Current Status of and Future Perspectives in Bacterial Degradation of Benzo[a]pyrene
Abstract
:1. Introduction
2. Studies on BaP Degradation
2.1. Degradation in Aerobic and Normal Conditions of Temperature and Salinity
2.2. Degradation under Extreme Conditions
2.2.1. Anaerobic Microorganisms
2.2.2. Thermophiles
2.2.3. Halophilic Conditions
3. General Findings, Gaps and Strategies to Improve the Isolation of Active BaP Degrading Bacteria
3.1. Biochemical Metabolic Pathways
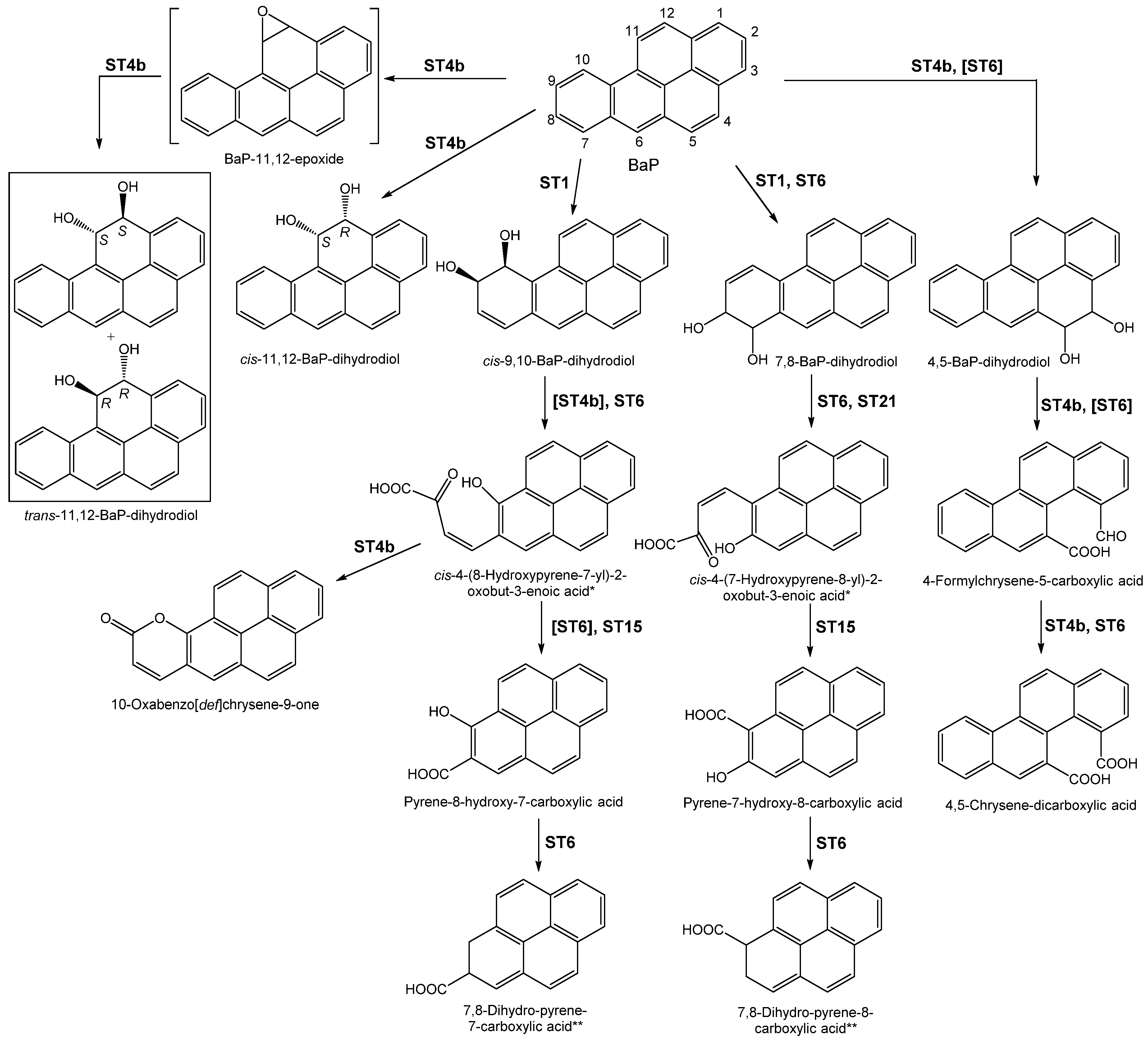
3.1.1. Aerobic Catabolism of BaP
3.1.2. Anaerobic Catabolism of BaP
3.2. Co-metabolism and Metabolic Inhibition
3.3. Biotransformation, Biomineralisation and Bioaugmentation
3.4. Use of BaP in Enrichment Experimental Setup
3.5. Genomes, Omics and Functional Metagenomics
4. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stogiannidis, E.; Laane, R. Source characterization of polycyclic aromatic hydrocarbons by using their molecular indices: An overview of possibilities. Rev. Environ. Contam. Toxicol. 2015, 234, 49–133. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Shafy, H.I.; Mansour, M.S.M. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egypt. J. Pet. 2016, 25, 107–123. [Google Scholar] [CrossRef] [Green Version]
- Nzila, A. Current status of the degradation of aliphatic and aromatic petroleum hydrocarbons by thermophilic microbes and future perspectives. Int. J. Environ. Res. Public Health 2018, 15, 2782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rentz, J.A.; Alvarez, P.J.; Schnoor, J.L. Benzo[a]pyrene degradation by Sphingomonas yanoikuyae JAR02. Environ. Pollut. 2008, 151, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Nisbet, I.C.T.; LaGoy, P.K. Toxic equivalency factors (TEFs) for polycyclic aromatic hydrocarbons (PAHs). Regul. Toxicol. Pharmacol. 1992, 16, 290–300. [Google Scholar] [CrossRef]
- Kasala, E.R.; Bodduluru, L.N.; Barua, C.C.; Sriram, C.S.; Gogoi, R. Benzo(a)pyrene induced lung cancer: Role of dietary phytochemicals in chemoprevention. Pharmacol. Rep. 2015, 67, 996–1009. [Google Scholar] [CrossRef]
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [Green Version]
- Petit, P.; Maitre, A.; Persoons, R.; Bicout, D.J. Lung cancer risk assessment for workers exposed to polycyclic aromatic hydrocarbons in various industries. Environ. Int. 2019, 124, 109–120. [Google Scholar] [CrossRef]
- Hood, D.B.; Ramesh, A.; Aschner, M. CHAPTER 17-Polycyclic Aromatic Hydrocarbons: Exposure from Emission Products and from Terrorist Attacks on US Targets–Implications for Developmental Central Nervous System Toxicity. In Handbook of Toxicology of Chemical Warfare Agents; Gupta, R.C., Ed.; Academic Press: San Diego, CA, USA, 2009; pp. 229–243. ISBN 978-0-12-374484-5. [Google Scholar]
- Haschek, W.M.; Rousseaux, C.G.; Wallig, M.A. Chapter 15-Immune System. In Fundamentals of Toxicologic Pathology, 2nd ed.; Haschek, W.M., Rousseaux, C.G., Wallig, M.A., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 451–489. ISBN 978-0-12-370469-6. [Google Scholar]
- Ramesh, A.; Archibong, A.E. Chapter 43-Reproductive toxicity of polycyclic aromatic hydrocarbons: Occupational relevance. In Reproductive and Developmental Toxicology; Gupta, R.C., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 577–591. ISBN 978-0-12-382032-7. [Google Scholar]
- Sen, S.; Field, J.M. Chapter Three-Genotoxicity of Polycyclic Aromatic Hydrocarbon Metabolites: Radical Cations and Ketones. In Advances in Molecular Toxicology; Fishbein, J.C., Heilman, J.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 7, pp. 83–127. ISBN 1872-0854. [Google Scholar]
- Verma, N.; Pink, M.; Rettenmeier, A.W.; Schmitz-Spanke, S. Review on proteomic analyses of benzo[a]pyrene toxicity. Proteomics 2012, 12, 1731–1755. [Google Scholar] [CrossRef]
- The Priority List of Hazardous Substances that Will Be the Subject of Toxicological Profiles. Available online: http://www.atsdr.cdc.gov/SPL/index.html (accessed on 15 October 2019).
- Tyagi, M.; da Fonseca, M.M.; de Carvalho, C.C. Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 2011, 22, 231–241. [Google Scholar] [CrossRef]
- Adams, G.O.; Fufeyin, P.T.; Okoro, S.E.; Ehinomen, I. Bioremediation, Biostimulation and Bioaugmention: A Review. Int. J. Environ. Bioremediat. Biodegrad. 2015, 3, 28–39. [Google Scholar] [CrossRef]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nzila, A. Update on the cometabolism of organic pollutants by bacteria. Environ. Pollut. 2013, 178, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Nzila, A. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons under anaerobic conditions: Overview of studies, proposed pathways and future perspectives. Environ. Pollut. 2018, 239, 788–802. [Google Scholar] [CrossRef]
- Seo, J.; Keum, Y.; Li, Q.X. Bacterial Degradation of Aromatic Compounds. Int. J. Environ. Res. Public Health 2009, 6, 278–309. [Google Scholar] [CrossRef]
- Ostrem Loss, E.M.; Yu, J.-H. Bioremediation and microbial metabolism of benzo(a)pyrene. Mol. Microbiol. 2018, 109, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.T.; Mahadevan, V.; Jerina, D.M.; Yogi, H.; Yeh, H.J. Oxidation of the carcinogens benzo[a]pyrene and benzo[a]anthracene to dihydrodiols by a bacterium. Science 1975, 189, 295–297. [Google Scholar] [CrossRef]
- Barnsley, E.A. The bacterial degradation of fluoranthene and benzo[a]pyrene. Can. J. Microbiol. 1975, 21, 1004–1008. [Google Scholar] [CrossRef]
- Heitkamp, M.A.; Cerniglia, C.E. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl. Environ. Microbiol. 1988, 54, 1612–1614. [Google Scholar] [CrossRef] [Green Version]
- Heitkamp, M.A.; Cerniglia, C.E. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl. Environ. Microbiol. 1989, 55, 1968–1973. [Google Scholar] [CrossRef] [Green Version]
- Moody, J.D.; Freeman, J.P.; Fu, P.P.; Cerniglia, C.E. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 2004, 70, 340–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosser, R.J.; Warshawsky, D.; Vestal, J.R. Indigenous and enhanced mineralization of pyrene, benzo[a]pyrene, and carbazole in soils. Appl. Environ. Microbiol. 1991, 57, 3462–3469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, J.; Grosser, R.; Jayasimhulu, K.; Xue, W.; Warshawsky, D. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl. Environ. Microbiol. 1996, 62, 13–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trzesicka-Mlynarz, D.; Ward, O.P. Degradation of polycyclic aromatic hydrocarbons (PAHs) by a mixed culture and its component pure cultures, obtained from PAH-contaminated soil. Can. J. Microbiol. 1995, 41, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Siddiqi, M.A.; Maccubin, A.E.; Kumar, S.; Sikka, H.C. Degradation of polynuclear aromatic hydrocarbons by Sphingomonas paucomobilis. Environ. Sci. Technol. 1996, 30, 136–142. [Google Scholar] [CrossRef]
- Juhasz, A.L.; Britz, M.L.; Stanley, G.A. Degradation of benzo[a]pyrene, dibenz[a,h]anthracene and coronene by Burkholderia cepacia. Water Sci. Technol. 1997, 36, 45–51. [Google Scholar] [CrossRef]
- Kanaly, R.; Bartha, R.; Fogel, S.; Findlay, M. Biodegradation of [(sup14)C]Benzo[a]pyrene Added in Crude Oil to Uncontaminated Soil. Appl. Environ. Microbiol. 1997, 63, 4511–4515. [Google Scholar] [CrossRef] [Green Version]
- Aitken, M.D.; Stringfellow, W.T.; Nagel, R.D.; Kazunga, C.; Chen, S.H. Characteristics of phenanthrene-degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can. J. Microbiol. 1998, 44, 743–752. [Google Scholar] [CrossRef]
- Chen, S.H.; Aitken, M.D. Salicylate stimulates the degradation of high-molecular weight polycyclic aromatic hydrocarbons by Pseudomonas saccharophila P15. Environ. Sci. Technol. 1999, 33, 435–439. [Google Scholar] [CrossRef]
- Kanaly, R.A.; Bartha, R. Cometabolic mineralization of benzo[a]pyrene caused by hydrocarbon additions to soil. Environ. Toxicol. Chem. 1999, 18, 2186–2190. [Google Scholar] [CrossRef]
- Hunter, R.D.; Ekunwe, S.I.; Dodor, D.E.; Hwang, H.M.; Ekunwe, L. Bacillus subtilis is a potential degrader of pyrene and benzo[a]pyrene. Int. J. Environ. Res. Public Heal. 2005, 2, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Boonchan, S.; Britz, M.L.; Stanley, G.A. Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl. Environ. Microbiol. 2000, 66, 1007–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, S.; Li, P.; Lian, B.; Stagnitti, F.; Stagnitti, M.; Chen, J.; Xie, L. Influence of Chemical Oxidant on Degradation of Benzo[a]Pyrene Metabolites by the Bacterium-Zoogloea sp. Environ. Eng. Sci. 2008, 25, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Lily, M.K.; Bahuguna, A.; Dangwal, K.; Garg, V. Degradation of Benzo[a]Pyrene by a novel strain Bacillus subtilis BMT4i (MTCC 9447). Braz. J. Microbiol. 2009, 40, 884–892. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; He, T.; Zhong, M.; Zhang, Y.; Li, E.; Huang, T.; Hu, Z. Isolation of marine benzo[a]pyrene-degrading Ochrobactrum sp. BAP5 and proteins characterization. J. Environ. Sci. 2009, 21, 1446–1451. [Google Scholar] [CrossRef]
- Luo, Y.R.; Tian, Y.; Huang, X.; Yan, C.L.; Hong, H.S.; Lin, G.H.; Zheng, T.L. Analysis of community structure of a microbial consortium capable of degrading benzo(a)pyrene by DGGE. Mar. Pollut. Bull. 2009, 58, 1159–1163. [Google Scholar] [CrossRef]
- Mohandass, R.; Rout, P.; Jiwal, S.; Sasikala, C. Biodegradation of benzo[a]pyrene by the mixed culture of Bacillus cereus and Bacillus vireti isolated from the petrochemical industry. J. Environ. Biol. 2012, 33, 985–989. [Google Scholar]
- Yessica, G.-P.; Alejandro, A.; Ronald, F.-C.; José, A.J.; Esperanza, M.-R.; Samuel, C.-S.J.; Remedios, M.-L.M.; Ormeño-Orrillo, E. Tolerance, growth and degradation of phenanthrene and benzo[a]pyrene by Rhizobium tropici CIAT 899 in liquid culture medium. Appl. Soil Ecol. 2013, 63, 105–111. [Google Scholar] [CrossRef]
- Ping, L.; Zhang, C.; Zhang, C.; Zhu, Y.; He, H.; Wu, M.; Tang, T.; Li, Z.; Zhao, H. Isolation and characterization of pyrene and benzo[a]pyrene-degrading Klebsiella pneumonia PL1 and its potential use in bioremediation. Appl. Microbiol. Biotechnol. 2014, 98, 3819–3828. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, S.N. Biodegradation of benzo(a)pyrene mediated by catabolic enzymes of bacteria. Int. J. Environ. Sci. Technol. 2014, 11, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Okai, M.; Kihara, I.; Yokoyama, Y.; Ishida, M.; Urano, N. Isolation and characterization of benzo[a]pyrene-degrading bacteria from the Tokyo Bay area and Tama River in Japan. FEMS Microbiol. Lett. 2015, 362, fnv143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guntupalli, S.; Thunuguntla, V.B.S.C.; Reddy, K.S.; Newton, M.I.; Rao, C.V.; Bondili, J.S. Enhanced Degradation of Carcinogenic PAHs Benzo(a)Pyrene and Benzo(k)Fluoranthene by a Microbial Consortia. Indian J. Sci. Technol. 2016, 9, 35. [Google Scholar] [CrossRef]
- Sowada, J.; Schmalenberger, A.; Ebner, I.; Luch, A.; Tralau, T. Degradation of benzo[a]pyrene by bacterial isolates from human skin. FEMS Microbiol. Ecol. 2014, 88, 129–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Song, X.; Kong, J.; Shen, C.; Huang, T.; Hu, Z. Anaerobic biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by a facultative anaerobe Pseudomonas sp. JP1. Biodegradation 2014, 25, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Fan, F.; Zhu, Y.; Wang, Y.; Liu, X.; Ding, A.; Dou, J. Comparative proteomic analysis and characterization of benzo(a)pyrene removal by Microbacterium sp. strain M.CSW3 under denitrifying conditions. Bioprocess Biosyst. Eng. 2017, 40, 1825–1838. [Google Scholar] [CrossRef]
- Qin, W.; Zhu, Y.; Fan, F.; Wang, Y.; Liu, X.; Ding, A.; Dou, J. Biodegradation of benzo(a)pyrene by Microbacterium sp. strain under denitrification: Degradation pathway and effects of limiting electron acceptors or carbon source. Biochem. Eng. J. 2017, 121, 131–138. [Google Scholar] [CrossRef]
- Qin, W.; Fan, F.; Zhu, Y.; Huang, X.; Ding, A.; Liu, X.; Dou, J. Anaerobic biodegradation of benzo(a)pyrene by a novel Cellulosimicrobium cellulans CWS2 isolated from polycyclic aromatic hydrocarbon-contaminated soil. Braz. J. Microbiol 2018, 49, 258–268. [Google Scholar] [CrossRef]
- Zhao, Z.; Wong, J.W.-C. Rapid Biodegradation of Benzo[a]pyrene by Bacillus subtilis BUM Under Thermophilic Condition. Environ. Eng. Sci. 2010, 27, 939–945. [Google Scholar] [CrossRef]
- Feitkenhauer, H.; Muller, R.; Markl, H. Degradation of polycyclic aromatic hydrocarbons and long chain alkanes at 60–70 degrees C by Thermus and Bacillus spp [corrected]. Biodegradation 2003, 14, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Luna, J.; Alvarez-Fitz, P.; Rios-Leal, E.; Acevedo-Quiroz, M.; Encarnacion-Guevara, S.; Moreno-Godinez, M.E.; Castellanos-Escamilla, M.; Toribio-Jimenez, J.; Romero-Ramirez, Y. Biotransformation of benzo[a]pyrene by the thermophilic bacterium Bacillus licheniformis M2-7. World J. Microbiol. Biotechnol. 2018, 34, 88. [Google Scholar] [CrossRef]
- Arulazhagan, P.; Vasudevan, N. Role of a moderately halophilic bacterial consortium in the biodegradation of polyaromatic hydrocarbons. Mar. Pollut. Bull. 2009, 58, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Arulazhagan, P.; Vasudevan, N. Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain Ochrobactrum sp. VA1. Mar. Pollut. Bull. 2011, 62, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, C.; Liu, H.; Jia, W.; Sun, H. Enzyme activities during Benzo[a]pyrene degradation by the fungus Lasiodiplodia theobromae isolated from a polluted soil. Sci. Rep. 2020, 10, 865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghattas, A.K.; Fischer, F.; Wick, A.; Ternes, T.A. Anaerobic biodegradation of (emerging) organic contaminants in the aquatic environment. Water Res. 2017, 116, 268–295. [Google Scholar] [CrossRef]
- Meckenstock, R.U.; Mouttaki, H. Anaerobic degradation of non-substituted aromatic hydrocarbons. Curr. Opin. Biotechnol. 2011, 22, 406–414. [Google Scholar] [CrossRef]
- Meckenstock, R.U.; Safinowski, M.; Griebler, C. Anaerobic degradation of polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 2004, 49, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Fathepure, B.Z. Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front. Microbiol. 2014, 5, 173. [Google Scholar] [CrossRef] [Green Version]
- Martins, L.F.; Peixoto, R.S. Biodegradation of petroleum hydrocarbons in hypersaline environments. Braz. J. Microbiol. 2012, 43, 865–872. [Google Scholar] [CrossRef] [Green Version]
- Sorokin, D.Y.; Janssen, A.J.H.; Muyzer, G. Biodegradation Potential of Halo(alkali)philic Prokaryotes. Crit. Rev. Environ. Sci. Technol. 2012, 42, 811–856. [Google Scholar] [CrossRef]
- Nzila, A. Mini review: Update on bioaugmentation in anaerobic processes for biogas production. Anaerobe 2017, 46, 3–12. [Google Scholar] [CrossRef]
- Mehta, R.; Singhal, P.; Singh, H.; Damle, D.; Sharma, A.K. Insight into thermophiles and their wide-spectrum applications. 3 Biotech 2016, 6, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, M.E.; Al-Dousary, M.; Hamzah, R.Y.; Fuchs, G. Isolation and characterization of indigenous thermophilic bacteria active in natural attenuation of bio-hazardous petrochemical pollutants. Int. Biodeterior. Biodegrad. 2006, 58, 213–223. [Google Scholar] [CrossRef]
- Kiel, M.; Engesser, K.-H. The biodegradation vs. biotransformation of fluorosubstituted aromatics. Appl. Microbiol. Biotechnol. 2015, 99, 7433–7464. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Ahn, C.K.; Woo, S.H.; Jung, G.Y.; Park, J.M. Synergic degradation of phenanthrene by consortia of newly isolated bacterial strains. J. Biotechnol. 2009, 144, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Janbandhu, A.; Fulekar, M.H. Biodegradation of phenanthrene using adapted microbial consortium isolated from petrochemical contaminated environment. J. Hazard. Mater. 2011, 187, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Semrany, S.; Favier, L.; Djelal, H.; Taha, S.; Amrane, A. Bioaugmentation: Possible solution in the treatment of Bio-Refractory Organic Compounds (Bio-ROCs). Biochem. Eng. J. 2012, 69, 75–86. [Google Scholar] [CrossRef]
- Herrero, M.; Stuckey, D.C. Bioaugmentation and its application in wastewater treatment: A review. Chemosphere 2015, 140, 119–128. [Google Scholar] [CrossRef]
- Nzila, A.; Razzak, S.A.; Zhu, J. Bioaugmentation: An emerging strategy of industrial wastewater treatment for reuse and discharge. Int. J. Environ. Res. Public Health 2016, 13, 846. [Google Scholar] [CrossRef]
- Chen, B.; Huang, J.; Yuan, K.; Lin, L.; Wang, X.; Yang, L.; Luan, T. Direct evidences on bacterial growth pattern regulating pyrene degradation pathway and genotypic dioxygenase expression. Mar. Pollut. Bull. 2016, 105, 73–80. [Google Scholar] [CrossRef]
- Szewczyk, R.; Kowalski, K. Metabolomics and Crucial Enzymes in Microbial Degradation of Contaminants. In Microbial Biodegradation: From Omics to Function and Application; Długoński, J., Ed.; Caister Academic Press: Norfolk, UK, 2016; pp. 43–66. ISBN 978-1-910190-45-6. [Google Scholar]
- DeBruyn, J.M.; Mead, T.J.; Sayler, G.S. Horizontal transfer of PAH catabolism genes in Mycobacterium: Evidence from comparative genomics and isolated pyrene-degrading bacteria. Environ. Sci. Technol. 2012, 46, 99–106. [Google Scholar] [CrossRef]
- Debruyn, J.M.; Chewning, C.S.; Sayler, G.S. Comparative quantitative prevalence of Mycobacteria and functionally abundant nidA, nahAc, and nagAc dioxygenase genes in coal tar contaminated sediments. Environ. Sci. Technol. 2007, 41, 5426–5432. [Google Scholar] [CrossRef] [PubMed]
- Nzila, A.; Ramirez, C.O.; Musa, M.M.; Sankara, S.; Basheer, C.; Li, Q.X. Pyrene biodegradation and proteomic analysis in Achromobacter xylosoxidans, PY4 strain. Int. Biodeterior. Biodegrad. 2018, 130, 40–47. [Google Scholar] [CrossRef]
- Yuan, K.; Xie, X.; Wang, X.; Lin, L.; Yang, L.; Luan, T.; Chen, B. Transcriptional response of Mycobacterium sp. strain A1-PYR to multiple polycyclic aromatic hydrocarbon contaminations. Environ. Pollut. 2018, 243, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Bhagowati, P.; Biswa, B.B.; Chanda, A.; Kalita, B. A comparative intracellular proteomic profiling of Pseudomonas aeruginosa strain ASP-53 grown on pyrene or glucose as sole source of carbon and identification of some key enzymes of pyrene biodegradation pathway. J. Proteom. 2017, 167, 25–35. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, K.; Ding, Y.; Situ, D.; Li, Y.; Long, Y.; Wang, L.; Ye, J. Metabolic and proteomic mechanism of benzo[a]pyrene degradation by Brevibacillus brevis. Ecotoxicol. Environ. Saf. 2019, 172, 1–10. [Google Scholar] [CrossRef]
- Ruan, M.-Y.; Liang, B.; Mbadinga, S.M.; Zhou, L.; Wang, L.-Y.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z. Molecular diversity of bacterial bamA gene involved in anaerobic degradation of aromatic hydrocarbons in mesophilic petroleum reservoirs. Int. Biodeterior. Biodegrad. 2016, 114, 122–128. [Google Scholar] [CrossRef]
- Porter, A.W.; Young, L.Y. Benzoyl-CoA, a universal biomarker for anaerobic degradation of aromatic compounds. Adv. Appl. Microbiol. 2014, 88, 167–203. [Google Scholar] [CrossRef]
- Porter, A.W.; Young, L.Y. The bamA gene for anaerobic ring fission is widely distributed in the environment. Front. Microbiol. 2013, 4, 302. [Google Scholar] [CrossRef] [Green Version]
- Róźalska, S.; Iwanicka-Nowicka, R. Organic Pollutants Degradation by Microorganisms: Genomics, Metagenomics and Metatranstriptomics Backgrounds. In Microbial Biodegradation: From Omics to Function and Application; Długoński, J., Ed.; Caister Academic Press: Norfolk, UK, 2016; pp. 1–12. [Google Scholar]
- Urbieta, M.S.; Donati, E.R.; Chan, K.-G.; Shahar, S.; Sin, L.L.; Goh, K.M. Thermophiles in the genomic era: Biodiversity, science, and applications. Biotechnol. Adv. 2015, 33, 633–647. [Google Scholar] [CrossRef]
- Ngara, T.R.; Zhang, H. Recent Advances in Function-based Metagenomic Screening. Genom. Proteom. Bioinform. 2018, 16, 405–415. [Google Scholar] [CrossRef]
- De Vasconcellos, S.P.; Angolini, C.F.F.; García, I.N.S.; Dellagnezze, B.M.; Da Silva, C.C.; Marsaioli, A.J.; dos Santos Neto, E.V.; De Oliveira, V.M. Screening for hydrocarbon biodegraders in a metagenomic clone library derived from Brazilian petroleum reservoirs. Org. Geochem. 2010, 41, 675–681. [Google Scholar] [CrossRef]
- Dellagnezze, B.M.; de Sousa, G.V.; Martins, L.L.; Domingos, D.F.; Limache, E.E.G.; de Vasconcellos, S.P.; da Cruz, G.F.; de Oliveira, V.M. Bioremediation potential of microorganisms derived from petroleum reservoirs. Mar. Pollut. Bull. 2014, 89, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, S.P.; Sierra-Garcia, I.N.; Dellagnezze, B.M.; Vicentini, R.; Midgley, D.; Silva, C.C.; Santos Neto, E.V.; Volk, H.; Hendry, P.; Oliveira, V.M. Functional and genetic characterization of hydrocarbon biodegrader and exopolymer-producing clones from a petroleum reservoir metagenomic library. Environ. Technol. (UK) 2017, 38, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.; Murrell, J. Stable isotope probing-linking microbial identity to function. Nat. Rev. Microbiol. 2005, 3, 499–504. [Google Scholar] [CrossRef]
- Fischer, A.; Manefield, M.; Bombach, P. Application of stable isotope tools for evaluating natural and stimulated biodegradation of organic pollutants in field studies. Curr. Opin. Biotechnol. 2016, 41, 99–107. [Google Scholar] [CrossRef]
- Song, M.; Luo, C.; Jiang, L.; Zhang, D.; Wang, Y.; Zhang, G. Identification of Benzo[a]pyrene-Metabolizing Bacteria in Forest Soils by Using DNA-Based Stable-Isotope Probing. Appl. Environ. Microbiol. 2015, 81, 7368–7376. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Murrell, J. When metagenomics meets stable-isotope probing: Progress and perspectives. Trends Microbiol. 2010, 18, 157–163. [Google Scholar] [CrossRef]
- Coyotzi, S.; Pratscher, J.; Murrell, J.C.; Neufeld, J.D. Targeted metagenomics of active microbial populations with stable-isotope probing. Curr. Opin. Biotechnol. 2016, 41, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Barnett, S.E.; Buckley, D.H. Simulating metagenomic stable isotope probing datasets with MetaSIPSim. BMC Bioinform. 2020, 21, 37. [Google Scholar] [CrossRef] [Green Version]
- Sinha, R.; Shukla, P. Current Trends in Protein Engineering: Updates and Progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar] [CrossRef]
- Zeymer, C.; Hilvert, D. Directed Evolution of Protein Catalysts. Annu. Rev. Biochem. 2018, 87, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Markel, U.; Essani, K.D.; Besirlioglu, V.; Schiffels, J.; Streit, W.R.; Schwaneberg, U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020, 49, 233–262. [Google Scholar] [CrossRef] [PubMed]

| Characteristics of Microorganisms | Study Number | Inoculum | Conditions of Microorganism Isolation | Name of Strain(s) | Co-Metabolite Substrate (Growth Substrate) | Main Results | References |
|---|---|---|---|---|---|---|---|
| Aerobic microorga-nisms and normal conditions of temperature and salinity | ST1 | Polluted stream | Enrichment in the presence of biphenyl | Beijerinckia B-836 | Biphenyl and succinic acid | - First evidence of BaP metabolite from bacteria. - Identification of BaP metabolites. | [22] |
| ST2 | Contaminated soil | Enrichment in the presence of succinate and salicylate | Pseudomonas NCIB 9816, Pseudomonas ATCC 17483, Pseudomonas putida PpG7 | succinate and salicylate | - Evidence of the utilisation of BaP by a bacterium. | [23] | |
| ST3 | Contaminated sediments | Enrichment in the presence of pyrene | PAH degrading bacteria | Peptone, yeast extract, and soluble starch | - Absence of mineralisation of BaP | [24] | |
| ST4a | Same as in ST3 | None | Mycobacterium sp. PYR-1 in the presence of contaminated sediment | Not defined (sediment) | - Absence of mineralisation of BaP in the presence of Mycopbacterium sp. strain alone, however, mineralisation in the presence of a sediment bioaugmented with Mycopbacterium sp. strain. | [25] | |
| ST4b | Same as in ST4a | Same as in ST4a | Same as in ST4a | BaP (but growth was not assessed) | - Identification of various BaP metabolites | [26] | |
| ST5 | Contaminated soil | Enrichment in the presence of pyrene | Mycobacterium sp. | phenanthrene | - Evidence of complete BaP biomineralisation (up to formation of 14C-CO2) from 14C-BaP | [27] | |
| ST6 | Same as ST5 | Same as ST5 | Same as ST5, strain name as Mycobacterium RJGII-135 | Same as ST5 | - Confirmation of BaP biomineralisation - Identification of BaP metabolites. | [28] | |
| ST7 | Contaminated soil | Enrichment in the presence of PAH mixture | Mixture of Pseudomonas putida, Flavobacterium sp., and Pseudomonas aeruginosa | glucose, yeast extract, and peptone | - Evidence of BaP degradation | [29] | |
| ST8 | Contaminated soil | Enrichment in the presence of fluoranthene | Sphingomonas paucimobilis, EPA 505 | Fluoranthene | - BaP degradation in the presence of fluoranthene - Confirmation of BaP biomineralisation | [30] | |
| ST9 | Contaminated soil | Enrichment in the presence of PAH mixture (including phenanthrene) | Burkholderia cepacia (3 strains: VUN 10 001, VUN 10 002 and VUN 10 003) | phenanthrene | - Degradation of BaP | [31] | |
| ST10 | Soil from cattle pasture | N/A | Use of soil natural consortium of microorganisms | Crude oil | - Degradation and biomineralisation of radiolabeled BaP to CO2 by soil consortium in the presence of crude oil. | [32] | |
| ST11 | Contaminated soil | Enrichment in the presence of phenanthrene | Agrobacterium tumefaciens, Pseudomonas saccharophila, Pseudomonas sp., Burkholderia cepacia, Pseudomonas saccharophila, Bacillus cereus, Sphingomonas paucimobilis | phenanthrene | Each bacterial strain could biomineralise BaP | [33] | |
| ST12 | Same as in ST11 | Same as in ST11 | P. saccharophila P15 | Salicylic acid | - Enhancement of BaP removal rate in the presence of salicylic acid - Evidence of induction of PAH dioxygenase by salicylate | [34] | |
| ST13 | Same as in ST10 | Same as in ST10 | Same as in ST10 | Various crude oil products | - Degradation of up to 60% 14C-BaP in the presence of co-substrates - Inhibition of degradation in the presence of diesel vapours | [35] | |
| ST14 | PAH contaminated soil sample | Enrichment in the presence of a mixture of pyrene, 1-aminopyrene, 1-hydroxypyrene and BaP | Bacillus subtilis Trg3 | None | Evidence of the BaP degradation as a sole substrate | [36] | |
| ST15 | Contaminated soil | Enrichment in the presence of phenanthrene | Sphingomonas yanoikuyae, JAR02 | Salicylate | - Mineralisation of BaP (to CO2) - Identification of BaP metabolites. - Inhibition of degradation by succinate | [4] | |
| ST16 | Gas plant-contaminated soils | Enrichment in the presence of BaP and pyrene | Stenotrophomonas maltophilia VUN 10,010; a bacterial consortium (VUN 10,009) and a fungus (Penicillium janthinellum VUO 10,201) | Pyrene | - BaP degradation in the presence of microbial mixture of bacteria and fungus - VUN 10,010, VUN 10,009 could slightly degrade BaP as a sole source of carbon. - Pronounced degradation with pyrene as growth substrate, and also when the co-culture of all microbes was used | [37] | |
| ST17 | oil-contaminated soils | None | Zoogloea sp. | None | - Zoogloea sp. could degrade BaP as a sole substrate. - Increase degradation when BaP was pre-oxidised by KMnO4 or H2O2 | [38] | |
| ST18 | Contaminated soil | Enrichment in the presence of BaP | Bacillus subtilis BMT4i (MTCC 9447) | None | - Degradation of BaP as a sole substrate - Strain could utilise other PAHs of lower molecular weight | [39] | |
| ST19 | Marine sediments | Enrichment in the presence of BaP | Ochrobactrum sp. BAP5 | None | - Evidence of degradation of BaP as a sole substrate - Expression of protein involved in aromatic degradation | [40] | |
| ST20 | Oil-contaminated water samples | Enrichment in the presence of phenanthrene, fluoranthene and pyrene | Ochrobactrum sp. alone or consortium of Ochrobactrum sp., Stenotrophomonas maltophilia and Pseudomonas fluorescens | None | BaP degradation alone or by the consortium | [41] | |
| ST21 | Oil-contaminated soil | Enrichment in the presence of BaP | Bacillus cereus and Bacillus vireti | None | - Degradation of BaP - Identification of a BaP metabolite. | [42] | |
| ST22 | NA | NA | Rhizobium tropici CIAT 899 (initially isolated from nodules of Phaseolus vulgaris plant) | Yeast extract and mannitol | Degradation of BaP, along with that of phenanthrene | [43] | |
| ST23 | Soil samples | Enrichment in the presence of pyrene and BaP | Klebsiella pneumonia PL1 | Pyrene | - Degradation of BaP and that of pyrene - Could also degrade BaP (and pyrene) in paddy soil | [44] | |
| ST24 | Sludge oil refinery | Enrichment in the presence of oil | Pseudomonas aeruginosa PSA5 and Rhodococcus sp. NJ2 | None | - Degradation BaP as a sole source of carbon - Evidence of expression of enzymes associated with aromatic degradation | [45] | |
| ST25 | Water and mud samples were collected from various sites (including oil refinery, a waste treatment plant, an oil plant) | Enrichment in the presence of peptone, beef extract and pyrene | * 9 bacterial strains (Mycobacterium fluoranthenivorans, Herbiconiux ginseng, Mycobacterium brisbanense, Bacillus megaterium, Sphingobium amiense, Mesorhizobium septentrionale, Vibrio rumoiensis, Olleya sp., Mesoflavibacter zeaxanthinifaciens, Mesoflavibacter zeaxanthinifaciens) | Salicylic acid | - Degradation of BaP by each bacterial strain - M. zeaxanthinifaciens was the most active in BaP degradation and could degrade BaP without salicylic acid | [46] | |
| ST26 | NA | NA | Consortium of bacterial strains Acinetobacter calcoaceticus, MTCC 2409, and MTCC 2289; Serrati amorcescens, MTCC 2645); Pseudomonas sp., MTCC 2445, Stenotrophomonas maltophilia, MTCC 2446) and the fungus Aspergillusterricolavar Americanus, MTCC 2739). | None | - Evidence of BaP degradation. - Identification of BaP metabolites | [47] | |
| ST27 | Swab samples from the different parts of human skin | Enrichment in the presence of BaP but dissolved in Dimethyl sulfoxide (DMSO) | Many bacterial species including Micrococcus, Bacillus, Pseudomonas, Staphylococcus | None | Evidence of BaP degradation, but it is not excluded that some bacteria may use DMSO as a source of carbon. | [48] | |
| Anaerobic bacteria | ST28 | Contaminated sediments | Enrichment in the presence of BaP, an using nitrate as an electron acceptor | Pseudomonas sp. JP1 | None | - Anaerobic degradation of BaP, along with phenanthrene and fluoranthene - Improved biodegradation in the presence of maltose and sucrose but not galactose and glucose - Identification of BaP metabolites | [49] |
| ST29 | Soil layer from a cooking plant | Enrichment in the presence of BaP, and sodium nitrate as an electron acceptor | Microbacterium sp. M.CSW3 | None | - Degradation of BaP - Improved degradation in the presence of phenanthrene and pyrene - Inhibition of degradation by glucose and lactose | [50] | |
| ST30 | Same as in ST29 | Same as in ST29 | Same as in ST29, and BaP degradation by Microbacterium sp. M.CSW3 was evaluated in the presence of various BaP/nitrate ratios | None | - BaP/nitrate ratio of 1:33 was associated with the highest BaP degradation - Identification of metabolites and proposed pathway | [51] | |
| ST31 | Same as in ST29 | Same as in ST29 | Cellulosimicrobium cellulans CWS2 | None | - Anaerobic degradation of BaP in the presence of nitrate - Improved biodegradation with the addition of glucose - Identification of metabolites | [52] | |
| Thermophiles | ST32 | Contaminated soil | Not provided | Bacillus subtilis BUM | Phenanthrene | BaP degradation at 55 °C in culture medium and in soil sample | [53] |
| ST33 | Hot springs, compost and Industrial wastewater | Not provided | Mixture of strains of Bacillus spp. and Thermus sp. | Hexadecane | Evidence of BaP degradation at 60–70 °C | [54] | |
| ST34 | Hot spring from Guerrero State, Mexico | Not provided | Bacillus licheniformis M2-7 at 50 °C | None | - Evidence of BaP degradation at 50 °C - Identification of phthalic acid as a metabolite | [55] | |
| Halophiles | ST35 | Contaminated soil sample | Enrichment in the presence of phenanthrene and 3% NaCl (wt/v) | Consortium of Ochrobactrum sp. VA1, Enterobacter cloacae VA2, Stenotrophomonas maltophilia VA3 | None | Degradation of BaP | [56] |
| ST36 | Contaminated soil sample | phenanthrene | Ochrobactrum sp. VA1 (isolated in ST35) | Yeast extract | Degradation of BaP | [57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nzila, A.; Musa, M.M. Current Status of and Future Perspectives in Bacterial Degradation of Benzo[a]pyrene. Int. J. Environ. Res. Public Health 2021, 18, 262. https://doi.org/10.3390/ijerph18010262
Nzila A, Musa MM. Current Status of and Future Perspectives in Bacterial Degradation of Benzo[a]pyrene. International Journal of Environmental Research and Public Health. 2021; 18(1):262. https://doi.org/10.3390/ijerph18010262
Chicago/Turabian StyleNzila, Alexis, and Musa M. Musa. 2021. "Current Status of and Future Perspectives in Bacterial Degradation of Benzo[a]pyrene" International Journal of Environmental Research and Public Health 18, no. 1: 262. https://doi.org/10.3390/ijerph18010262





