissemination of extended spectrum beta-lactamase and carbapenemase encoding genes: A scoping review. Water Res. 2020, 180, 115880. [Google Scholar] [CrossRef] [PubMed]
- Eftim, S.E.; Hong, T.; Soller, J.; Boehm, A.; Warren, I.; Ichida, A.; Nappier, S.P. Occurrence of norovirus in raw sewage—A systematic literature review and meta-analysis. Water Res. 2017, 111, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Soller, J.A.; Eftim, S.E.; Nappier, S.P. Direct potable reuse microbial risk assessment methodology: Sensitivity analysis and application to state log credit allocations. Water Res. 2018, 128, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Jury, K.L.; Khan, S.J.; Vancov, T.; Stuetz, R.M.; Ashbolt, N.J. Are sewage treatment plants promoting antibiotic resistance? Crit. Rev. Environ. Sci. Technol. 2011, 41, 243–270. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency (USEPA). 40 CFR Part 431 Effluent Limitations Guidelines and New Source Performance Standards for the Meat and Poultry Products Point Source Category; Final Rule; USEPA: Washington, DC, USA, 2004; pp. 54476–54555.
- U.S. Environmental Protection Agency (USEPA). Keeping Raw Sewage & Contaminated Stormwater Out of the Public’s Water; United States Environmental Protection Agency: New York, NY, USA, 2011.
- U.S. Environmental Protection Agency (USEPA). Sanitary Sewer Overflows; USEPA: Washington, DC, USA, 2019. Available online: https://www.epa.gov/npdes/sanitary-sewer-overflows-ssos (accessed on 18 May 2020).
- Pouillot, R.; Van Doren, J.M.; Woods, J.; Plante, D.; Smith, M.; Goblick, G.; Roberts, C.; Locas, A.; Hajen, W.; Stobo, J.; et al. Meta-analysis of the reduction of norovirus and male-specific coliphage concentrations in wastewater treatment plants. Appl. Environ. Microbiol. 2015, 81, 4669–4681. [Google Scholar] [CrossRef] [Green Version]
- Honda, R.; Tachi, C.; Yasuda, K.; Hirata, T.; Noguchi, M.; Hara-Yamamura, H.; Yamamoto-Ikemoto, R.; Watanabe, T. Estimated discharge of antibiotic-resistant bacteria from combined sewer overflows of urban sewage system. NPJ Clean Water 2020, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Hua, J.; An, P.; Winter, J.; Gallert, C. Elimination of COD, microorganisms and pharmaceuticals from sewage by trickling through sandy soil below leaking sewers. Water Res. 2003, 37, 4395–4404. [Google Scholar] [CrossRef]
- Paul, M.; Wolf, L.; Fund, K.; Held, I.; Winter, J.; Eiswirth†, M.; Gallert, C.; Hötzl, H. Microbiological condition of urban groundwater in the vicinity of leaky sewer systems. Acta Hydrochim. Hydrobiol. 2004, 32, 351–360. [Google Scholar] [CrossRef]
- Rosi-Marshall, E.J.; Kelly, J.J. Antibiotic stewardship should consider environmental fate of antibiotics. Environ. Sci. Technol. 2015, 49, 5257–5258. [Google Scholar] [CrossRef] [Green Version]
- Gallert, C.; Fund, K.; Winter, J. Antibiotic resistance of bacteria in raw and biologically treated sewage and in groundwater below leaking sewers. Appl. Microbiol. Biotechnol. 2005, 69, 106–112. [Google Scholar] [CrossRef]
- Sigala, J.; Unc, A. A PCR-DGGE approach to evaluate the impact of wastewater source on the antibiotic resistance diversity in treated wastewater effluent. Water Sci. Technol. 2012, 65, 1323–1331. [Google Scholar] [CrossRef]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divyashree, M.; Mani, M.K.; Shama Prakash, K.; Vijaya Kumar, D.; Veena Shetty, A.; Shetty, A.K.; Karunasagar, I. Hospital wastewater treatment reduces NDM-positive bacteria being discharged into water bodies. Water Environ. Res. 2020, 92, 562–568. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Parnanen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [Green Version]
- U.S. Environmental Protection Agency (USEPA). Five-Year Review of the 2012 Recreational Water Quality Criteria; EPA 823-R-18-001; United States Environmental Protection Agency, Office of Water: Washington, DC, USA, 2018.
- Reinthaler, F.F.; Posch, J.; Feierl, G.; Wust, G.; Haas, D.; Ruckenbauer, G.; Mascher, F.; Marth, E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef]
- Pauwels, B.; Verstraete, W. The treatment of hospital wastewater: An appraisal. J. Water Health 2006, 4, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital wastewater releases of carbapenem-resistance pathogens and genes in urban India. Environ. Sci. Technol. 2017, 51, 13906–13912. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.B. Antibacterial household products: Cause for concern. Emerg. Infect. Dis. 2001, 7, 512–515. [Google Scholar] [CrossRef]
- Aiello, A.E.; Larson, E. Antibacterial cleaning and hygiene products as an emerging risk factor for antibiotic resistance in the community. Lancet Infect. Dis. 2003, 3, 501–506. [Google Scholar] [CrossRef]
- Aiello, A.E.; Marshall, B.; Levy, S.B.; Della-Latta, P.; Lin, S.X.; Larson, E. Antibacterial cleaning products and drug resistance. Emerg. Infect. Dis. 2005, 11, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Hutinel, M.; Huijbers, P.M.C.; Fick, J.; Ahren, C.; Larsson, D.G.J.; Flach, C.F. Population-level surveillance of antibiotic resistance in Escherichia coli through sewage analysis. Eurosurveillance 2019, 24. [Google Scholar] [CrossRef] [Green Version]
- McArdell, C.S.; Molnar, E.; Suter, M.J.F.; Giger, W. Occurrence and fate of macrolide antibiotics in wastewater treatment plants and in the Glatt Valley Watershed, Switzerland. Environ. Sci. Technol. 2003, 37, 5479–5486. [Google Scholar] [CrossRef]
- Voigt, A.M.; Zacharias, N.; Timm, C.; Wasser, F.; Sib, E.; Skutlarek, D.; Parcina, M.; Schmithausen, R.M.; Schwartz, T.; Hembach, N.; et al. Association between antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in anthropogenic wastewater—An evaluation of clinical influences. Chemosphere 2019, 241, 125032. [Google Scholar] [CrossRef]
- Scott, T.-M.; Phillips, P.J.; Kolpin, D.W.; Colella, K.M.; Furlong, E.T.; Foreman, W.T.; Gray, J.L. Pharmaceutical manufacturing facility discharges can substantially increase the pharmaceutical load to U.S. wastewaters. Sci. Total Environ. 2018, 636, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, M.; Hu, J.; Ren, L.; Zhang, Y.; Li, K. Determination and fate of oxytetracycline and related compounds in oxytetracycline production wastewater and the receiving river. Environ. Toxicol. Chem. 2008, 27, 80–86. [Google Scholar] [CrossRef]
- Larsson, D.G.; de Pedro, C.; Paxeus, N. Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J. Hazard. Mater. 2007, 148, 751–755. [Google Scholar] [CrossRef]
- LaPara, T.M.; Burch, T.R.; McNamara, P.J.; Tan, D.T.; Yan, M.; Eichmiller, J.J. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ. Sci. Technol. 2011, 45, 9543–9549. [Google Scholar] [CrossRef] [Green Version]
- Czekalski, N.; Gascón Díez, E.; Bürgmann, H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014, 8, 1381–1390. [Google Scholar] [CrossRef]
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Castillo, G.; Callejas, L.; López, H.; Olmos, J. Frequency of Transferable Multiple Antibiotic Resistance Amongst Coliform Bacteria Isolated from a Treated Sewage Effluent in Antofagasta; Pontificia Universidad Católica de Valparaíso: Valparaíso, Chile, 2006. [Google Scholar]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef]
- Amador, P.P.; Fernandes, R.M.; Prudencio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of class A and class C beta-lactamases. J. Environ. Sci. Health 2015, 50, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Blaak, H.; Lynch, G.; Italiaander, R.; Hamidjaja, R.A.; Schets, F.M.; de Roda Husman, A.M. Multidrug-resistant and extended spectrum Beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLoS ONE 2015, 10, e0127752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Brechet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum beta-lactamase-producing Escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef]
- Kim, S.; Park, H.; Chandran, K. Propensity of activated sludge to amplify or attenuate tetracycline resistance genes and tetracycline resistant bacteria: A mathematical modeling approach. Chemosphere 2010, 78, 1071–1077. [Google Scholar] [CrossRef]
- McKinney, C.W.; Pruden, A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 2012, 46, 13393–13400. [Google Scholar] [CrossRef]
- Pruden, A.; Larsson, D.G.J.; Amézquita, A.; Collignon, P.; Brandt, K.K.; Graham, D.W.; Lazorchak, J.M.; Suzuki, S.; Silley, P.; Snape, J.R.; et al. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 2013, 121, 878–885. [Google Scholar] [CrossRef]
- Hembach, N.; Schmid, F.; Alexander, J.; Hiller, C.; Rogall, E.T.; Schwartz, T. Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Front. Microbiol. 2017, 8, 1282. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Shin, H.; Han, D.; Hur, H.G.; Unno, T. Metagenomic analysis reveals the prevalence and persistence of antibiotic- and heavy metal-resistance genes in wastewater treatment plant. J. Microbiol. 2018, 56, 408–415. [Google Scholar] [CrossRef]
- Marano, R.B.M.; Zolti, A.; Jurkevitch, E.; Cytryn, E. Antibiotic resistance and class 1 integron gene dynamics along effluent, reclaimed wastewater irrigated soil, crop continua: Elucidating potential risks and ecological constraints. Water Res. 2019, 164, 114906. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.B.; Farrah, S.R.; Harwood, V.J.; Levine, A.D.; Lukaskik, J.; Menendez, P.; Scott, T.M. Reduction of Pathogens, Indicator Bacteria, and Alternative Indicators by Wastewater Treatment and Reclamation Processes; Final Report No. 00-PUM-2T; Water Environment Research Foundation (WERF): Alexandria, VA, USA, 2004. [Google Scholar]
- Asano, T.; Burton, F.; Leverenz, H.; Tsuchihashi, R.; Tchobanoglous, G. Water Reuse: Issues, Technologies, and Applications; McGraw-Hill: New York, NY, USA, 2007. [Google Scholar]
- Mitch, A.A.; Gasner, K.C.; Mitch, W.A. Fecal coliform accumulation within a river subject to seasonally-disinfected wastewater discharges. Water Res. 2010, 44, 4776–4782. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Vaz-Moreira, I.; Varela Della Giustina, S.; Llorca, M.; Barceló, D.; Schubert, S.; Berendonk, T.U.; Michael-Kordatou, I.; Fatta-Kassinos, D.; Martinez, J.L.; et al. Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ. Int. 2020, 140, 105733. [Google Scholar] [CrossRef] [PubMed]
- Gogoi, A.; Mazumder, P.; Tyagi, V.K.; Tushara Chaminda, G.G.; An, A.K.; Kumar, M. Occurrence and fate of emerging contaminants in water environment: A review. Groundw. Sustain. Dev. 2018, 6, 169–180. [Google Scholar] [CrossRef]
- Karthikeyan, K.G.; Meyer, M.T. Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total Environ. 2006, 361, 196–207. [Google Scholar] [CrossRef]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Fatta-Kassinos, D.; Cytryn, E.; Donner, E.; Zhang, T. Challenges related to antimicrobial resistance in the framework of urban wastewater reuse. Water Res. 2020, 170, 115308. [Google Scholar] [CrossRef]
- Li, N.; Sheng, G.-P.; Lu, Y.-Z.; Zeng, R.J.; Yu, H.-Q. Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res. 2017, 111, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Y.; Ahmed, Y.; Jin, M.; Li, J. Control strategies to combat dissemination of antibiotic resistance in urban water systems. In Antibiotic Resistance in the Environment: A Worldwide Overview; Manaia, C.M., Donner, E., Vaz-Moreira, I., Hong, P., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 147–187. [Google Scholar]
- Mantilla-Calderon, D.; Plewa, M.J.; Michoud, G.; Fodelianakis, S.; Daffonchio, D.; Hong, P.Y. Water disinfection byproducts increase natural transformation rates of environmental DNA in Acinetobacter baylyi ADP1. Environ. Sci. Technol. 2019, 53, 6520–6528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augsburger, N.; Mantilla-Calderon, D.; Daffonchio, D.; Hong, P.-Y. Acquisition of extracellular DNA by Acinetobacter baylyi ADP1 in response to solar and UV-C254nm disinfection. Environ. Sci. Technol. 2019, 53, 10312–10319. [Google Scholar] [CrossRef] [Green Version]
- Subirats, J.; Di Cesare, A.; Varela Della Giustina, S.; Fiorentino, A.; Eckert, E.M.; Rodriguez-Mozaz, S.; Borrego, C.M.; Corno, G. High-quality treated wastewater causes remarkable changes in natural microbial communities and intI1 gene abundance. Water Res. 2019, 167, 114895. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency (USEPA). Profile of the Healthcare Industry: EPA Office of Compliance Sector Notebook Project; EPA 310-R-05-002; United States Environmental Protection Agency, Office of Compliance: Washington, DC, USA, 2005.
- U.S. Environmental Protection Agency (USEPA). NPDES Permit Writers’ Manual; EPA-8330K-10-001; United States Environmental Protection Agency, Office of Wastewater Management, Water Permits Division, State and Regional Branch: Washington, DC, USA, 2010.
- U.S. Environmental Protection Agency (USEPA). Biosolids Technology Fact Sheet Land Application of Biosolids; EPA 832-F-00-064; USEPA: Washington, DC, USA, 2000.
- Brooks, J.P.; Maxwell, S.L.; Rensing, C.; Gerba, C.P.; Pepper, I.L. Occurrence of antibiotic-resistant bacteria and endotoxin associated with the land application of biosolids. Can. J. Microbiol. 2007, 53, 616–622. [Google Scholar] [CrossRef] [Green Version]
- Munir, M.; Xagoraraki, I. Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil. J. Environ. Qual. 2011, 40, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Diehl, D.L.; LaPara, T.M. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ. Sci. Technol. 2010, 44, 9128–9133. [Google Scholar] [CrossRef]
- Ma, Y.; Wilson, C.A.; Novak, J.T.; Riffat, R.; Aynur, S.; Murthy, S.; Pruden, A. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environ. Sci. Technol. 2011, 45, 7855–7861. [Google Scholar] [CrossRef] [PubMed]
- Reinthaler, F.F.; Feierl, G.; Galler, H.; Haas, D.; Leitner, E.; Mascher, F.; Melkes, A.; Posch, J.; Winter, I.; Zarfel, G.; et al. ESBL-producing E. coli in Austrian sewage sludge. Water Res. 2010, 44, 1981–1985. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Blaak, H.; de Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; de Roda Husman, A.M. Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Neilson, J.W.; Josephson, K.L.; Pepper, I.L.; Arnold, R.B.; Di Giovanni, G.D.; Sinclair, N.A. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl. Environ. Microbiol. 1994, 60, 4053–4058. [Google Scholar] [CrossRef] [Green Version]
- U.S. Environmental Protection Agency (USEPA). Literature review of Contaminants in Livestock and Poultry Manure and Implications for Water Quality; EPA 820-R-13-002; United States Environmental Protection Agency, Office of Water: Washington, DC, USA, 2013.
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Animal Health Institute (AHI). Survey Indicates Most Antibiotics Used in Animals are Used for Treating and Preventing Disease; Animal Health Institute: Washington, DC, USA, 2000. [Google Scholar]
- Done, H.Y.; Venkatesan, A.K.; Halden, R.U. Does the recent growth of aquaculture create antibiotic resistance threats different from those associated with land animal production in agriculture? AAPS J. 2015, 17, 513–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, S.B. The challenge of antibiotic resistance. Sci. Am. 1998, 278, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, V.O.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Technol. 2012, 31, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Florini, K.; Denison, R.; Stiffler, T.; Fitzgerald, T.; Goldburg, R. Resistant Bugs and Antibiotic Drugs: State and County Estimates of Antibiotics in Agricultural Feed and Animal Waste; Environmental Defense Fund: New York, NY, USA, 2005. [Google Scholar]
- Hoelzer, K.; Wong, N.; Thomas, J.; Talkington, K.; Jungman, E.; Coukell, A. Antimicrobial drug use in food-producing animals and associated human health risks: What, and how strong, is the evidence? BMC Vet. Res. 2017, 13, 211. [Google Scholar] [CrossRef]
- Peak, N.; Knapp, C.W.; Yang, R.K.; Hanfelt, M.M.; Smith, M.S.; Aga, D.S.; Graham, D.W. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 2007, 9, 143–151. [Google Scholar] [CrossRef]
- Sapkota, A.R.; Curriero, F.C.; Gibson, K.E.; Schwab, K.J. Antibiotic-resistant enterococci and fecal indicators in surface water and groundwater impacted by a concentrated Swine feeding operation. Environ. Health Perspect. 2007, 115, 1040–1045. [Google Scholar] [CrossRef]
- Bernot, M.J.; Smith, L.; Frey, J. Human and veterinary pharmaceutical abundance and transport in a rural central Indiana stream influenced by confined animal feeding operations (CAFOs). Sci. Total Environ. 2013, 445–446, 219–230. [Google Scholar] [CrossRef]
- Hubbard, L.E.; Givens, C.E.; Griffin, D.W.; Iwanowicz, L.R.; Meyer, M.T.; Kolpin, D.W. Poultry litter as potential source of pathogens and other contaminants in groundwater and surface water proximal to large-scale confined poultry feeding operations. Sci. Total Environ. 2020, 735, 139459. [Google Scholar] [CrossRef]
- Graham, J.P.; Nachman, K.E. Managing waste from confined animal feeding operations in the United States: The need for sanitary reform. J. Water Health 2010, 8, 646–670. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency (USEPA). Detecting and Mitigating the Environmental Impact of Fecal Pathogens Originating from Confined Animal Feeding Operations: Review; EPA 600/R-06/021; United States Environmental Protection Agency, Office of Research and Development, National Risk Management Research Laboratory: Washington, DC, USA, 2005.
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. MBio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Suttner, B.; Johnston, E.R.; Orellana, L.H.; Rodriguez, R.L.; Hatt, J.K.; Carychao, D.; Carter, M.Q.; Cooley, M.B.; Konstantinidis, K.T. Metagenomics as a public health risk assessment tool in a study of natural creek sediments influenced by agricultural and livestock runoff: Potential and limitations. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Abraham, S.; Sahibzada, S.; Hewson, K.; Laird, T.; Abraham, R.; Pavic, A.; Truswell, A.; Lee, T.; O’Dea, M.; Jordan, D. Emergence of fluoroquinolone-resistant Campylobacter jejuni and Campylobacter coli among Australian chickens in the absence of fluoroquinolone use. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [Green Version]
- Lerma, L.L.; Benomar, N.; Knapp, C.W.; Correa Galeote, D.; Galvez, A.; Abriouel, H. Diversity, distribution and quantification of antibiotic resistance genes in goat and lamb slaughterhouse surfaces and meat products. PLoS ONE 2014, 9, e114252. [Google Scholar] [CrossRef]
- Svanström, P. Pathogens and Antibiotic Resistant Bacteria in Abattoir Waste and Animals—A Study Involving Abattoir Wastewater, Earthworms and Marabou Storks; Swedish University of Agricultural Sciences: Uppsala, Sweden, 2014. [Google Scholar]
- Onuoha, S. Distribution and antibiogram of bacterial species in effluents from abattoirs in Nigeria. J. Environ. Occup. Sci. 2018, 7, 1–8. [Google Scholar] [CrossRef]
- Onuoha, S.; Okafor, C.; Aduo, B.; Nwaka, F. Distribution of antibiotic resistant bacteria from abattoir wastes and its receiving waters at Nkwo-ezzamgbo, Ebonyi State, Nigeria. World J. Med. Sci. 2016, 13, 242–250. [Google Scholar] [CrossRef]
- Afsharnia, M.; Naraghi, B.; Mardaneh, J.; Kianmehr, M.; Biglari, H. The data of Escherichia coli strains genes in different types of wastewater. Data Brief 2018, 21, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, K.; Bernhardt, C.; Pelton, T.; Schaeffer, E.; Phillips, A. Water Pollution from Slaughterhouses: Three Quarters of U.S. Meat Processing Plants that Discharge into Waterways Violated their Permits, 2016–2018; Environmental Integrity Project: Washington, DC, USA, 2018. [Google Scholar]
- Chattopadhyay, S. Exposure Assessment of Livestock Carcass Management Options during a Foreign Animal Disease Outbreak; EPA/600/R-18/074; U.S. Environmental Protection Agency, Office of Research and Development, Homeland Security Research Program: Cincinnati, OH, USA, 2018.
- U.S. Environmental Protection Agency (USEPA). Pesticide Registration: What are Antimicrobial Pesticides? United States Environmental Protection Agency: Washington, DC, USA, 2017.
- Kellogg, R.L.; Nehring, R.; Grube, A.; Goss, D.W.; Plotkin, S. Environmental indicators of pesticide leaching and runoff from farm fields. Agric Prod 2000. [Google Scholar] [CrossRef]
- Burr, T.J.; Norelli, J.L.; Katz, B.; Wilcox, W.F.; Hoying, S.A. Streptomycin resistance of Pseudomonas syringae pv. papulans in apple orchards and its association with a conjugative plasmid. Phytopathology 1988, 78, 410–413. [Google Scholar] [CrossRef]
- Rodríguez, C.; Lang, L.; Wang, A.; Altendorf, K.; García, F.; Lipski, A. Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline- and gentamicin-resistant bacteria. Appl. Environ. Microbiol. 2006, 72, 5870. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Sánchez, C.; Altendorf, K.; Smalla, K.; Lipski, A. Spraying of oxytetracycline and gentamicin onto field-grown coriander did not affect the abundance of resistant bacteria, resistance genes, and broad host range plasmids detected in tropical soil bacteria. Biol. Fertil. Soils 2008, 44, 589–596. [Google Scholar] [CrossRef]
- Yashiro, E.; McManus, P.S. Effect of streptomycin treatment on bacterial community structure in the apple phyllosphere. PLoS ONE 2012, 7, e37131. [Google Scholar] [CrossRef]
- Norelli, J.L.; Burr, T.J.; Lo Cicero, A.M.; Gilbert, M.T.; Katz, B.H. Homologous streptomycin resistance gene present among diverse gram-negative bacteria in New York State apple orchards. Appl. Environ. Microbiol. 1991, 57, 486–491. [Google Scholar] [CrossRef] [Green Version]
- U.S. Department of Agriculture (USDA). 2013 Census of Agriculture; USDA: Washington, DC, USA, 2018. Available online: https://www.nass.usda.gov/Publications/AgCensus/2012/Online_Resources/Aquaculture/ (accessed on 18 May 2020).
- Food and Agricultural Organization (FAO). Aquaculture Systems and Practices: A Selected Review; United Nations Development Programme, Food and Agriculture Organization of the United Nations: Rome, Italy, 1989. [Google Scholar]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [Green Version]
- Baquero, F.; Martínez, J.-L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Muziasari, W.I.; Parnanen, K.; Johnson, T.A.; Lyra, C.; Karkman, A.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 2016, 92, fiw052. [Google Scholar] [CrossRef] [Green Version]
- Reverter, M.; Sarter, S.; Caruso, D.; Avarre, J.-C.; Combe, M.; Pepey, E.; Pouyaud, L.; Vega-Heredía, S.; de Verdal, H.; Gozlan, R.E. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 2020, 11, 1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, D.; Drum, D.J.; Stalknecht, D.E.; White, D.G.; Lee, M.D.; Ayers, S.; Sobsey, M.; Maurer, J.J. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 2005, 11, 935–938. [Google Scholar] [CrossRef]
- Hower, S.; Phillips, M.C.; Brodsky, M.; Dameron, A.; Tamargo, M.A.; Salazar, N.C.; Jackson, C.R.; Barrett, J.B.; Davidson, M.; Davis, J.; et al. Clonally related methicillin-resistant Staphylococcus aureus isolated from short-finned pilot whales (Globicephala macrorhynchus), human volunteers, and a bayfront cetacean rehabilitation facility. Microb. Ecol. 2013, 65, 1024–1038. [Google Scholar] [CrossRef]
- Alm, E.W.; Daniels-Witt, Q.R.; Learman, D.R.; Ryu, H.; Jordan, D.W.; Gehring, T.M.; Santo Domingo, J. Potential for gulls to transport bacteria from human waste sites to beaches. Sci. Total Environ. 2018, 615, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Literak, I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Dolejska, M.; Papagiannitsis, C.C. Plasmid-mediated resistance is going wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Ramey, A.M.; Ahlstrom, C.A. Antibiotic Resistant Bacteria in Wildlife: Perspectives on Trends, Acquisition and Dissemination, Data Gaps, and Future Directions. J. Wildl. Dis. 2020, 56, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Vogt, N.A.; Stevens, C.P.G.; Pearl, D.L.; Taboada, E.N.; Jardine, C.M. Generalizability and comparability of prevalence estimates in the wild bird literature: Methodological and epidemiological considerations. Anim. Health Res. Rev. 2020. [Google Scholar] [CrossRef]
- Rogers, S.W.; Shaffer, C.E.; Langen, T.A.; Jahne, M.; Welsh, R. Antibiotic-resistant genes and pathogens shed by wild deer correlate with land application of residuals. EcoHealth 2018, 15, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Ewers, C.; Wieler, L.H. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef] [Green Version]
- Sjolund, M.; Bonnedahl, J.; Hernandez, J.; Bengtsson, S.; Cederbrant, G.; Pinhassi, J.; Kahlmeter, G.; Olsen, B. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg. Infect. Dis. 2008, 14, 70–72. [Google Scholar] [CrossRef]
- Fenlon, D.R. Seagulls (Larus spp.) as vectors of salmonellae: An investigation into the range of serotypes and numbers of salmonellae in gull faeces. J. Hyg. 1981, 86, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Chu, W.; Brown, J.; Becker, S.J.; Harwood, V.J.; Jiang, S.C. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar. Pollut. Bull. 2003, 46, 748–755. [Google Scholar] [CrossRef]
- Fogarty, L.R.; Haack, S.K.; Wolcott, M.J.; Whitman, R.L. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 2003, 94, 865–878. [Google Scholar] [CrossRef]
- Simoes, R.R.; Poirel, L.; Da Costa, P.M.; Nordmann, P. Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg. Infect. Dis. 2010, 16, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Bonnedahl, J.; Drobni, M.; Gauthier-Clerc, M.; Hernandez, J.; Granholm, S.; Kayser, Y.; Melhus, A.; Kahlmeter, G.; Waldenstrom, J.; Johansson, A.; et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 2009, 4, e5958. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H.; et al. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS ONE 2012, 7, e53039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wade, T.J.; Calderon, R.L.; Brenner, K.P.; Sams, E.; Beach, M.; Haugland, R.; Wymer, L.; Dufour, A.P. High sensitivity of children to swimming-associated gastrointestinal illness: Results using a rapid assay of recreational water quality. Epidemiology 2008, 19, 375–383. [Google Scholar] [CrossRef]
- Wade, T.J.; Sams, E.; Brenner, K.P.; Haugland, R.; Chern, E.; Beach, M.; Wymer, L.; Rankin, C.C.; Love, D.; Li, Q.; et al. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: A prospective cohort study. Environ. Health 2010, 9, 66. [Google Scholar] [CrossRef] [Green Version]
- O’Flaherty, E.; Solimini, A.; Pantanella, F.; Cummins, E. The potential human exposure to antibiotic resistant-Escherichia coli through recreational water. Sci. Total Environ. 2019, 650, 786–795. [Google Scholar] [CrossRef]
- Blackburn, B.G.; Craun, G.F.; Yoder, J.S.; Hill, V.; Calderon, R.L.; Chen, N.; Lee, S.H.; Levy, D.A.; Beach, M.J. Surveillance for waterborne-disease outbreaks associated with drinking water—United States, 2001–2002. MMWR Surveill Summ. 2004, 53, 23–45. [Google Scholar]
- Grisey, E.; Belle, E.; Dat, J.; Mudry, J.; Aleya, L. Survival of pathogenic and indicator organisms in groundwater and landfill leachate through coupling bacterial enumeration with tracer tests. Desalination 2010, 261, 162–168. [Google Scholar] [CrossRef]
- Gerba, C.P. Assessment of enteric pathogen shedding by bathers during recreational activity and its impact on water quality. Quant. Microbiol. 2000, 2, 55–68. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Flach, C.-F.; Larsson, D.G.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef]
- Chiao, T.H.; Clancy, T.M.; Pinto, A.; Xi, C.; Raskin, L. Differential resistance of drinking water bacterial populations to monochloramine disinfection. Environ. Sci. Technol. 2014, 48, 4038–4047. [Google Scholar] [CrossRef]
- Knight, G.M.; Davies, N.G.; Colijn, C.; Coll, F.; Donker, T.; Gifford, D.R.; Glover, R.E.; Jit, M.; Klemm, E.; Lehtinen, S.; et al. Mathematical modelling for antibiotic resistance control policy: Do we know enough? BMC Infect. Dis. 2019, 19, 1011. [Google Scholar] [CrossRef]
- Ashbolt, N.J.; Amezquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human Health Risk Assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- Amos, G.C.A.; Gozzard, E.; Carter, C.E.; Mead, A.; Bowes, M.J.; Hawkey, P.M.; Zhang, L.; Singer, A.C.; Gaze, W.H.; Wellington, E.M.H. Validated predictive modelling of the environmental resistome. ISME J. 2015, 9, 1467–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef]
- Ovejero, C.M.; Delgado-Blas, J.F.; Calero-Caceres, W.; Muniesa, M.; Gonzalez-Zorn, B. Spread of mcr-1-carrying Enterobacteriaceae in sewage water from Spain. J. Antimicrob. Chemother. 2017, 72, 1050–1053. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef] [Green Version]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef]
- He, L.Y.; Liu, Y.S.; Su, H.C.; Zhao, J.L.; Liu, S.S.; Chen, J.; Liu, W.R.; Ying, G.G. Dissemination of antibiotic resistance genes in representative broiler feedlots environments: Identification of indicator ARGs and correlations with environmental variables. Environ. Sci. Technol. 2014, 48, 13120–13129. [Google Scholar] [CrossRef] [PubMed]
- Vital, P.G.; Zara, E.S.; Paraoan, C.E.M.; Dimasupil, M.; Angela, Z.; Abello, J.J.M.; Santos, I.T.G.; Rivera, W.L. Antibiotic resistance and extended-spectrum Beta-Lactamase production of Escherichia coli isolated from irrigation waters in selected urban farms in metro Manila, Philippines. Water 2018, 10, 548. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.; Jahne, M.; Keely, S.; Brinkman, N.; Haugland, R.; Leibowitz, S.; Wheaton, E.; Garland, J.; Martin, R. Modeling and Predicting the Occurrences of Antibiotic Resistance Genes in US Rivers and Streams; Annual Meeting of the Society for Freshwater Science: Salt Lake City, UT, USA, 2019. [Google Scholar]
- Stachler, E.; Crank, K.; Bibby, K. Co-occurrence of crAssphage with antibiotic resistance genes in an impacted urban watershed. Environ. Sci. Technol. Lett. 2019, 6, 216–221. [Google Scholar] [CrossRef]
- Li, J.; Cheng, W.; Xu, L.; Strong, P.J.; Chen, H. Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ. Sci. Pollut. Res. Int. 2015, 22, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Pholwat, S.; Liu, J.; Taniuchi, M.; Chinli, R.; Pongpan, T.; Thaipisutikul, I.; Ratanakorn, P.; Platts-Mills, J.A.; Fleece, M.; Stroup, S.; et al. Genotypic antimicrobial resistance assays for use on E. coli isolates and stool specimens. PLoS ONE 2019, 14, e0216747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, P.F.; Tyson, G.H.; Kabera, C.; Chen, Y.; Li, C.; Folster, J.P.; Ayers, S.L.; Lam, C.; Tate, H.P.; Zhao, S. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 2016, 60, 5515–5520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention (CDC). National Antimicrobial Resistance Monitoring System (NARMS) Now: Human Data; U.S. Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020.
- Matheu, J.; Aidara-Kane, A.; Andremont, A. The ESBL Tricycle AMR Surveillance Project: A Simple, One Health Approach to Global Surveillance; The World Alliance Against Antibiotic Resistance (WAAAR): Suffolk, UK, 2017. [Google Scholar]
- World Health Organization (WHO). Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2017–2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- World Health Organization (WHO). Monitoring and Evaluation of the Global Action Plan on Antimicrobial Resistance: Framework and Recommended Indicators; World Health Organization, Food and Agriculture Organization of the United Nations, World Organisation for Animal Health: Geneva, Switzerland, 2019. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic Resistance in Recreational Waters: State of the Science. Int. J. Environ. Res. Public Health 2020, 17, 8034. https://doi.org/10.3390/ijerph17218034
Nappier SP, Liguori K, Ichida AM, Stewart JR, Jones KR. Antibiotic Resistance in Recreational Waters: State of the Science. International Journal of Environmental Research and Public Health. 2020; 17(21):8034. https://doi.org/10.3390/ijerph17218034
Chicago/Turabian StyleNappier, Sharon P., Krista Liguori, Audrey M. Ichida, Jill R. Stewart, and Kaedra R. Jones. 2020. "Antibiotic Resistance in Recreational Waters: State of the Science" International Journal of Environmental Research and Public Health 17, no. 21: 8034. https://doi.org/10.3390/ijerph17218034
APA StyleNappier, S. P., Liguori, K., Ichida, A. M., Stewart, J. R., & Jones, K. R. (2020). Antibiotic Resistance in Recreational Waters: State of the Science. International Journal of Environmental Research and Public Health, 17(21), 8034. https://doi.org/10.3390/ijerph17218034





