Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Microorganisms
2.2. Matrix-Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) Identification of Isolates
2.3. Antimicrobial Resistance Characterization
2.4. Phylogenetic Group and Multilocus Sequence Typing (MLST) Analysis
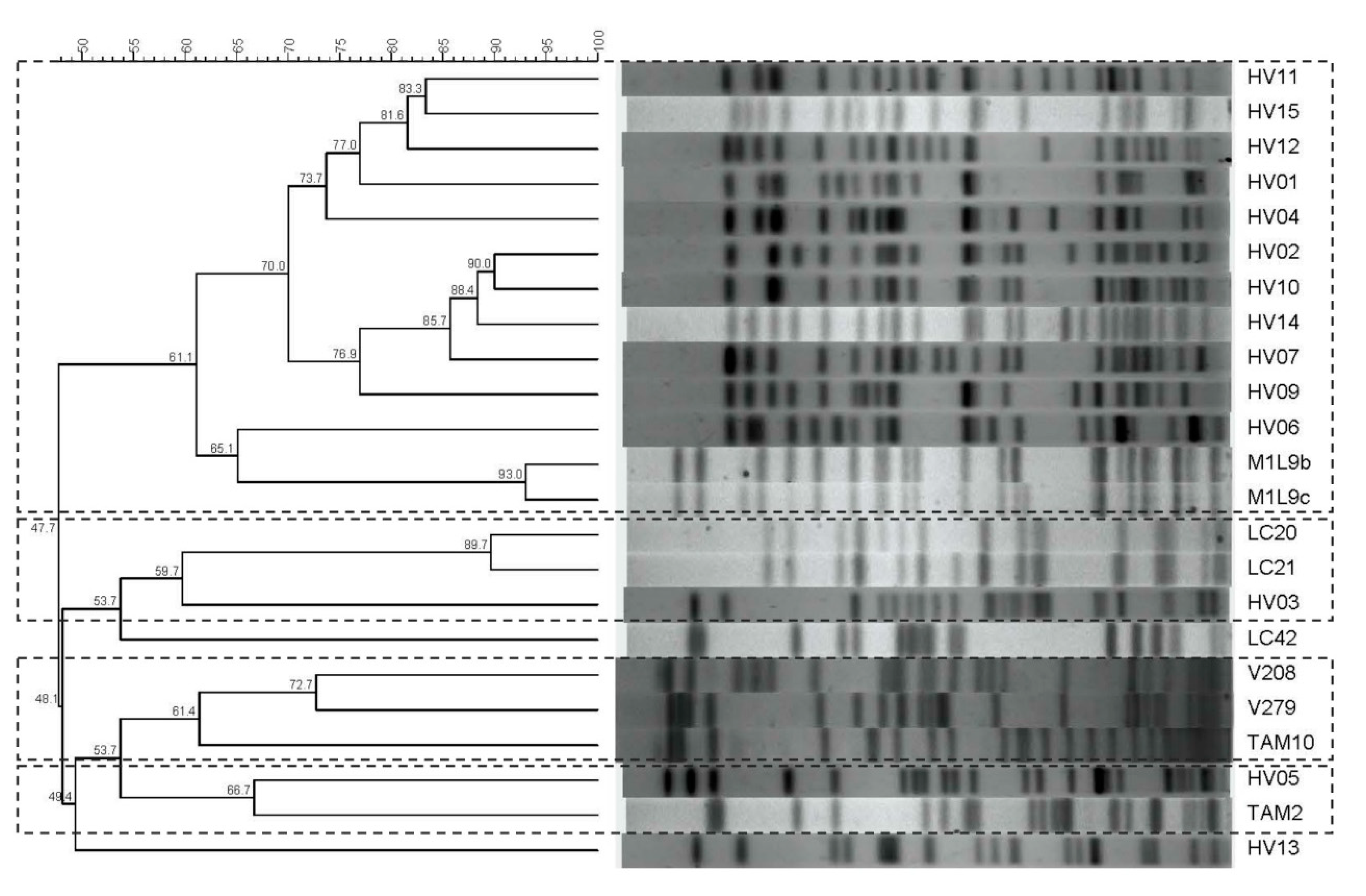
2.5. Pulse Field Gel Electrophoresis (PFGE)
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- EFSA. BIOHAZ Panel Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 2011, 9, 2322. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Liebana, E.; Carattoli, A.; Coque, T.M.; Hasman, H.; Magiorakos, A.-P.; Mevius, D.; Peixe, L.; Poirel, L.; Schuepbach-Regula, G.; Torneke, K.; et al. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food producing animals: An EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 2013, 56, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Van Hoek, A.H.A.M.; Veenman, C.; van Overbeek, W.M.; Lynch, G.; de Roda Husman, A.M.; Blaak, H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Odenthal, S.; Akineden, Ö.; Usleber, E. Extended-spectrum β-lactamase producing Enterobacteriaceae in bulk tank milk from German dairy farms. Int. J. Food Microbiol. 2016, 238, 72–78. [Google Scholar] [CrossRef]
- EFSA. ECDC The European union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, 5598. [Google Scholar]
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.M.; Cole, L.; Daignault, D.; Desruisseau, A.; et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010, 16, 48–54. [Google Scholar] [CrossRef]
- Geser, N.; Stephan, R.; Hächler, H. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 2012, 8, 21. [Google Scholar] [CrossRef]
- Egea, P.; López-Cerero, L.; Torres, E.; del Carmen Gómez-Sánchez, M.; Serrano, L.; Navarro Sánchez-Ortiz, M.D.; Rodriguez-Baño, J.; Pascual, A. Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. Int. J. Food Microbiol. 2012, 159, 69–73. [Google Scholar] [CrossRef]
- Randall, L.P.; Lodge, M.P.; Elviss, N.C.; Lemma, F.L.; Hopkins, K.L.; Teale, C.J.; Woodford, N. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Escherichia coli. Int. J. Food Microbiol. 2017, 241, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Skočková, A.; Bogdanovičová, K.; Koláčková, I.; Karpíšková, R. Antimicrobial-resistant and extended-spectrum β-lactamase-producing Escherichia coli in raw cow’s milk. J. Food Prot. 2015, 78, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Torpdahl, M.; Nielsen, E.M.; Scheutz, F.; Olesen, B.; Hansen, D.S.; Hasman, H. Detection of a Shiga toxin- and extended-spectrum-β-lactamase-producing Escherichia coli O157:H7 human clinical isolate. J. Antimicrob. Chemother. 2013, 68, 1203–1204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ishii, Y.; Kimura, S.; Alba, J.; Shiroto, K.; Otsuka, M.; Hashizume, N.; Tamura, K.; Yamaguchi, K. Extended-spectrum beta-lactamase-producing Shiga toxin gene (Stx1)-positive Escherichia coli O26:H11: A new concern. J. Clin. Microbiol. 2005, 43, 1072–1075. [Google Scholar] [CrossRef][Green Version]
- Dorado-García, A.; Smid, J.H.; van Pelt, W.; Bonten, M.J.M.; Fluit, A.C.; van den Bunt, G.; Wagenaar, J.A.; Hordijk, J.; Dierikx, C.M.; Veldman, K.T.; et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: A pooled analysis. J. Antimicrob. Chemother. 2018, 73, 339–347. [Google Scholar] [CrossRef]
- Ojer-Usoz, E.; González, D.; Vitas, A. Clonal Diversity of ESBL-Producing Escherichia coli Isolated from Environmental, Human and Food Samples. Int. J. Environ. Res. Public Health 2017, 14, 676. [Google Scholar] [CrossRef]
- Mesa, R.J.; Blanc, V.; Blanch, A.R.; Cortés, P.; González, J.J.; Lavilla, S.; Miró, E.; Muniesa, M.; Saco, M.; Tórtola, M.T.; et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage). J. Antimicrob. Chemother. 2006, 58, 211–215. [Google Scholar] [CrossRef]
- Müller, A.; Stephan, R.; Nüesch-Inderbinen, M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 2016, 541, 667–672. [Google Scholar] [CrossRef]
- Day, M.J.; Rodríguez, I.; van Essen-Zandbergen, A.; Dierikx, C.; Kadlec, K.; Schink, A.-K.; Wu, G.; Chattaway, M.A.; DoNascimento, V.; Wain, J.; et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 2016, 71, 1178–1182. [Google Scholar] [CrossRef]
- Monstein, H.J.; Östholm-Balkhed, Å.; Nilsson, M.V.; Nilsson, M.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- NCBI, AMRFinderPlus—Pathogen Detection—NCBI. Available online: https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/ (accessed on 21 January 2020).
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Otero, V.; Rodríguez-Calleja, J.M.; Otero, A.; García-López, M.L.; Santos, J.A. Genetic characterization of atypical enteropathogenic Escherichia coli (EPEC) strains isolated from ewes’ milk, sheep farms environment and humans by multilocus sequence typing and pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 2013, 79, 5864–5869. [Google Scholar] [CrossRef] [PubMed]
- Briñas, L.; Moreno, M.A.; Zarazaga, M.; Porrero, C.; Sáenz, Y.; García, M.; Dominguez, L.; Torres, C. Detection of CMY-2, CTX-M-14, and SHV-12 beta-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 2003, 47, 2056–2058. [Google Scholar]
- Sudarwanto, M.; Akineden, Ö.; Odenthal, S.; Gross, M.; Usleber, E. Extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae in bulk tank milk from dairy farms in Indonesia. Foodborne Pathog. Dis. 2015, 12, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Coque, T.M. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Bou, G.; Cartelle, M.; Tomas, M.; Canle, D.; Molina, F.; Moure, R.; Eiros, J.M.; Guerrero, A. Identification and broad dissemination of the CTX-M-14 β-lactamase in different Escherichia coli strains in the Northwest Area of Spain. J. Clin. Microbiol. 2002, 40, 4030–4036. [Google Scholar] [CrossRef]
- Costa, D.; Vinué, L.; Poeta, P.; Coelho, A.C.; Matos, M.; Sáenz, Y.; Somalo, S.; Zarazaga, M.; Rodrigues, J.; Torres, C. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli isolates in faecal samples of broilers. Vet. Microbiol. 2009, 138, 339–344. [Google Scholar] [CrossRef]
- Snow, L.C.; Wearing, H.; Stephenson, B.; Teale, C.J.; Coldham, N.G. Investigation of the presence of ESBL-producing Escherichia coli in the North Wales and West Midlands areas of the UK in 2007 to 2008 using scanning surveillance. Vet. Rec. 2011, 169, 656. [Google Scholar] [CrossRef]
- Shah, A.A.; Hasan, F.; Ahmed, S.; Hameed, A. Extended-spectrum β-lactamases (ESBLs): Characterization, epidemiology and detection. Crit. Rev. Microbiol. 2004, 30, 25–32. [Google Scholar] [CrossRef]
- Stefani, S.; Giovanelli, I.; Anacarso, I.; Condò, C.; Messi, P.; De Niederhäusern, S.; Bondi, M.; Iseppi, R.; Sabia, C. Prevalence and characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae in food-producing animals in Northern Italy. New Microbiol. 2014, 37, 551–555. [Google Scholar] [PubMed]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef] [PubMed]
- Mora, A.; Blanco, M.; López, C.; Mamani, R.; Blanco, J.E.; Alonso, M.P.; García-Garrote, F.; Dahbi, G.; Herrera, A.; Fernández, A.; et al. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int. J. Antimicrob. Agents 2011, 37, 16–21. [Google Scholar] [CrossRef] [PubMed]
| ESBL Genes | Antimicrobial Susceptibilities (µg/mL) 1,2 | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Source | TEM | SHV | CTX | CTX Group | AMP | AMC | TZP | CEF | CTX | C/C | FOX | AZT | CIP | GM | TO | AK | TIG | TET | ETP | IMP | NAL | FOF | SXT |
| HV1 | Human | 15 | 1 | >16 | S | S | >16 | >32 | S | S | >16 | >2 | S | S | S | S | >8 | S | S | >16 | S | >4/76 | ||
| HV2 | Human | 15 | 1 | >16 | 16/8 | S | >16 | >32 | S | S | >16 | >2 | >8 | >8 | S | S | S | S | S | >16 | S | S | ||
| HV3 | Human | + | 1 | 1 | >16 | >16/8 | 16 | >16 | >32 | S | S | 16 | >2 | >8 | >8 | S | S | >8 | S | S | >16 | S | >4/76 | |
| HV4 | Human | 14 | 9 | >16 | S | S | >16 | >32 | S | S | >16 | >2 | S | S | S | S | S | S | S | >16 | S | S | ||
| HV5 | Human | + | 15 | 1 | >16 | S | S | >16 | >32 | S | S | 4 | S | S | S | S | S | >8 | S | S | S | S | S | |
| HV6 | Human | + | 15 | 1 | >16 | 16/8 | S | >16 | >32 | S | S | >16 | >2 | S | >8 | S | S | S | S | S | >16 | >64 | >4/76 | |
| HV7 | Human | 15 | 1 | >16 | 16/8 | S | >16 | >32 | S | S | >16 | >2 | S | >8 | S | S | S | S | S | >16 | S | >4/76 | ||
| HV8 | Human | + | + | >16 | S | S | >16 | 16 | S | S | >16 | >2 | S | S | S | S | >8 | S | S | >16 | S | >4/76 | ||
| HV9 | Human | 15 | 1 | >16 | >16/8 | 16 | >16 | >32 | S | S | >16 | >2 | S | >8 | 16 | S | >8 | S | S | >16 | S | 4/76 | ||
| HV10 | Human | 15 | 1 | >16 | 16/8 | S | >16 | >32 | S | S | >16 | >2 | >8 | >8 | S | S | >8 | S | S | >16 | S | >4/76 | ||
| HV11 | Human | + | 15 | 1 | >16 | S | S | >16 | >32 | S | S | >16 | >2 | S | S | S | S | S | S | S | >16 | S | S | |
| HV12 | Human | + | 14 | 9 | >16 | 16/8 | S | >16 | >32 | S | S | 16 | >2 | >8 | >8 | S | S | >8 | S | S | >16 | S | ≥2/38 | |
| HV13 | Human | 1 | 1 | >16 | 8/4 | S | >16 | >32 | S | S | >16 | >2 | S | S | S | S | >8 | S | S | >16 | S | S | ||
| HV14 | Human | 15 | 1 | >16 | >16/8 | >64 | >16 | >32 | S | S | >16 | >2 | >8 | S | S | S | S | S | S | >16 | 64 | S | ||
| HV15 | Human | 15 | 1 | >16 | 16/8 | S | >16 | >32 | S | S | >16 | >2 | S | >8 | S | S | S | S | S | >16 | S | >4/76 | ||
| LC20 | Goat’s milk | 14 | 9 | >16 | S | S | >16 | >32 | S | S | 8 | S | S | S | S | S | S | S | S | S | S | S | ||
| LC21 | Goat’s milk | 14 | 9 | >16 | S | S | >16 | >32 | S | S | 4 | S | S | S | S | S | S | S | S | S | S | S | ||
| LC42 | Goat’s milk | + | 14 | 9 | >16 | S | S | >16 | >32 | S | S | 16 | S | S | S | S | S | >8 | S | S | S | S | S | |
| M1L9B | Ewe’s milk | + | 9 | 9 | >16 | S | S | >16 | >32 | S | 16 | 8 | >2 | 4 | S | S | S | >8 | S | S | >16 | S | >4/76 | |
| M1L9C | Ewe’s milk | + | 9 | 9 | >16 | S | S | >16 | >32 | S | 16 | 4 | >2 | S | S | S | S | >8 | S | S | >16 | S | >4/76 | |
| V208 | Ewe’s milk | + | S | S | S | S | S | S | S | S | S | S | S | S | S | >8 | S | S | S | S | S | |||
| V279 | Ewe’s milk | + | >16 | S | S | 16 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |||
| V298 | Ewe’s milk | + | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |||
| TAM2 | Chicken meat | + | + | >16 | S | S | >16 | 8 | S | S | >16 | S | S | S | S | S | >8 | S | S | S | S | S | ||
| TAM10 | Chicken meat | + | + | >16 | 16/8 | S | >16 | >32 | S | 16 | >16 | >2 | S | S | S | S | >8 | S | S | >16 | S | S | ||
| Antibiotic 1 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | TZP | CEF | CTX | C/C | FOX | AZT | CIP | GM | TO | AK | TIG | TET | ETP | IMP | NAL | FOF | SXT | |
| Human | 100 | 60 | 20 | 100 | 100 | 0 | 0 | 100 | 93.3 | 33.3 | 53.3 | 6,6 | 0 | 53.3 | 0 | 0 | 93.3 | 13.3 | 60 |
| Food | 80 | 10 | 0 | 80 | 70 | 0 | 30 | 70 | 30 | 10 | 0 | 0 | 0 | 60 | 0 | 0 | 30 | 0 | 20 |
| Strain | Source | Phylogenetic Group | Sequence Type | Clonal Complex | PFGE |
|---|---|---|---|---|---|
| HV1 | Human | B2 | ST131 | 131 | 1 |
| HV2 | Human | B2 | ST131 | 131 | 1 |
| HV3 | Human | A | ST88 | 23 | 2 |
| HV4 | Human | B1 | ST86 related | 86 | 1 |
| HV5 | Human | B2 | ST3136 | 131 | 4 |
| HV6 | Human | B2 | ST7519 | 131 | 1 |
| HV7 | Human | B2 | ST131 | 131 | 1 |
| HV8 | Human | D | ST648 related | 648 | NT |
| HV9 | Human | B2 | ST131 | 131 | 1 |
| HV10 | Human | B2 | ST131 related | 131 | 1 |
| HV11 | Human | D | ST131 | 131 | 1 |
| HV12 | Human | D | ST131 | 131 | 1 |
| HV13 | Human | D | ST117 | ||
| HV14 | Human | B2 | ST131 | 131 | 1 |
| HV15 | Human | A | ST131 | 131 | 1 |
| LC20 | Goat’s milk | D | ST59 related | 59 | 2 |
| LC21 | Goat’s milk | D | ST59 | 59 | 2 |
| LC42 | Goat’s milk | B1 | ST155 | 155 | |
| M1L9B | Ewe’s milk | D | ST57 | 350 | 1 |
| M1L9C | Ewe’s milk | D | ST57 | 350 | 1 |
| V208 | Ewe’s milk | D | ST57 | 350 | 3 |
| V279 | Ewe’s milk | D | ST57 | 350 | 3 |
| V298 | Ewe’s milk | B1 | ST447 | NT | |
| TAM2 | Chicken meat | A | ST373 | 1689 | 4 |
| TAM10 | Chicken meat | B1 | ST345 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alegría, Á.; Arias-Temprano, M.; Fernández-Natal, I.; Rodríguez-Calleja, J.M.; García-López, M.-L.; Santos, J.A. Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients. Int. J. Environ. Res. Public Health 2020, 17, 1312. https://doi.org/10.3390/ijerph17041312
Alegría Á, Arias-Temprano M, Fernández-Natal I, Rodríguez-Calleja JM, García-López M-L, Santos JA. Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients. International Journal of Environmental Research and Public Health. 2020; 17(4):1312. https://doi.org/10.3390/ijerph17041312
Chicago/Turabian StyleAlegría, Ángel, Marta Arias-Temprano, Isabel Fernández-Natal, Jose M. Rodríguez-Calleja, María-Luisa García-López, and Jesús A. Santos. 2020. "Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients" International Journal of Environmental Research and Public Health 17, no. 4: 1312. https://doi.org/10.3390/ijerph17041312
APA StyleAlegría, Á., Arias-Temprano, M., Fernández-Natal, I., Rodríguez-Calleja, J. M., García-López, M.-L., & Santos, J. A. (2020). Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients. International Journal of Environmental Research and Public Health, 17(4), 1312. https://doi.org/10.3390/ijerph17041312







