Occurrence of KPC-Producing Escherichia coli in Psittaciformes Rescued from Trafficking in Paraíba, Brazil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Sampling
2.2. Microbial Isolation
2.3. Bacterial Identification
2.4. Antimicrobial Susceptibility Testing and ESBL Screening
2.5. Detection of Genes Encoding for Resistance by PCR and Genetic Relatedness Analysis by REP-PCR
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sousa, A.T.H.I.; Makino, H.; Bruno, V.C.M.; Candido, S.L.; Nogueira, B.S.; Menezes, I.G.; Nakazato, L.; Dutra, V. Perfil de resistência antimicrobiana de Klebsiella pneumoniae isoladas de animais domésticos e silvestres. Arq. Bras. Med. Vet. Zootec. 2019, 71, 584–593. [Google Scholar] [CrossRef] [Green Version]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. 2018, 285, 20180332. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.L.; Palácio, S.B.; Garcia, J.E.; Cavalcanti, I.M.F. Dissemination of multidrug-resistant bacteria in birds. Appro. Poult. Dairy Vet. Sci. 2018, 3, 257–259. [Google Scholar]
- Guenther, S.; Grobbel, M.; Lübke-Becker, A.; Goedecke, A.; Friedrich, N.D.; Wieler, L.H.; Ewers, C. Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet. Microbiol. 2010, 144, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.E.; Williams, N.J.; Bennett, M. Disperse abroad in the land: The role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 2016, 12, 20160137. [Google Scholar] [CrossRef] [Green Version]
- Swift, B.M.C.; Bennett, M.; Waller, K.; Dodd, C.; Murray, A.; Gomes, R.L.; Humphreys, B.; Hobman, J.L.; Jones, M.A.; Whitlock, S.E.; et al. Anthropogenic environmental drivers of antimicrobial resistance in wildlife. Sci. Total Environ. 2019, 649, 12–20. [Google Scholar] [CrossRef]
- Trindade, L.C.; Figueira, P.T. Perfil de susceptibilidade antibacteriana e produção de hemolisina de enterobactérias de psitacídeos em cativeiro. Pubvet 2018, 12, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Stoesser, N.; Sheppard, A.E.; Peirano, G.; Anson, L.W.; Pankhurs, T.L.; Sebra, R.; Phan, H.T.T.; Kasarskis, A.; Mathers, A.J.; Peto, T.E.A.; et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)—Producing Escherichia Coli. Sci. Rep. 2017, 7, 5917. [Google Scholar] [CrossRef]
- Gibson, L.F.; Khoury, J.T. Storage and survival of bacteria by ultra-freeze. Lett. Appl. Microbiol. 1986, 3, 127–129. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute—CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. In CLSI Document M07-A9; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical and Laboratory Standards Institute—CLSI. Performance Standards for Antimicrobial Susceptibility Testing. In CLSI Document M100-S27; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Jarlier, V.; Nicolas, M.-H.; Fournier, G.; Philippon, A. Extended broadspectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning. A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Van Der Zee, U.; Verbakel, H.; Van Zon, J.C.; Frenay, I.; Van Belkum, A.; Peeters, M.; Buiting, U.; Bergmans, U. Molecular genotyping of Staphylococcus aureus strains: Comparison of repetitive elemento sequence-based PCR with various typing methods and isolation of a novel epidemicity marker. J. Clin. Microbiol. 1999, 37, 342–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet. Infect. Dis. 2015, 16, 161–168. [Google Scholar] [CrossRef]
- Poirel, L.; Schrenzel, J.; Cherkaoui, A.; Bernabeu, S.; Renzi, G.; Nordmann, P. Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J. Antimicrob. Chemother. 2011, 66, 1730–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, A.; Ferraro, M.J.; Pino, R.M.; Dew, R.B.; Moland, E.S.; Lockhart, T.J.; Thomson, K.S.; Goering, R.V.; Hanson, N.D. Plasmid Mediated Carbapenem-Hydrolyzing Enzyme KPC-2 in an Enterobacter sp. Antimicrob. Agents. Chemother. 2004, 48, 4438–4440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, E.S.; Maciel, W.C.; Albuquerque, A.H.; Machado, D.N.; Bezerra, W.G.A.; Vasconcelos, R.H.; Lima, B.P.; Gonçalves, G.A.M.; Teixeira, R.S.C. Prevalence and antimicrobial resistance profile of Enterobacteria isolated from Psittaciformes of illegal wildlife trade. Acta. Sci. Vet. 2016, 43, 1313. [Google Scholar]
- Sanches, L.A.; Gomes, M.S.; Texeira, R.H.F.; Cunha, M.P.V.; Oliveira, M.G.X.; Vieira, M.A.M.; Gomes, T.A.T.; Knobl, T. Captive wild birds as reservoirs of enteropathogenic E. coli (EPEC) and Shiga-toxin producing E. coli (STEC). Braz. J. Microbiol. 2017, 48, 760–763. [Google Scholar] [CrossRef]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robison, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kang’ethe, E.K.; et al. Clinically relevant antimicrobial resistance at the wildlife-livestock-human interface in Nairobi: An epidemiological study. Lancet. Planet. Health 2019, 3, 259–269. [Google Scholar] [CrossRef] [Green Version]
- Dolejska, M.; Literak, I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents. Chomother. 2019, 63, e01167-19. [Google Scholar] [CrossRef] [Green Version]
- Ahlstrom, C.A.; Ramey, A.M.; Woksepp, H.; Bonnedahl, J. Repeated detection of carbapenemase-producing Escherichia coli in gulls inhabiting Alaska. Antimicrob. Agents Chemother. 2019, 63, e00758-19. [Google Scholar] [CrossRef] [Green Version]
- Sigirci, B.D.; Celik, B.; Halac, B.; Adiguzel, M.C.; Kekec, I.; Metiner, K.; Ikiz, S.; Bagcigil, A.F.; Ozgur, N.Y.; Ak, S.; et al. Antimicrobial resistance profiles of Escherichia coli isolated from companion birds. J. King. Saud. Univ. Sci. 2020, 32, 1069–1073. [Google Scholar] [CrossRef]
- Pontes, O.S.; Coutinho, D.A.S.; Iovine, R.O.; Cunha, M.P.V.; Knöbl, T.; Carvalho, V.M. Survey on pathogenic Escherichia coli and Salmonella spp. in captive cockatiels (Nymphicus hollandicus). Braz. J. Microbiol. 2019, 49, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.N.; Lopes, E.S.; Albuquerque, Á.H.; Bezerra, W.G.A.; Horn, R.V.; Lima, S.V.G.; Siqueira, R.A.S.; Beleza, A.J.F.; Oliveira, F.R.; Cardoso, W.N.; et al. Detecção e avaliação do perfil de sensibilidade antibacteriana de enterobactérias isoladas de periquitos cara-suja (Pyrrhura griseipectus) em cativeiro. Arq. Bras. Med. Vet. Zootec. 2016, 68, 1732–1736. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ma, Z.-B.; Zeng, Z.-L.; Yang, X.-W.; Huang, Y.; Liu, J.-H. The role of wildlife (wid birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017, 38, 55–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Species | Popular Name | Number of Individuals |
|---|---|---|
| Amazona aestiva | Turquoise-fronted parrot | 10 |
| Amazona amazonica | Orange-winged parrot | 2 |
| Amazona festiva | Festive parrot | 1 |
| Ara ararauna | Blue-and-yellow macaw | 3 |
| Ara chloropterus | Red-and-green macaw | 3 |
| Ara macaw | Scarlet macaw | 1 |
| Diopsittaca nobilis | Red-shouldered macaw | 5 |
| Eupsittula aurea | Peach-fronted parakeet | 1 |
| Eupsittula cactorum | Cactus parakeet | 1 |
| Forpus xanthopterygius | Blue-winged parrotlet | 1 |
| Psittacara leucophthalmus | White-eyed parakeet | 1 |
| Thectocercus acuticaudatus | Blue-crowned parakeet | 1 |
| Total: 30 |
| Genes Encoding for Beta-Lactamase Resistance | ||||
|---|---|---|---|---|
| Gene | Sequences (5-3′) | Amplicon size (BP 1) | TC2 | Reference |
| blaCTX-M F | SCSATGTGCAGYACCAGTAA | 554 | 1 | [15] |
| blaCTX-M R | CCGCRATATGRTTGGTGGTG | |||
| Genes Encoding for Carbapenems Resistance | ||||
| Gene | Sequences (5-3′) | Amplicon (BP 1) | TC2 | Reference |
| blaNDM F | GGTTTGGCGATCTGGTTTTC | 621 | 2 | [16] |
| blaNDM R | CGGAATGGCTCATCACGATC | |||
| blaKPC-2 F | TCGCCGTCTAGTTCTGCTGTCTTG | 800 | 3 | [17] |
| blaKPC-3 R | CAATCCCTCGAGCGCGAGTC | |||
| Genes Encoding for Colistin Resistance | ||||
| Gene | Sequences (5-3′) | Amplicon (BP 1) | TC2 | References |
| MCR-1 F | GATCGGATTGGAGAACCAGA | 343 | 4 | [15] |
| MCR-1 R | ATTTCTGACCGCATTTCCAT | |||
| Broth | Growth | Antimicrobial Supplementation | Number of Isolates | Confirmed E. coli -Positive Isolates | ||
|---|---|---|---|---|---|---|
| Cef1 | Imi2 | Poly B3 | ||||
| MacConkey | 28/90 | 05/28 | 7/28 | 16/28 | 36/71 | 25/71 |
| STGG 4 | 13/90 | 03/13 | 0/13 | 10/13 | 24/71 | 19/71 |
| Antimicrobials | Initials | Susceptibility Profile | |||||
|---|---|---|---|---|---|---|---|
| S 1 | % | I 2 | % | R 3 | % | ||
| Sulfamethoxazole + trimethoprim (25MCG) | SUT | 40 | 93.0 | 1 | 2.4 | 2 | 4.6 |
| Ertapenem (10MCG) | ETP | 36 | 83.7 | 3 | 7.0 | 4 | 9.3 |
| Meropenem (10MCG) | MER | 38 | 88.4 | 3 | 7.0 | 2 | 4.6 |
| Imipenem (10MCG) | IPM | 40 | 93.0 | 3 | 7.0 | 0 | 0.0 |
| Amikacin (30MCG) | AMI | 40 | 93.0 | 3 | 7.0 | 0 | 0.0 |
| Ciprofloxacin (5MCG) | CIP | 31 | 72.0 | 7 | 16.3 | 5 | 11.7 |
| Tetracycline (30MCG) | TET | 40 | 93.0 | 1 | 2.4 | 2 | 4.6 |
| Chloramphenicol (30MCG) | CLO | 40 | 93.0 | 1 | 2.4 | 2 | 4.6 |
| Ceftriaxone (30MCG) | CRO | 40 | 93.0 | 2 | 4.6 | 1 | 2.4 |
| Ceftazidime (30MCG) | CAZ | 37 | 86.1 | 2 | 4.6 | 4 | 9.3 |
| Ampicillin + sulbactam (20MCG) | APS | 38 | 88.4 | 2 | 4.6 | 3 | 7.0 |
| Aztreonam (30MCG) | ATM | 41 | 95.2 | 1 | 2.4 | 1 | 2.4 |
| Amoxicillin/acid-clavulanic (30MCG) | ACM | 39 | 90.6 | 1 | 2.4 | 3 | 7.0 |
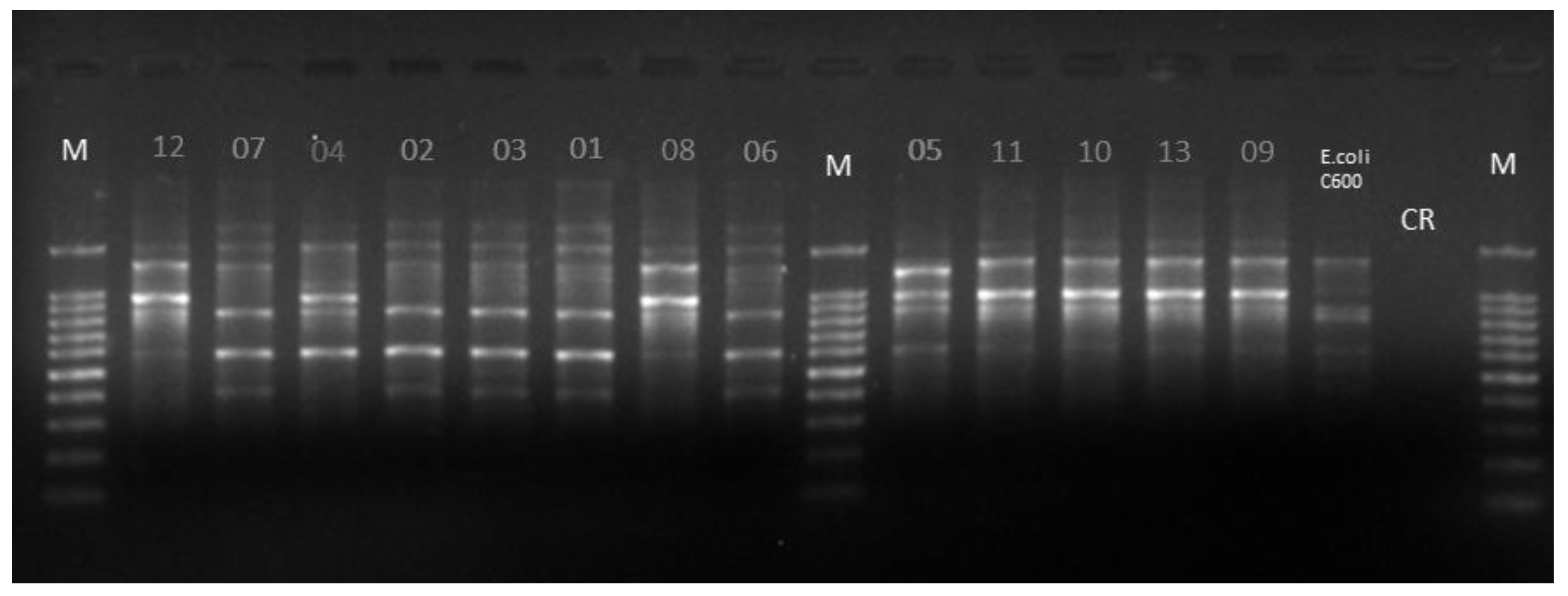
| Sample Number | Species | Animal Id 1 | Enrichment Broth 2 | ATB Supplementation 3 | Genotype 4 |
|---|---|---|---|---|---|
| 01 | Ara ararauna | ARA 2 | STGG | Poli B | II |
| 02 | Ara ararauna | ARA 2 | STGG | Poli B | II |
| 03 | Ara ararauna | ARA 2 | STGG | Poli B | II |
| 04 | Ara macao | ARA 4 | STGG | Poli B | III |
| 05 | Ara chloropterus | ARA 5 | STGG | Poli B | IV |
| 06 | Ara chloropterus | ARA 5 | STGG | Poli B | II |
| 07 | Ara chloropterus | ARA 5 | STGG | Poli B | II |
| 08 | Amazona aestiva | PAP 8 | STGG | Poli B | I |
| 09 | Amazona aestiva | PAP 8 | MAC | Imi | I |
| 10 | Amazona festiva | PAP 9 | STGG | Poli B | I |
| 11 | Amazona amazonica | PAP 11 | STGG | Poli B | I |
| 12 | Amazona amazonica | PAP 11 | STGG | Poli B | I |
| 13 | Amazona aestiva | PAP 12 | MAC | Imi | I |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, G.G.d.C.; Campana, E.H.; Vasconcelos, P.C.; Silva, N.M.V.d.; Santos Filho, L.; Leite, E.L.; Givisiez, P.E.N.; Gebreyes, W.A.; Oliveira, C.J.B.d. Occurrence of KPC-Producing Escherichia coli in Psittaciformes Rescued from Trafficking in Paraíba, Brazil. Int. J. Environ. Res. Public Health 2021, 18, 95. https://doi.org/10.3390/ijerph18010095
Silva GGdC, Campana EH, Vasconcelos PC, Silva NMVd, Santos Filho L, Leite EL, Givisiez PEN, Gebreyes WA, Oliveira CJBd. Occurrence of KPC-Producing Escherichia coli in Psittaciformes Rescued from Trafficking in Paraíba, Brazil. International Journal of Environmental Research and Public Health. 2021; 18(1):95. https://doi.org/10.3390/ijerph18010095
Chicago/Turabian StyleSilva, Gedean Galdino da Cruz, Eloiza Helena Campana, Priscylla Carvalho Vasconcelos, Núbia Michelle Vieira da Silva, Lauro Santos Filho, Elma Lima Leite, Patrícia Emília Naves Givisiez, Wondwossen Abebe Gebreyes, and Celso José Bruno de Oliveira. 2021. "Occurrence of KPC-Producing Escherichia coli in Psittaciformes Rescued from Trafficking in Paraíba, Brazil" International Journal of Environmental Research and Public Health 18, no. 1: 95. https://doi.org/10.3390/ijerph18010095





