Toxicity, Physiological, and Ultrastructural Effects of Arsenic and Cadmium on the Extremophilic Microalga Chlamydomonas acidophila
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algae Isolation and Cultivation
2.2. Cytotoxicity Assays. Evaluation of ROS Generation
2.3. Ultrastructural analysis by Transmission Electron Microscopy (TEM)
2.4. TEM–Energy Dispersive X-Ray Microanalysis (XEDS)
2.5. Isolation of Total RNA. Primer Design and Quantitative Real-Time RT–PCR (qRT–PCR)
2.6. Statistics
3. Results
3.1. Cell Viability and ROS Generation Assessments
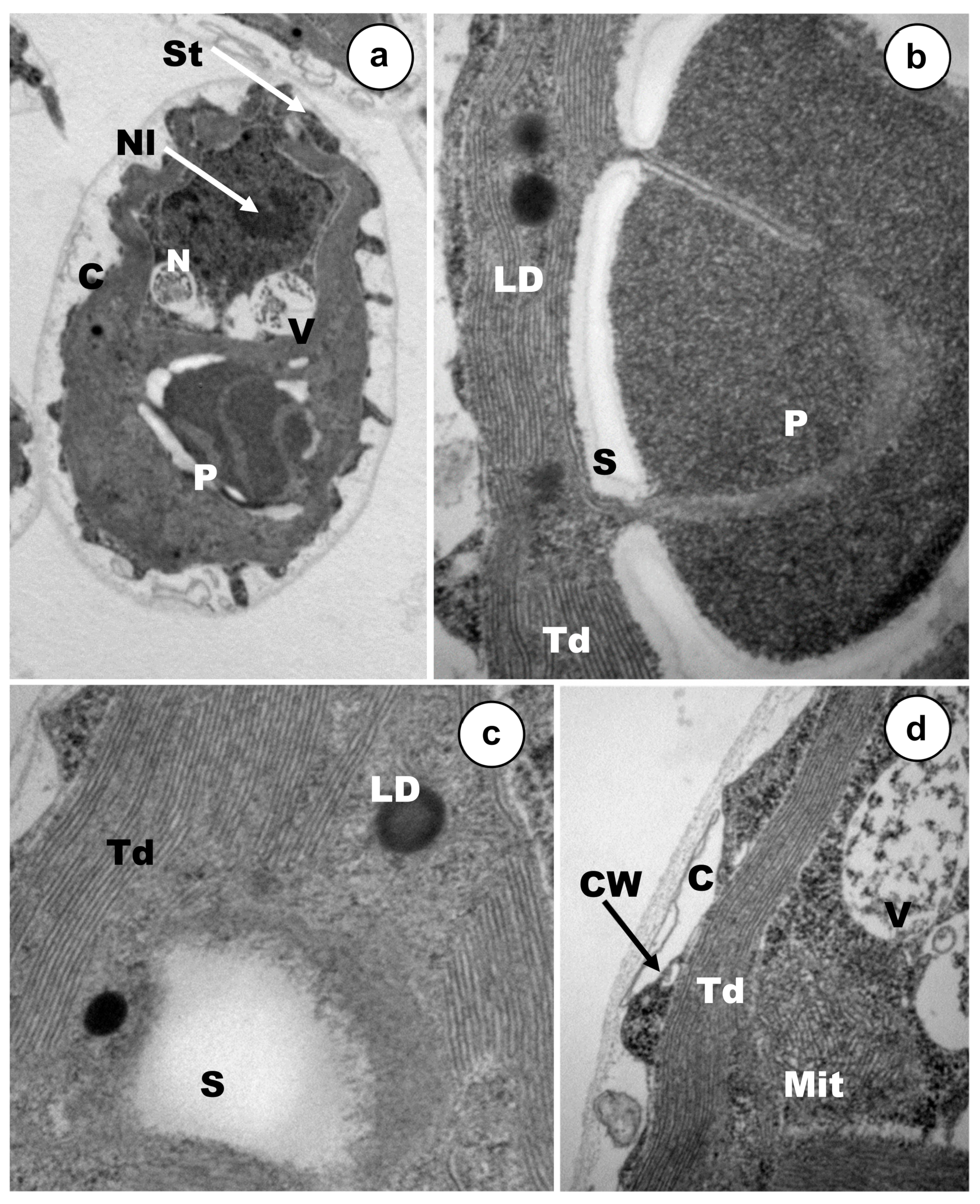
3.2. Ultrastructural Alterations under Cadmium and Arsenic Treatments
3.3. TEM Energy Dispersive X-Ray Microanalysis (XEDS)
3.4. Analysis of CaPCS2 Gene Expression by Quantitative RT-PCR
4. Discussion
4.1. Cytotoxicity and ROS Induction
4.2. Ultrastructural Changes
4.3. Analysis of Mechanisms Involved in Tolerance
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pacheco, F.; Varandas, S.G.; Sanches-Fernandes, L.F.; Valle, R.F., Jr. Soil losses in rural watershed with environmental land use conflicts. Sci. Total Environ. 2014, 1, 110–120. [Google Scholar] [CrossRef]
- Oremo, J.; Orata, F.; Owino, J.; Shivoga, W. Assessment of heavy metals in benthic macroinvertebrates, water and sediments in River Isiukhu. Environ. Monit. Assess. 2019, 191, 11. [Google Scholar] [CrossRef]
- Ayandiran, T.A.; Fawole, O.O.; Adewoye, S.O.; Ogundiran, M.A. Bioconcentration of metals in the body muscle and gut of Clarias gariepinus exposed to sublethal concentrations of soap and detergent effluent. J. Cell Anim. Biol. 2009, 8, 113–118. [Google Scholar]
- Agah, H.; Leermakers, M.; Elskens, M.; Fatemi, S.M.; Baeyens, W. Accumulation of trace metals in the muscle and liver tissues of five fish species from the Persian Gulf. Environ. Monit. Assess. 2009, 157, 499–514. [Google Scholar] [CrossRef]
- Miazek, K.; Iwanek, W.; Remacle, C.; Richel, A.; Goffin, D. Effect of metals, metalloids and metallic nanoparticles on microalgae growth and industrial product biosynthesis: a review. Int. J. Mol. Sci. 2015, 16, 23929–23969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, S.; Parray, J.A. Approaches to Heavy Metal Tolerance in Plants; Springer: Singapore, 2016. [Google Scholar]
- Singh, N.; Raghubanshi, A.; Upadhyay, A.; Rai, U. Arsenic and other heavy metal accumulation in plants and algae growing naturally in contaminated area of West Bengal, India. Ecotoxicol. Environ. Saf. 2016, 130, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.Z.; Jenkins, K.D. Metal Speciation and Bioavailability in Aquatic Ecosystems; John Wiley & Sons Ltd.: Chichester, UK, 1995; p. 236. [Google Scholar]
- Schultze, M. Limnology of pit lakes. In Acidic Pit Lakes: The Legacy of Coal and Metal Surface Mines; Geller, W., Schultze, M., Kleinmann, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 23–224. [Google Scholar]
- Smucker, N.J.; Drerup, S.A.; Vis, M.L. Roles of benthic algae in the structure, function, and assessment of stream ecosystems affected by acid mine drainage. J. Phycol. 2014, 50, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, D.K. Advances in the hydrogeochemistry and microbiology of acid mine waters. Int. Geol. Rev. 2000, 42, 499–515. [Google Scholar] [CrossRef]
- Johnson, D.B. Chemical and microbiological characteristics of mineral spoils and drainage waters at abandoned coal and metal mines. Water Air Soil Pollut. Focus 2003, 3, 47–66. [Google Scholar] [CrossRef]
- Johnson, B.; Aguilera, A. Environmental microbiology in acidophilic environments. In Manual of Environmental Microbiology, 4th ed.; ASM Press: Washington, DC, USA, 2016; pp. 243–260. [Google Scholar]
- Dean, A.P.; Hartley, A.; McIntosh, O.A.; Smith, A.; Feord, H.K.; Holmberg, N.H.; King, T.; Yardley, E.; White, K.N.; Pittman, J.K. Metabolic adaptation of a Chlamydomonas acidophila strain isolated from acid mine drainage ponds with low eukaryotic diversity. Sci. Total Environ. 2019, 647, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Cid, C.; Garcia-Descalzo, L.; Casado-Lafuente, V.; Amils, R.; Aguilera, A. Proteomic analysis of the response of an acidophilic strain of Chlamydomonas sp. (Chlorophyta) to natural metal-rich water. Proteomics 2010, 10, 2026–2036. [Google Scholar] [CrossRef] [PubMed]
- Gerloff-Elias, A.; Barua, D.; Mölich, A.; Spijkerman, E. Temperature-and pH dependent accumulation of heat-shock proteins in the acidophilic green alga Chlamydomonas acidophila. FEMS Microbiol. Ecol. 2006, 56, 345–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirooka, S.; Hirose, Y.; Kanesaki, Y.; Higuchi, S.; Fujiwara, T.; Onuma, R.; Era, A.; Ohbayashi, R.; Uzuka, A.; Nozaki, H.; et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc. Natl. Acad. Sci. USA 2017, 114, E8304–E8313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schönknecht, G.; Chen, W.H.; Ternes, C.M.; Barbier, G.G.; Shrestha, R.P.; Stanke, M.; Bräutigam, A.; Baker, B.J.; Banfield, J.F.; Garavito, R.M.; et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 2013, 339, 1207–1210. [Google Scholar] [CrossRef]
- Olsson, S.; Penacho, V.; Puente-Sánchez, F.; Díaz, S.; Gonzalez-Pastor, J.E.; Aguilera, A. Horizontal gene transfer of phytochelatin synthases from bacteria to extremophilic green algae. Microb. Ecol. 2017, 73, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Aguilera, A.; Souza, V.; Gómez, F.; Amils, R. Development and structure of eukaryotic biofilms in an extreme acidic environment, Río Tinto. Microb. Ecol. 2007, 53, 294–305. [Google Scholar] [CrossRef]
- García-Moyano, A.; González-Toril, E.; Aguilera, A.; Amils, R. Comparative microbial ecology study of the sediments and the water column of the Río Tinto, an extreme acidic environment. FEMS Microbiol. Ecol. 2012, 81, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Cuypers, A.; Plusquin, M.; Remans, T.; Jozefczak, M.; Keunen, E.; Gielen, H.; Opdenakker, K.; Nair, A.R.; Munters, E.; Artois, T.; et al. Cadmium stress: an oxidative challenge. Biometals 2010, 23, 927–940. [Google Scholar] [CrossRef]
- Rani, A.; Kumar, A.; Lal, A.; Pant, M. Cellular mechanisms of cadmium-induced toxicity: A review. Int. J. Environ. Health Res. 2014, 24, 378–399. [Google Scholar] [CrossRef]
- Jomova, K.; Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 2011, 283, 65–87. [Google Scholar] [CrossRef]
- Rahman, M.T.; De Ley, M. Arsenic induction of metallothionein and metallothionein induction against arsenic cytotoxicity. Rev. Environ. Contam. Toxicol. 2017, 240, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Morris, H.; Cronin, M.T.D. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, S.; Xu, P.; Liu, C.; Liu, M.; Wang, Y.; Wang, C.; Zhang, C.; Ge, Y. Review of arsenic speciation, toxicity and metabolism in microalgae. Rev. Environ. Sci. Biotechol. 2015, 14, 427–451. [Google Scholar] [CrossRef]
- Roggenbeck, B.A.; Banerjee, M.; Leslie, E.M. Cellular arsenic transport pathways in mammals. J. Environ. Sci. 2016, 49, 38–58. [Google Scholar] [CrossRef]
- Rippka, J.; Deruelles, J.B.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Guillard, R.R.L.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana and Detonula confervacea. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Bliss, C. The calculation of the dosage-mortality curve. Ann. Appl. Biol. 1935, 22, 134–167. [Google Scholar] [CrossRef]
- Benov, L.; Sztejnberg, L.; Fridovich, I. Critical evaluation of the use of hydroethidine as a measure of superoxide anion radical. Free Radic. Biol. Med. 1998, 25, 826–831. [Google Scholar] [CrossRef]
- Dentler, W. Fixation of Tetrahymena cells for electron microscopy. Methods Cell Biol. 2000, 62, 323–331. [Google Scholar] [CrossRef]
- Díaz, S.; Martín-González, A.; Cubas, L.; Ortega, R.; Amaro, F.; Rodríguez-Martín, D.; Gutiérrez, J.C. High resistance of Tetrahymena thermophila to paraquat: Mitochondrial alterations, oxidative stress and antioxidant genes expression. Chemosphere 2016, 144, 900–917. [Google Scholar] [CrossRef]
- Larionov, A.; Krause, A.; Miller, W. Standard curve-based method for relative real time PCR data processing. BMC Bioinform. 2005, 6, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acid Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Behrenfeld, M.J.; O’Malley, R.T.; Siegel, D.A.; McClain, C.R.; Sarmiento, J.L.; Feldman, G.C.; Milligan, A.J.; Falkowski, P.G.; Letelier, R.M.; Boss, E.S. Climate-Driven trends in contemporary ocean productivity. Nature 2006, 444, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Suárez, S.; Torres, E.; Pérez-Rama, M.; Herrero, C.; Abalde, J. Cadmium toxicity on the freshwater microalga Chlamydomonas moewusii Gerloff: Biosynthesis of thiol compounds. Environ. Toxicol. Chem. 2010, 29, 2009–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, X.; Liu, R.; Xu, S.; Bai, F.; Xu, J.; Yang, Y.; Shi, J.; Wu, Z. Assessment of growth rate, chlorophyll a fluorescence, lipid peroxidation and antioxidant enzyme activity in Aphanizomenon flos-aquae, Pediastrum simplex and Synedraacus exposed to cadmium. Ecotoxicology 2015, 24, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Franklin, N.M.; Stauber, J.L.; Lim, R.P.; Petocz, P. Toxicity of metal mixtures to a tropical freshwater alga (Chlorella sp): The effect of interactions between copper, cadmium, and zinc on metal cell binding and uptake. Environ. Toxicol. Chem. 2002, 21, 2412–2422. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.K.; Ismail, A.; Omar, H.; Tan, S.G. Toxicities and tolerances of Cd, Cu, Pb and Zn in a primary producer (Isochrysis galbana) and in a primary consumer (Perna viridis). Environ. Int. 2004, 29, 1097–1104. [Google Scholar] [CrossRef]
- Monteiro, C.; Fonseca, S.C.; Castro, P.M.L.; Malcata, F.-X. Toxicity of cadmium and zinc on two Microalgae, Scenedesmus obliquus and Desmodesmus pleiomorphus, from Northern Portugal. J. Appl. Phycol. 2011, 23, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Branco, D.; Lima, A.; Almeida, S.F.; Figueira, E. Sensitivity of biochemical markers to evaluate cadmium stress in the freshwater diatom Nitzschia palea. Aquat. Toxicol. 2010, 99, 109–117. [Google Scholar] [CrossRef]
- Rodgher, S.; Gaeta Espíndola, E.L.; Fonseca, F.C.; Toniett, A.E. Cadmium and chromium toxicity to Pseudokirchneriella subcapitata and Microcystis aeruginosa. Braz. Arch. Biol. Technol. 2012, 55, 161–169. [Google Scholar] [CrossRef]
- Aguilera, A.; Amils, R. Tolerance to cadmium in Chlamydomonas sp. (Chlorophyta) strains isolated from an extreme acidic environment, the Tinto River (SW, Spain). Aquat. Toxicol. 2005, 75, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, K.; Tominaga, N. Isolation, growth, ultrastructure and metal tolerance of the green alga, Chlamydomonas acidophila (Chlorophyta). Biosci. Biotechnol. Biochem. 2001, 65, 2650–2656. [Google Scholar] [CrossRef] [PubMed]
- Adamo, G.M.; Brocca, S.; Passolunghi, S.; Salvato, B.; Lotti, M. Laboratory evolution of copper tolerant yeast strains. Microb. Cell Factories 2012, 11, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Francisco, P.; Martín-González, A.; Turkewitz, A.P.; Gutiérrez, J.C. Extreme metal adapted, knockout and knockdown strains reveal a coordinated gene expression among different Tetrahymena thermophila metallothionein isoforms. PLoS ONE 2017, 12, e0189076. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tran, I.; Agrawal, A.F. Does genetic variation maintained by environmental heterogeneity facilitate adaptation to novel selection? Am. Nat. 2016, 188, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parro, V.; Moreno-Paz, M.; González-Toril, E. Analysis of environmental transcriptomes by DNA microarrays. Environ. Microbiol. 2007, 9, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Spijkerman, E.; Behrend, H.; Fach, B.; Gaedke, U. Decreased phosphorus incorporation explains the negative effect of high iron concentrations in the green microalga Chlamydomonas acidophila. Sci. Total Environ. 2018, 626, 1342–1349. [Google Scholar] [CrossRef]
- Kumar, K.; Dahms, H.U.; Won, E.J.; Lee, J.S.; Shin, K.H. Microalgae—A promising tool for heavy metal remediation. Ecotoxicol. Environ. Saf. 2015, 113, 329–352. [Google Scholar] [CrossRef]
- Levy, J.L.; Stauber, J.L.; Adams, M.S.; Maher, W.A.; Kirby, J.K.; Jolley, D.F. Toxicity, biotransformation, and mode of action of arsenic in two freshwater microalgae (Chlorella sp. and Monoraphidium arcuatum). Environ. Toxicol. Chem. 2005, 24, 2630–2639. [Google Scholar] [CrossRef]
- Rahman, M.A.; Hogan, B.; Duncan, D.; Doyle, C.; Krassoi, R.; Rahman, M.M.; Naidu, R.; Lim, R.P.; Maher, W.; Hassler, C. Toxicity of arsenic species to three freshwater organisms and biotransformation of inorganic arsenic by freshwater phytoplankton (Chlorella sp. CE-35). Ecotoxicol. Environ. Saf. 2014, 106, 126–135. [Google Scholar] [CrossRef]
- Wang, N.X.; Li, Y.; Deng, X.H.; Miao, A.J.; Ji, R.; Yang, L.Y. Toxicity and bioaccumulation kinetics of arsenate in two freshwater green algae under different phosphate regimes. Water Res. 2013, 47, 2497–2506. [Google Scholar] [CrossRef] [PubMed]
- Casiot, C.; Bruneel, O.; Personné, J.C.; Leblanc, M.; Elbaz-Poulichet, F. Arsenic oxidation and bioaccumulation by the acidophilic protozoan, Euglena mutabilis, in acid mine drainage (Carnoulès, France). Sci. Total Environ. 2004, 320, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Fryzova, R.; Pohanka, M.; Martinkova, P.; Cihlarova, H.; Brtnicky, M.; Hladky, J.; Kynicky, J. Oxidative stress and heavy metals in plants. Rev. Environ. Contam. Toxicol. 2017, 245, 129–156. [Google Scholar] [CrossRef]
- Karadjova, I.B.; Slaveykova, V.I.; Tsalev, D.L. The biouptake and toxicity of arsenic species on the green microalga Chlorella salina in seawater. Aquat. Toxicol. 2008, 87, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Pawlik-Skowrónska, B.; Pirszel, J.; Kalinowska, R.; Skowroński, T. Arsenic availability, toxicity and direct role of GSH and phytochelatins in As detoxification in the green alga Stichococcus bacillaris. Aquat. Toxicol. 2004, 70, 201–212. [Google Scholar] [CrossRef]
- Miot, J.; Morin, G.; Skouri-Panet, F.; Férard, C.; Poitevin, A.; Aubry, E.; Ona-Nguema, G.; Juillot, F.; Guyot, F.; Brown, G.E., Jr. Speciation of arsenic in Euglena gracilis cells exposed to As (V). Environ. Sci. Technol. 2009, 43, 3315–3321. [Google Scholar] [CrossRef]
- Wang, N.X.; Huang, B.; Xu, S.; Wei, Z.B.; Miao, A.J.; Ji, R.; Yang, L.Y. Effects of nitrogen and phosphorus on arsenite accumulation, oxidation, and toxicity in Chlamydomonas reinhardtii. Aquat. Toxicol. 2014, 157, 167–174. [Google Scholar] [CrossRef]
- Valko, M.; Jomova, K.; Rhodes, C.J.; Kuča, K.; Musílek, K. Redox-and non-redox-metal-induced formation of free radicals and their role in human disease. Arch. Toxicol. 2016, 90, 1–37. [Google Scholar] [CrossRef]
- Jamers, A.; Lenjou, M.; Deraedt, P.; Bockstaele, D.V.; Blust, R.; Coen, W. Flow cytometric analysis of the cadmium exposed green alga Chlamydomonas reinhardtii (Chlorophyceae). Eur. J. Phycol. 2009, 44, 541–550. [Google Scholar] [CrossRef]
- Watanabe, M.; Suzuki, T. Involvement of reactive oxygen stress in cadmium-induced cellular damage in Euglena gracilis. Comp. Biochem. Physiol. Part C 2002, 131, 491–500. [Google Scholar] [CrossRef]
- Watanabe, M.; Henmib, K.; Ogawa, K.; Suzuki, T. Cadmium-Dependent generation of reactive oxygen species and mitochondrial DNA breaks in photosynthetic and non-photosynthetic strains of Euglena gracilis. Comp. Biochem. Physiol. Part C 2003, 134, 227–234. [Google Scholar] [CrossRef]
- Jomova, K.; Vondrakova, D.; Lawson, M.; Valko, M. Metals, oxidative stress and neurodegenerative disorders. Mol. Cell. Biochem. 2010, 345, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Szivák, I.; Behra, R.; Sigg, L. Metal-Induced reactive oxygen species production in Chlamydomonas reinhardtii (Chlorophyceae). J. Phycol. 2009, 45, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Ning, Z.; Wang, Y.; Zheng, Y.; Zhang, C.; Figeys, D. Quantitative proteomic analysis of Dunaliella salina upon acute arsenate exposure. Chemosphere 2016, 145, 112–118. [Google Scholar] [CrossRef]
- Nassiri, Y.; Mansot, J.L.; Wéry, J.; Ginsburger-Vogel, T.; Amiard, J.C. Ultrastructure and electron energy loss spectroscopy studies of sequestration mechanisms of Cd and Cu in the marine diatom Skeletonema costatum. Arch. Environ. Contam. Toxicol. 1997, 33, 147–155. [Google Scholar] [CrossRef]
- Nishikawa, K.; Yamakoshi, Y.; Uemura, I.; Tominaga, N. Ultrastructural changes in Chlamydomonas acidophila (Chrolophyta) induced by heavy metals and polyphosphate metabolism. FEMS Microbiol. Ecol. 2003, 44, 253–259. [Google Scholar] [CrossRef]
- Samadani, M.; Dewez, D. Cadmium accumulation and toxicity affect the extracytoplasmic polyphosphate level in Chlamydomonas Reinhardtii. Ecotoxicol. Environ. Saf. 2018, 166, 200–206. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Sun, G.X.; Rensing, C.; Zhu, Y.G. Biomethylation and volatilization of arsenic by the marine microalgae Ostreococcus tauri. Chemosphere 2013, 93, 47–53. [Google Scholar] [CrossRef]
- Carfagna, S.; Lanza, N.; Salbitani, G.; Basile, A.; Sorbo, S.; Vona, V. Physiological and morphological responses of lead or cadmium exposed Chlorella sorokiniana 211-8K (Chlorophyceae). SpringerPlus 2013, 2, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Souza, L.D.; Simioni, C.; Bouzon, Z.L.; Schneider, R.C.; Gressler, P.; Miotto, M.C.; Rossi, M.J.; Rörig, L.R. Morphological and ultrastructural characterization of the acidophilic and lipid-producer strain Chlamydomonas acidophila LAFIC-004 under different culture conditions. Protoplasma 2017, 254, 1385–1398. [Google Scholar] [CrossRef]
- Vitova, M.; Bisova, K.; Kawano, S.; Zachleder, V. Accumulation of energy reserves in algae: From cell cycles to biotechnological applications. Biotechnol. Adv. 2015, 33, 1204–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallego, A.; Martín-González, A.; Ortega, R.; Gutiérrez, J.C. Flow cytometry assessment of cytotoxicity and reactive oxygen species generation by single and binary mixtures of cadmium, zinc and copper on populations of the ciliated protozoan Tetrahymena thermophila. Chemosphere 2007, 68, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Franklin, N.M.; Stauber, J.L.; Lim, R.P. Development of flow cytometry-based algal bioassays for assessing toxicity of copper in natural waters. Environ. Toxicol. Chem. 2001, 20, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.C.; Barsanti, L.; Passarelli, V.; Evangelista, V.; Conforti, V.; Gualtieri, P. Effects of chromium on photosynthetic and photoreceptive apparatus of the alga Chlamydomonas reinhardtii. Environ. Res. 2007, 105, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Juarez, A.B.; Barsanti, L.; Passarelli, V.; Evangelista, V.; Vesentini, N.; Conforti, V.; Gualtieri, P. In Vivo microspectroscopy monitoring of chromium effects on the photosynthetic and photoreceptive apparatus of Eudorina unicocca and Chlorella kessleri. J. Environ. Monit. 2008, 10, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cobbina, S.J.; Mao, G.; Xu, H.; Zhang, Z.; Yang, L. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ. Sci. Pollut. Res. 2016, 23, 8244–8259. [Google Scholar] [CrossRef]
- García-García, J.D.; Sánchez-Thomas, R.; Moreno-Sánchez, R. Bio-Recovery of non-essential heavy metals by intra-and extracellular mechanisms in free-living microorganisms. Biotechnol. Adv. 2016, 34, 859–873. [Google Scholar] [CrossRef]
- Bräutigam, A.; Schaumlöffel, D.; Preud’homme, H.; Thondorf, I.; Wesenberg, D. Physiological characterization of cadmium-exposed Chlamydomonas reinhardtii. Plant Cell Environ. 2011, 34, 2071–2082. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Zheng, Y.; Ge, Y. Phytochelatin synthesis in Dunaliella salina induced by arsenite and arsenate under various phosphate regimes. Ecotoxicol. Environ. Saf. 2017, 139, 150–160. [Google Scholar] [CrossRef]
- Hernández, L.E.; Sobrino-Plata, J.; Montero-Palmero, M.B.; Carrasco-Gil, S.; Flores-Cáceres, M.L.; Ortega-Villasante, C.; Escobar, C. Contribution of glutathione to the control of cellular redox homeostasis under toxic metal and metalloid stress. J. Exp. Bot. 2015, 66, 2901–2911. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.S.; Dillon, C.T.; Vogt, S.; Lai, B.; Stauber, J.; Jolley, D.F. Copper uptake, intracellular localization, and speciation in marine microalgae measured by synchrotron radiation X-ray fluorescence and absorption microspectroscopy. Environ. Sci. Technol. 2016, 50, 8827–8839. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Jagadevan, S. Genetic identification of arsenate reductase and arsenite oxidase in redox transformations carried out by arsenic metabolising prokaryotes—A comprehensive review. Chemosphere 2016, 163, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, L.; Duan, G.; Sun, G. Characterization of arsenate transformation and identification of arsenate reductase in a green alga Chlamydomonas reinhardtii. Environ. Sci. 2011, 23, 1186–1193. [Google Scholar] [CrossRef]






| PCR Primer | Gene | Sequence (5′ to 3′) | F/R |
|---|---|---|---|
| CaPCH2_3F | CaPCS2 (target gene) | TGGGATTGGGATATTGTGCT | F |
| CaPCH2_2R | ATCTGTTTATGCCCCTGCAC | R | |
| 18SrRNAF | 18S rRNA (housekeeping) | TCAACTTTCGATGGTAGGATAGTG | F |
| 18SrRNAR | CCGTGTCAGGATTGGGTAATTT | R | |
| RT46_ACT1 | Actin (housekeeping) | CTCACTCTCAACATTCCAGCAA | F |
| RT46_ACT2 | GGGCCCGCTCTCATCATACTC | R |
| Gene | E | S | R2 |
|---|---|---|---|
| CaPCS2 | 1.95 | −3.43 | 0.99 |
| 18S rRNA | 2.09 | −3.12 | 0.98 |
| Actin | 2.10 | −3.2 | 0.98 |
| Treatment | Exposure (h) | Fold Induction (±SD) | Author |
|---|---|---|---|
| As(III) | 1 | 14 ± 5 | This study |
| 3 | 526 ± 317 | ||
| 24 | 63 ± 20.4 | ||
| As(V) | 1 | 585.4 ± 239 | This study |
| 3 | 2959 ± 649 | ||
| 24 | 402 ± 116 | ||
| Cd | 1 | 2.0 ± 1.1 | Olsson et al. [19] |
| 3 | 1275.3 ± 218.8 | ||
| 24 | 47.8 ± 24.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz, S.; De Francisco, P.; Olsson, S.; Aguilera, Á.; González-Toril, E.; Martín-González, A. Toxicity, Physiological, and Ultrastructural Effects of Arsenic and Cadmium on the Extremophilic Microalga Chlamydomonas acidophila. Int. J. Environ. Res. Public Health 2020, 17, 1650. https://doi.org/10.3390/ijerph17051650
Díaz S, De Francisco P, Olsson S, Aguilera Á, González-Toril E, Martín-González A. Toxicity, Physiological, and Ultrastructural Effects of Arsenic and Cadmium on the Extremophilic Microalga Chlamydomonas acidophila. International Journal of Environmental Research and Public Health. 2020; 17(5):1650. https://doi.org/10.3390/ijerph17051650
Chicago/Turabian StyleDíaz, Silvia, Patricia De Francisco, Sanna Olsson, Ángeles Aguilera, Elena González-Toril, and Ana Martín-González. 2020. "Toxicity, Physiological, and Ultrastructural Effects of Arsenic and Cadmium on the Extremophilic Microalga Chlamydomonas acidophila" International Journal of Environmental Research and Public Health 17, no. 5: 1650. https://doi.org/10.3390/ijerph17051650
APA StyleDíaz, S., De Francisco, P., Olsson, S., Aguilera, Á., González-Toril, E., & Martín-González, A. (2020). Toxicity, Physiological, and Ultrastructural Effects of Arsenic and Cadmium on the Extremophilic Microalga Chlamydomonas acidophila. International Journal of Environmental Research and Public Health, 17(5), 1650. https://doi.org/10.3390/ijerph17051650





