Comparative Study on Different Remediation Strategies Applied in Petroleum-Contaminated Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Artificially PHs-Contaminated Soils
2.2. Microorganisms and Culture Conditions
2.3. Remediation Strategies for PH Contaminated soils
2.4. Determination of the Content of Alkanes (ALKs) and Polycyclic Aromatic Hydrocarbons (PAHs)
2.5. Extraction of Bacterial DNA, PCR Amplification, and Sequencing
2.6. Quantitative PCR Analysis of the AlkB and Nah Genes
2.7. Detection of Microbial Populations and Enzyme Activity in the Soil
2.8. Determination of Physicochemical Soil Characteristics
2.9. Bioinformatic Analysis and Data Availability
3. Results
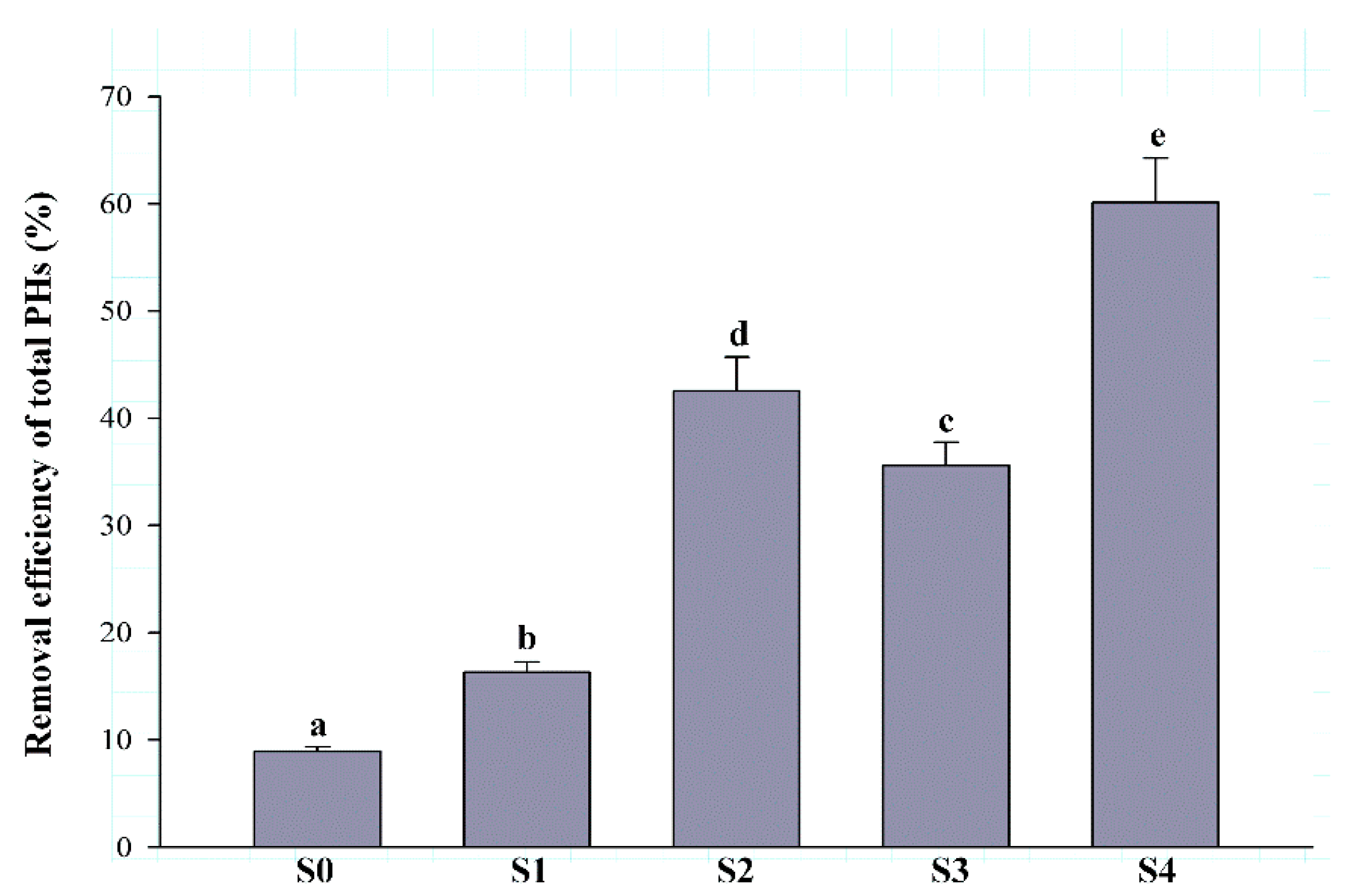
3.1. Degradation Characteristics of PH-Contaminated Soils with Different Remediation Strategies
3.2. Bacterial Community Structure in Soils Subjected to Different Remediation Strategies
3.2.1. Operational Taxonomic Units (OTUs) and Diversity of Bacteria
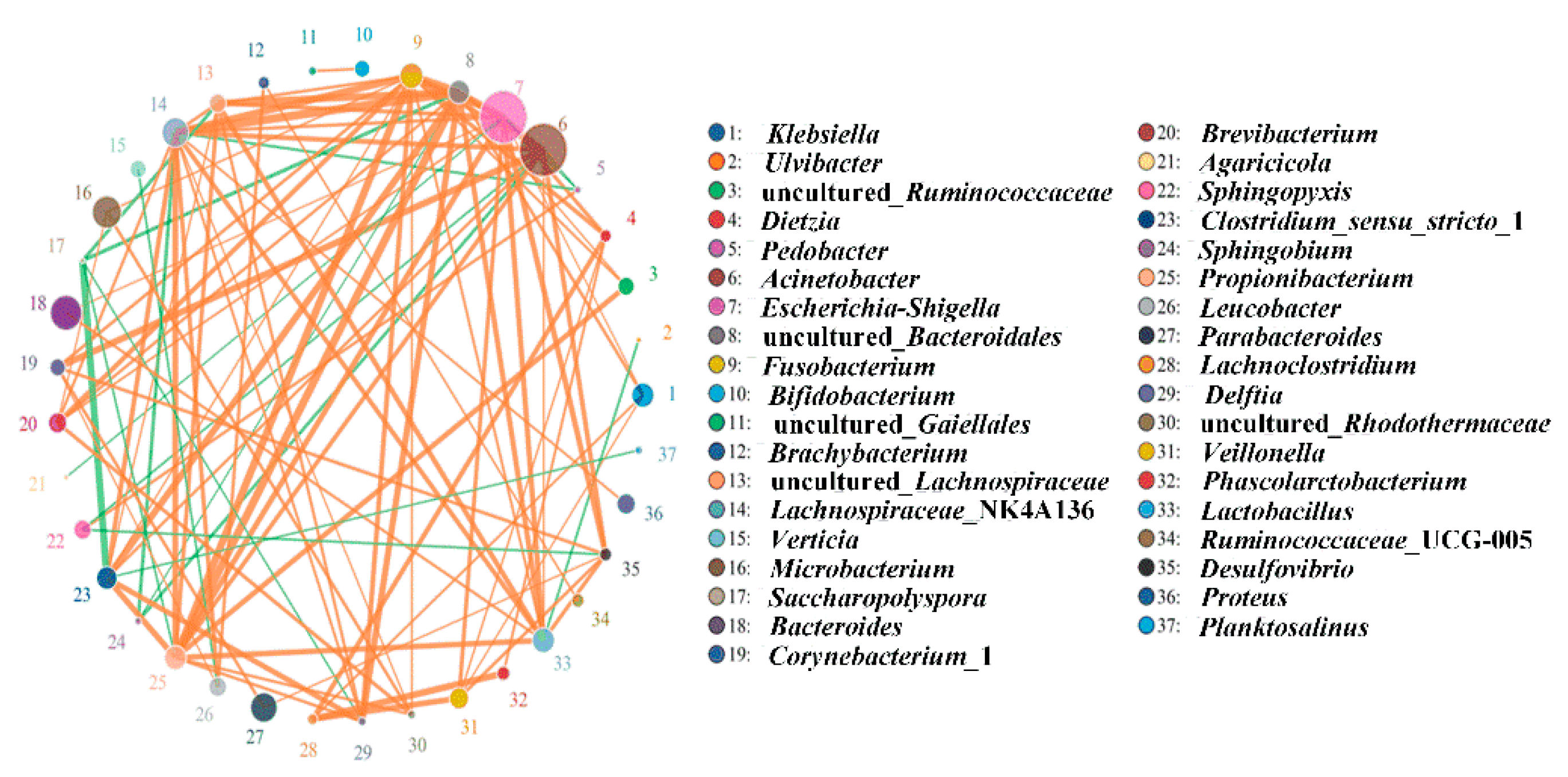
3.2.2. Bacterial Community Composition
3.3. Contribution to and Correlation of Bacteria with the Remediation Process of Contaminated Soils
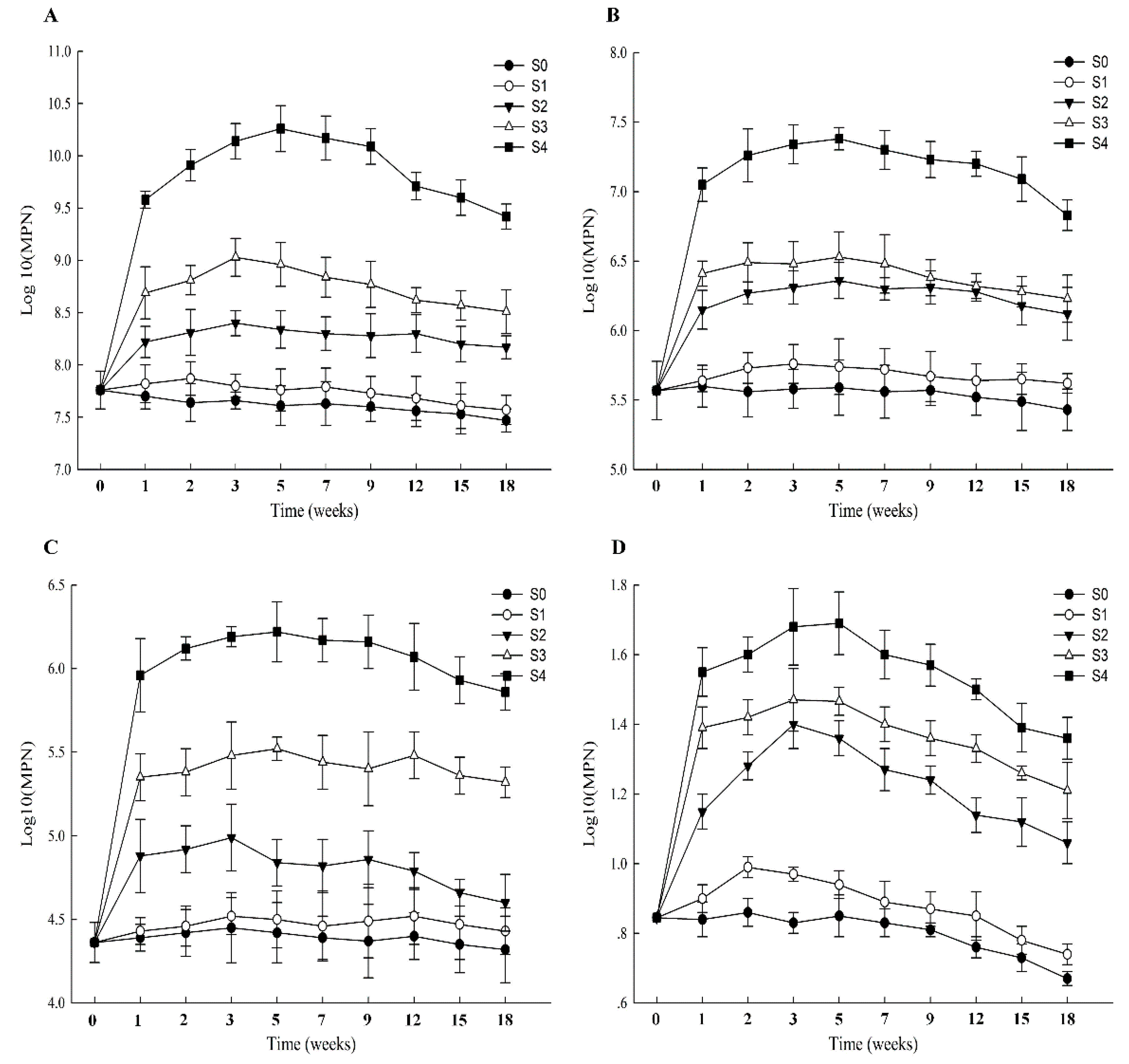
3.4. Numbers of PH-Degraders in Soil Samples from Different Remediation Strategies
3.5. Abundance of alkB and Nah Genes in Soil Samples Treated with Different Remediation Strategies
3.6. Enzyme Activities in the Soils Subjected to Different Remediation Strategies
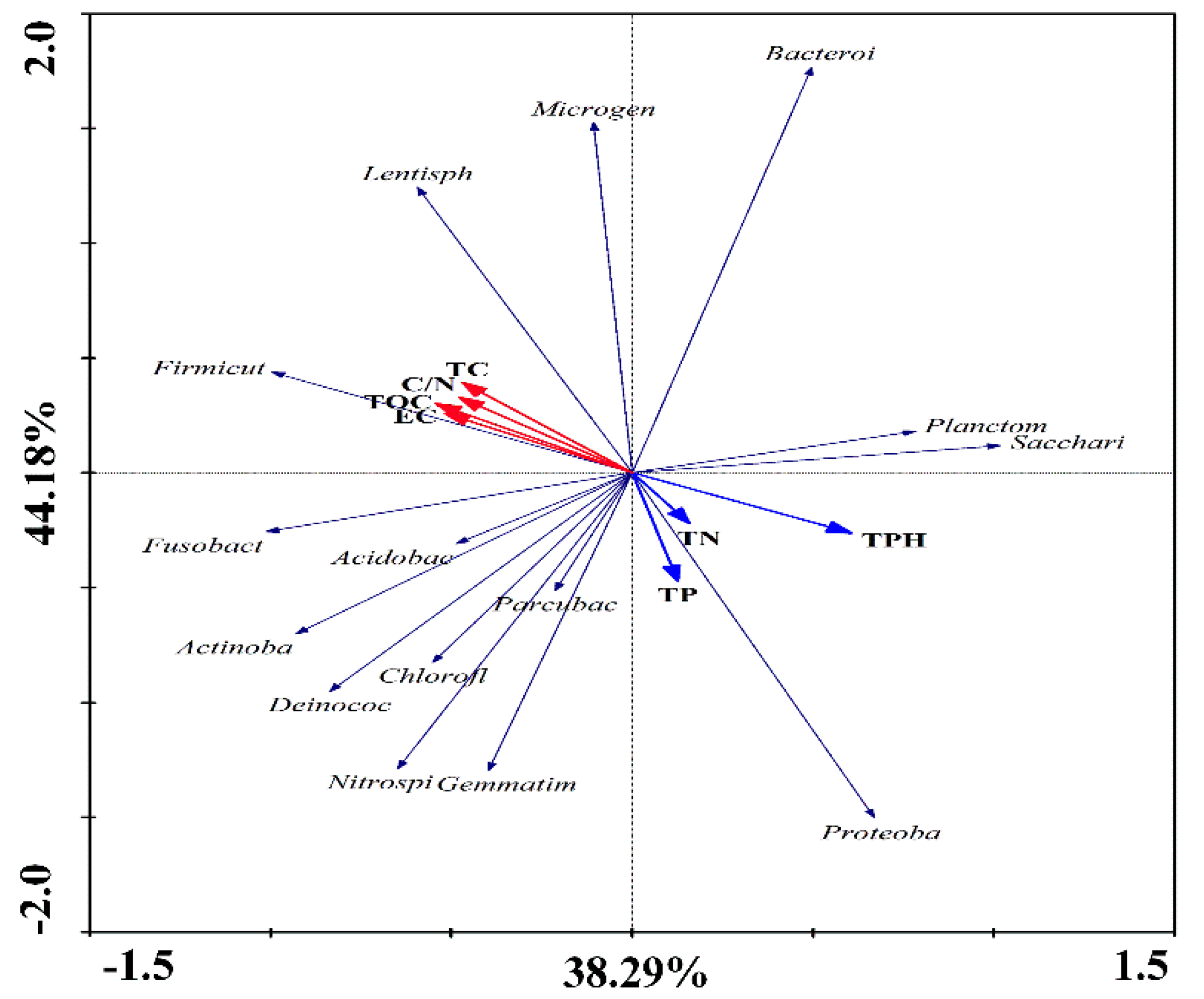
3.7. Correlation between the Bacterial Community Structure and Environmental Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gao, Y.; Guo, S.; Wang, J.; Li, D.; Wang, H.; Zeng, D.H. Effects of different remediation treatments on crude oil contaminated saline soil. Chemosphere 2014, 117, 486–493. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Test Method for Evaluating Solid Waste, SW-846, 3rd ed.; U.S. EPA: Washington, DC, USA, 1986; Volume 1A.
- Hussain, I.; Puschenreiter, M.; Gerhard, S.; Schoftner, P.; Yousaf, S.; Wang, A.; Syed, J.H.; Reichenauer, T.G. Rhizoremediation of petroleum hydrocarbon-contaminated soils: Improvement opportunities and field applications. Environ. Exp. Bot. 2018, 147, 202–219. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Sharma, R.; Singh, K.; Sharma, A. Application of bioremediation technology in the environment contaminated with petroleum hydrocarbon. Ann. Microbiol. 2016, 63, 417–431. [Google Scholar] [CrossRef]
- Ron, E.Z.; Rosenberg, E. Enhanced bioremediation of oil spills in the sea. Curr. Opin. Biotechnol. 2014, 27, 191–194. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int. Biodeterior. Biodegrad. 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Roy, A.; Dutta, A.; Pal, S.; Gupta, A.; Sarkar, J.; Chatterjee, A.; Saha, A.; Sarkar, P.; Sar, P.; Kazy, S.K. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour. Technol. 2018, 253, 22–32. [Google Scholar] [CrossRef]
- Dueholm, M.S.; Marques, I.G.; Karst, S.M.; D’Imperio, S.; Tale, V.P.; Lewis, D.; Nielsen, P.H.; Nielsen, J.L. Survival and activity of individual bioaugmentation strains. Bioresour. Technol. 2015, 186, 192–199. [Google Scholar] [CrossRef]
- Wu, M.L.; Ye, X.; Chen, K.; Li, W.; Yuan, J.; Jiang, X. Bacterial community shift and hydrocarbon transformation during bioremediation of short-term petroleum-contaminated soil. Environ. Pollut. 2017, 223, 657–664. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Al-Sabahi, J.; Al-Maqrashi, F.; Al-Habsi, A.; Al-Hinai, M. Characterization of hydrocarbon-degrading bacteria isolated from oil-contaminated sediments in the Sultanate of Oman and evaluation of bioaugmentation and biostimulation approaches in microcosm experiments. Int. Biodeterior. Biodegrad. 2014, 89, 58–66. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Thavamani, P.; Venkateswarlu, K.; Lee, Y.B.; Naidu, R.; Megharaj, M. Remediation approaches for polycyclic aromatic hydrocarbons (pahs) contaminated soils: Technological constraints, emerging trends and future directions. Chemosphere 2016, 168, 944–968. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Roy, A.; Pal, S.; Mohapatra, B.; Kazy, S.K.; Maiti, M.K.; Sar, P. Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour. Technol. 2017, 242, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, B.M.; Rees, D. Bioremediation of oil spills: A review of challenges for research advancement. Ann. Environ. Sci. 2014, 8, 9–37. [Google Scholar]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef]
- Wilkes, H.; Buckel, W.; Golding, B.T.; Rabus, R. Metabolism of hydrocarbons in n-Alkane utilizing anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 2016, 26, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Meckenstock, R.U.; Boll, M.; Mouttaki, H.; Koelschbach, J.S.; Tarouco, P.C.; Weyrauch, P.; Dong, X.; Himmelberg, A.M. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons. J. Mol. Microbiol. Biotechnol. 2016, 26, 118. [Google Scholar] [CrossRef] [PubMed]
- Brooijmans, R.J.W.; Pastink, M.I.; Siezen, R.J. Hydrocarbon degrading bacteria: The oil spill clean-up crew. Microb. Biotechnol. 2009, 2, 587–594. [Google Scholar] [CrossRef]
- Liu, P.W.G.; Wang, S.Y.; Huang, S.G.; Wang, M.Z. Effects of soil organic matter and ageing on remediation of diesel oil-contaminated soil and on the microbial ecology. Environ. Technol. 2012, 33, 2661–2672. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, B.I.; Kim, J.G. Effect of various amendments on heavy mineral oil bioremediation and soil microbial activity. Bioresour. Technol. 2008, 99, 2578–2587. [Google Scholar] [CrossRef]
- Trindade, P.V.O.; Sobral, L.G.; Rizzo, A.C.L.; Leite, S.G.F.; Soriano, A.U. Bioremediation of a weathered and a recently oil-contaminated soils from Brazil: A comparison study. Chemosphere 2005, 58, 515–522. [Google Scholar] [CrossRef]
- Wu, M.L.; Dick, W.A.; Li, W.; Wang, X.C.; Yang, Q.; Wang, T.T.; Xu, L.; Zhang, M.H.; Chen, L.M. Bioaugmentation and biostimulation of hydrocarbon degradation and the microbial community in a petroleum-contaminated soil. Appl. Biochem. Biotechnol. 2016, 107, 158–164. [Google Scholar]
- Mori, H.; Maruyama, F.; Kato, H.; Toyoda, A.; Dozono, A.; Ohtsubo, Y.; Nagata, Y.; Fujiyama, A.; Tsuda, M.; Kurokawa, K. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Wang, J.; Liao, J.Q.S.; Xie, G.; Huang, Y. Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Appl. Microbiol. Biotechnol. 2015, 99, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Perez-de-Mora, A.; Engel, M.; Schloter, M. Abundance and diversity of n-alkane-degrading bacteria in a forest soil co-contaminated with hydrocarbons and metals: A molecular study on alkB homologous genes. Microb. Ecol. 2011, 62, 959–972. [Google Scholar] [CrossRef]
- Rodriguez-Kabana, R.; Truelove, B. Effects of crop rotation and fertilization on catalase activity in a soil of the southeastern United States. Plant. Soil 1982, 69, 97–104. [Google Scholar] [CrossRef]
- Kuhnert-Finkernagel, R.; Kandeler, E. Enzymes Involved in Carbon Metabolism. In Methods in Soil Biology; Schinner, F., Ohlinger, R., Kandeler, E., Margesin, R., Eds.; Springer: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- Casida, L.E.; Klein, D.A.; Santoro, T. Soil dehydrogenase activity. Soil Sci. Soc. Am. J. 1964, 47, 599–603. [Google Scholar] [CrossRef]
- Bandick, A.K.; Dick, R.P. Field management effects on soil enzyme activities. Soil Biol. Biochem. 1999, 31, 1471–1479. [Google Scholar] [CrossRef]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis, 2nd ed.; Agronomy Series No. 9, Part 2; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; Soil Science Society of America, Inc.: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 60. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Cai, Z.; Zhou, Q.; Zhang, Z.; Chen, C. Bioelectrochemical stimulation of petroleum hydrocarbon degradation in saline soil using U-tube microbial fuel cells. Biotechnol. Bioeng. 2012, 109, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Schjonning, P.; Thomsen, I.K.; Petersen, S.O.; Kristensen, K.; Christensen, B.T. Relating soil microbial activity to water content and tillage-induced differences in soil structure. Geoderma 2011, 163, 256–264. [Google Scholar] [CrossRef]
- U.S. EPA. Environmental Protection Agency. Application, Performance, and Costs of Biotreatment Technologies for Contaminated Soils; EPA/600/R-03/037, Battelle Contract No. 68-D-00-185; U.S. EPA: Washington, DC, USA, 2002.
- Maila, M.P.; Cloete, T.E. The use of biological activities to monitor the removal of fuel contaminants-perspective for monitoring hydrocarbon contamination: A review. Int. Biodeterior. Biodegrad. 2005, 55, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Qu, Y.; Ma, Q.; Zhang, X.; Zhou, J.; Ma, F. Performance and microbial community dynamics in bioaugmented aerated filter reactor treating with coking wastewater. Bioresour. Technol. 2015, 190, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wei, L.; Zhang, H.; Yang, K.; Wang, H. Removal performance and microbial communities in a sequencing batch reactor treating hypersaline phenol-laden wastewater. Bioresour. Technol. 2016, 218, 146–152. [Google Scholar] [CrossRef]
- Zhuang, X.L.; Han, Z.; Bai, Z.H.; Zhang, G.Q.; Shim, H. Progress in decontamination by halophilic microorganisms in saline wastewater and soil. Environ. Pollut. 2010, 158, 1119–1126. [Google Scholar] [CrossRef]
- Wu, M.L.; Li, W.; Dick, W.A.; Ye, X.Q.; Chen, K.L.; Kost, D.; Chen, L.M. Bioremediation of hydrocarbon degradation in a petroleum-contaminated soil and microbial population and activity determination. Chemosphere 2017, 169, 124–130. [Google Scholar] [CrossRef]
- Pascual, J.A.; Hernandez, T.; Garcia, C.; Ayuso, M. Enzymatic activities in an arid soil amended with urban organic wastes: Laboratory experiment. Bioresour. Technol. 1998, 64, 131–138. [Google Scholar] [CrossRef]
- Margesin, R.; Zimmerbauer, A.; Schinner, F. Monitoring of bioremediation by soil biological activities. Chemosphere 2000, 40, 339–346. [Google Scholar] [CrossRef]
- Rogers, J.A.; Tedaldi, D.J.; Kavanaugh, M.C. A screening protocol for bioremediation of contaminated soil. Environ. Prog. 1993, 12, 146–149. [Google Scholar] [CrossRef]


| Petroleum Hydrocarbon | Content (mg/kg) | Soil Characteristics | Values | Microbiological Characteristics | Values |
|---|---|---|---|---|---|
| Tridecane | 926.32 ± 35.96 | Moisture content (%) | 12.84 ± 1.36 | Heterotrophic bacterial numbers (cell/g) | 5.75 ± 0.37 × 107 |
| Tetradecane | 912.92 ± 48.75 | pH | 6.21 ± 0.20 | TPH degraders (MPN/g) | 3.70 ± 0.21 × 105 |
| Pentadecane | 936.57 ± 62.31 | Salinity (mg/kg) | 73.64 ± 6.26 | ALKs degraders (MPN/g) | 2.30 ± 0.12 × 104 |
| Hexadecane | 915.18 ± 68.55 | Electrical conductivity (uS/cm) | 134.83 ± 4.66 | PAHs degraders (MPN/g) | ND |
| Heptadecane | 925.73 ± 74.21 | Total carbon (mg/kg) | 462.24 ± 18.96 | - | - |
| Octadecane | 936.81 ± 47.98 | Total organic carbon (mg/kg) | 382.97 ± 12.47 | - | - |
| Eicosane | 928.66 ± 53.29 | Total nitrogen (mg/kg) | 18.65 ± 0.85 | - | - |
| Heneicosane | 944.57 ± 67.85 | Total phosphorus (mg/kg) | 2.57 ± 0.14 | - | - |
| Naphthalene | 935.26 ± 74.32 | Total potassium (mg/kg) | 15.21 ± 0.49 | - | - |
| Phenanthrene | 954.39 ± 53.27 | C/N ratio | 20.59 ± 0.77 | - | - |
| Total | 9316.41 ± 625.87 | - | - | - | - |
| Method | Remediation Strategy |
|---|---|
| S0 | control conditions, no treatment |
| S1 | soil moisture content: 25–30% |
| S2 | 0.455 g (NH4)2SO4 and 0.035 g K2HPO4 were added to contaminated soil; soil moisture content: 25–30% |
| S3 | both 1 × 108 cells/g soil of a microbial consortium (Microbacterium sp. CICC 10762, K. marina CICC 23948, M. luteus CICC 10680, K. rosea CGMCC 1.15046, S. capitis CICC 21722, and B. odysseyi DSM 18869.) were added to contaminated soil; soil moisture content: 25–30% |
| S4 | 0.455 g (NH4)2SO4 and 0.035 g K2HPO4 were added to contaminated soil; a microbial consortium (same to S3) were added to contaminated oil; soil moisture content: 25–30% |
| Method | ALKs (mg/kg) | PAHs (mg/kg) |
|---|---|---|
| S0 | 6685.73 ± 428.71 | 1797.95 ± 125.74 |
| S1 | 6082.68 ± 518.85 | 1713.92 ± 94.58 |
| S2 | 4125.43 ± 367.22 | 1228.59 ± 141.52 |
| S3 | 4690.04 ± 589.11 | 1310.85 ± 175.48 |
| S4 | 2827.54 ± 308.45 | 885.70 ± 96.89 |
| Index | S0 | S1 | S2 | S3 | S4 |
|---|---|---|---|---|---|
| Coverage (%) | 99.92 ± 0.02a | 99.97 ± 0.01b | 99.96 ± 0.02b | 99.95 ± 0.02ab | 99.95 ± 0.02ab |
| OTUs | 428.00 ± 29.46a | 443.33 ± 33.71bc | 499.67 ± 54.64bc | 491.33 ± 25.01c | 650.67 ± 29.94d |
| ACE | 456.23 ± 40.30a | 470.48 ± 31.12ab | 544.87 ± 50.55bc | 542.85 ± 28.80c | 672.48 ± 35.55d |
| Chao1 | 443.38 ± 48.98a | 466.40 ± 45.51ab | 552.37 ± 55.79b | 536.63 ± 37.14b | 671.82 ± 33.95c |
| Shannon | 4.01 ± 0.03a | 4.25 ± 0.07a | 4.98 ± 0.06b | 4.97 ± 0.16b | 5.51 ± 0.11b |
| Gene | S0 | S1 | S2 | S3 | S4 |
|---|---|---|---|---|---|
| alkB | 3.25 ± 0.14 × 104a | 4.03 ± 0.29 × 104a | 4.29 ± 0.16 × 105b | 7.26 ± 0.64 × 104a | 4.20 ± 0.29 × 106c |
| nah | 1.62 ± 0.05 × 103a | 3.55 ± 0.18 × 103b | 8.91 ± 0.51 × 103c | 4.17 ± 0.33 × 103b | 2.58 ± 0.14 × 104d |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, J.-Q.; He, Q.-S.; Liu, M.-H.; Chen, H.; Sun, M.-B.; Wen, J.-P. Comparative Study on Different Remediation Strategies Applied in Petroleum-Contaminated Soils. Int. J. Environ. Res. Public Health 2020, 17, 1606. https://doi.org/10.3390/ijerph17051606
Cui J-Q, He Q-S, Liu M-H, Chen H, Sun M-B, Wen J-P. Comparative Study on Different Remediation Strategies Applied in Petroleum-Contaminated Soils. International Journal of Environmental Research and Public Health. 2020; 17(5):1606. https://doi.org/10.3390/ijerph17051606
Chicago/Turabian StyleCui, Jia-Qi, Qing-Sheng He, Ming-Hui Liu, Hong Chen, Ming-Bo Sun, and Jian-Ping Wen. 2020. "Comparative Study on Different Remediation Strategies Applied in Petroleum-Contaminated Soils" International Journal of Environmental Research and Public Health 17, no. 5: 1606. https://doi.org/10.3390/ijerph17051606




