Association of Microbiome with Oral Squamous Cell Carcinoma: A Systematic Review of the Metagenomic Studies
Abstract
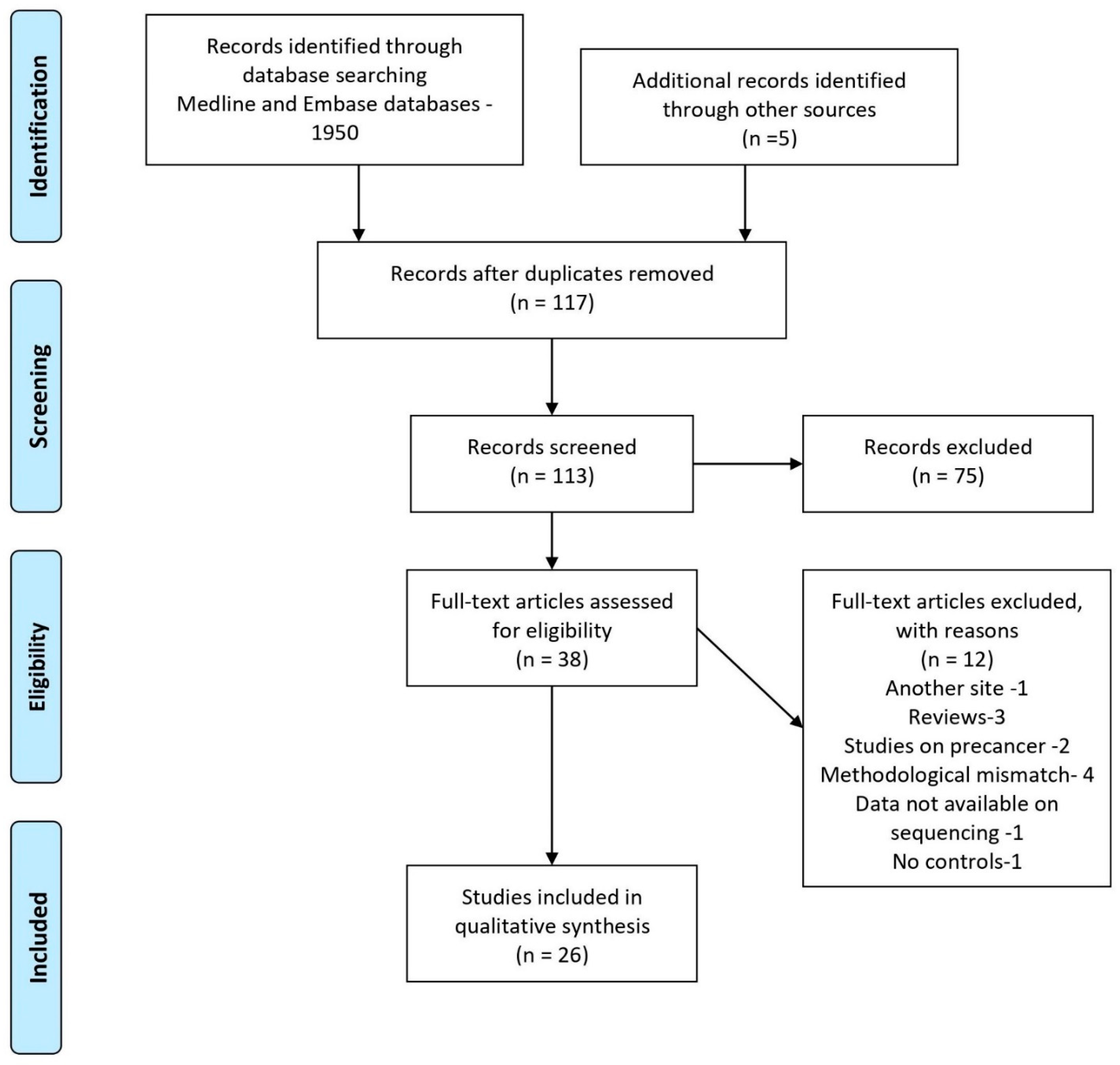
:1. Introduction
2. Materials and Methods
2.1. Design
2.2. Data Sources and Search Strategy
2.3. Eligibility Criteria
2.4. Study Selection and Data Extraction
- study characteristics—author, year, country, study design, sample details;
- outcomes—diversity and richness, relative abundances of various taxa and microbial functions;
- methodology—DNA extraction, amplification, sequencing platforms, and reference.
2.5. Risk of Bias Assessment
3. Results
3.1. Study Characteristics
3.2. Sample Collection and Measurement
3.3. Techniques of DNA Extraction and Sequencing
3.3.1. DNA Extraction
3.3.2. DNA Amplification, Sequencing, and Reference Databases
3.4. Microbial Diversity and Abundance
3.5. Microbial Abundance
3.5.1. Phyla
3.5.2. Classes and Family
3.5.3. Genera
3.5.4. Species
3.6. Microbial Association with other Clinical Factors
3.7. Microbial Functions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fitzsimonds, Z.; Rodriguez-Hernandez, C.; Bagaitkar, J.; Lamont, R. From Beyond the Pale to the Pale Riders: The Emerging Association of Bacteria with Oral Cancer. J. Dent. Res. 2020, 99, 604–612. [Google Scholar] [CrossRef]
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer Statistics, 2010. CA Cancer J. Clin. 2010, 70, 7–30. [Google Scholar] [CrossRef]
- Conway, D.I.; Purkayastha, M.; Chestnutt, I.G. The changing epidemiology of oral cancer: Definitions, trends, and risk factors. Br. Dent. J. 2018, 225, 867–873. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, D.; Kunnath Menon, R.K.; Veettil, S.; George Botelho, M.; Johnson, N.W. Periodontal Diseases as Putative Risk Factors for Head and Neck Cancer: Systematic Review and Meta-Analysis. Cancers 2020, 12, 1893. [Google Scholar] [CrossRef]
- Gopinath, D.; Thannikunnath, B.V.; Neermunda, S.F. Prevalence of Carcinomatous Foci in Oral Leukoplakia: A Clinicopathologic Study of 546 Indian Samples. J. Clin. Diagn. Res. 2016, 10, ZC78–ZC83. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S.; Kujan, O.; Aguirre-Urizar, J.M.; Bagan, J.V.; González-Moles, M.Á.; Kerr, A.R.; Lodi, G.; Mello, F.W.; Monteiro, L.; Ogden, G.R.; et al. Oral potentially malignant disorders: A consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2020. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Yeh, Y.-M.; Yu, H.-Y.; Chin, C.-Y.; Hsu, C.-W.; Liu, H.; Huang, P.-J.; Hu, S.-N.; Liao, C.-T.; Chang, K.-P.; et al. Oral Microbiota Community Dynamics Associated with Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpiński, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsland, D.; Neefjes, J. Bacterial infections and cancer. EMBO Rep. 2018, 19, e46632. [Google Scholar] [CrossRef]
- Gopinath, D.; Menon, R.K.; Banerjee, M.; Yuxiong, R.S.; Botelho, M.G.; Johnson, N.W. Culture-independent studies on bacterial dysbiosis in oral and oropharyngeal squamous cell carcinoma: A systematic review. Crit. Rev. Oncol. 2019, 139, 31–40. [Google Scholar] [CrossRef]
- Bjerre, R.; Bandier, J.; Skov, L.; Engstrand, L.; Johansen, J.D. The role of the skin microbiome in atopic dermatitis: A systematic review. Br. J. Dermatol. 2017, 177, 1272–1278. [Google Scholar] [CrossRef]
- Pushalkar, S.; Mane, S.; Ji, X.; Li, Y.; Evans, C.; Crasta, O.R.; Morse, D.; Meagher, R.; Singh, A.; Saxena, D. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol. Med. Microbiol. 2011, 61, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, B.L.; Kuczynski, J.; Bhattacharya, A.; Huey, B.; Corby, P.M.; Queiroz, E.L.S.; Nightingale, K.; Kerr, A.R.; DeLacure, M.D.; Veeramachaneni, R.; et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE 2014, 9, e98741. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Godoy-Vitorino, F.; Jedlicka, A.; Rodríguez-Hilario, A.; González, H.; Bondy, J.; Lawson, F.; Folawiyo, O.; Michailidi, C.; Dziedzic, A.; et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget 2016, 7, 51320–51334. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Preston, R.; White, J.R.; Godoy-Vitorino, F.; Rodriguez-Hilario, A.; Navarro, K.; Gonzalez, H.; Michailidi, C.; Jedlicka, A.; Canapp, S.; Bondy, J.; et al. High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget 2017, 8, 110931–110948. [Google Scholar]
- Banerjee, S.; Tian, T.; Wei, Z.; Peck, K.N.; Shih, N.; Chalian, A.A.; O’Malley, B.W.; Weinstein, G.S.; Feldman, M.D.; Alwine, J.; et al. Microbial Signatures Associated with Oropharyngeal and Oral Squamous Cell Carcinomas. Sci. Rep. 2017, 7, 4036. [Google Scholar] [CrossRef] [Green Version]
- Bornigen, D.; Ren, B.; Pickard, R.; Li, J.; Ozer, E.; Hartmann, E.M.; Xiao, W.; Tickle, T.; Rider, J.; Gevers, D.; et al. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci. Rep. 2017, 7, 17686. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.B.; Ahn, J.; Fan, X.; Peters, B.A.; Ma, Y.; Yang, L.; Agalliu, I.; Burk, R.D.; Ganly, I.; Purdue, M.P.; et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018, 4, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.M.; Luo, T.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Microbial Communities Associated with Primary and Metastatic Head and Neck Squamous Cell Carcinoma—A High Fusobacterial and Low Streptococcal Signature. Sci. Rep. 2017, 7, 9934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganly, I.; Yang, L.; Giese, R.A.; Hao, Y.; Nossa, C.W.; Morris, L.G.T.; Rosenthal, M.; Migliacci, J.; Kelly, D.; Tseng, W.; et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int. J. Cancer 2019, 145, 775–784. [Google Scholar] [CrossRef]
- Sharma, A.K.; DeBusk, W.T.; Stepanov, I.; Gomez, A.; Khariwala, S.S. Oral Microbiome Profiling in Smokers with and without Head and Neck Cancer Reveals Variations Between Health and Disease. Cancer Prev. Res. 2020, 13, 463–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef]
- Chang, C.; Geng, F.; Shi, X.; Li, Y.; Zhang, X.; Zhao, X.; Pan, Y. The prevalence rate of periodontal pathogens and its association with oral squamous cell carcinoma. Appl. Microbiol. Biotechnol. 2018, 103, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zheng, H.J.; Zhang, C.P. The Oral Microbiota May Have Influence on Oral Cancer. Front. Cell. Infect. Microbiol. 2020, 9, 476. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, L.; Yuan, R.; Yu, X.; Chen, Z.; Yang, F.; Sun, G.; Dong, Q. Signatures of Mucosal Microbiome in Oral Squamous Cell Carcinoma Identified Using a Random Forest Model. Cancer Manag. Res. 2020, 12, 5353–5363. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-H.; Chen, H.-M.; Yang, S.-F.; Wen-Liang, C.; Peng, C.-Y.; Tzu-Ling, Y.; Tsai, L.-L.; Wu, B.-C.; Hsin, C.-H.; Huang, C.-N.; et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Sci. Rep. 2017, 7, 16540. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, J.-R.; Chang, C.-C.; Lee, W.-T.; Huang, C.-C.; Ou, C.-Y.; Tsai, S.-T.; Chen, K.-C.; Huang, J.-S.; Wong, T.-Y.; Lai, Y.-H.; et al. The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogens 2018, 39, 778–787. [Google Scholar] [CrossRef] [Green Version]
- Panda, M.; Rai, A.K.; Rahman, T.; Das, A.; Das, R.; Sarma, A.; Kataki, A.C.; Chattopadhyay, I. Alterations of salivary microbial community associated with oropharyngeal and hypopharyngeal squamous cell carcinoma patients. Arch. Microbiol. 2019, 202, 785–805. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Panda, M.; Das, A.K.; Rahman, T.; Das, R.; Das, K.; Sarma, A.; Kataki, A.C.; Chattopadhyay, I. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch. Microbiol. 2021, 203, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Al-Hebshi, N.N.; Nasher, A.T.; Maryoud, M.Y.; Homeida, H.E.; Chen, T.; Idris, A.M.; Johnson, N.W. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 2017, 7, 1834. [Google Scholar] [CrossRef] [Green Version]
- Mok, S.F.; Karuthan, C.; Cheah, Y.K.; Ngeow, W.C.; Rosnah, Z.; Yap, S.F.; Ong, H.K.A. The oral microbiome community variations associated with normal, potentially malignant disorders and malignant lesions of the oral cavity. Malays. J. Pathol. 2017, 39, 1–15. [Google Scholar]
- Lim, Y.; Fukuma, N.; Totsika, M.; Kenny, L.; Morrison, M.; Punyadeera, C. The Performance of an Oral Microbiome Biomarker Panel in Predicting Oral Cavity and Oropharyngeal Cancers. Front. Cell. Infect. Microbiol. 2018, 8, 267. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, K.; Shimizu, D.; Hirabayashi, S.; Ueda, S.; Miyabe, S.; Oh-Iwa, I.; Nagao, T.; Shimozato, K.; Nomoto, S. Changes in oral microbial profiles associated with oral squamous cell carcinoma vs leukoplakia. J. Investig. Clin. Dent. 2019, 10, e12445. [Google Scholar] [CrossRef]
- Takahashi, Y.; Park, J.; Hosomi, K.; Yamada, T.; Kobayashi, A.; Yamaguchi, Y.; Iketani, S.; Kunisawa, J.; Mizuguchi, K.; Maeda, N.; et al. Analysis of oral microbiota in Japanese oral cancer patients using 16S rRNA sequencing. J. Oral Biosci. 2019, 61, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.; Al-Hebshi, N.; Perera, I.; Ipe, D.; Ulett, G.; Speicher, D.; Chen, T.; Johnson, N. Inflammatory Bacteriome and Oral Squamous Cell Carcinoma. J. Dent. Res. 2018, 97, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Vesty, A.; Gear, K.; Biswas, K.; Radcliff, F.J.; Taylor, M.W.; Douglas, R.G. Microbial and inflammatory-based salivary biomarkers of head and neck squamous cell carcinoma. Clin. Exp. Dent. Res. 2018, 4, 255–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Jagannathan, N. Oral field cancerization: An update on current concepts. Oncol. Rev. 2014, 8, 244. [Google Scholar] [CrossRef] [Green Version]
- Whisner, C.; Aktipis, C.A. The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One Another’s Growth. Curr. Nutr. Rep. 2019, 8, 42–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.-H.; Li, B.-B.; Wang, B.; Zhao, J.; Zhang, X.-Y.; Li, T.-T.; Li, W.-B.; Tang, D.; Qiu, M.-J.; Wang, X.-C.; et al. The role of Fusobacterium nucleatum in colorectal cancer: From carcinogenesis to clinical management. Chronic Dis. Transl. Med. 2019, 5, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, D.; Menon, R.K. Unravelling the molecular signatures in HNSCC: Is the homogenous paradigm becoming obsolete? Oral Oncol. 2018, 82, 195. [Google Scholar] [CrossRef]
- Gopinath, D.; Menon, R.K.; Wie, C.C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; Botelho, M.G.; et al. Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome. J. Oral Microbiol. 2021, 13, 1857998. [Google Scholar] [CrossRef]
- Gopinath, D.; Menon, R.K.; Wie, C.C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; Botelho, M.G.; et al. Differences in the bacteriome of swab, saliva, and tissue biopsies in oral cancer. Sci. Rep. 2021, 11, 1181. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, D.; Menon, R.K. Increasing Reproducibility in Oral Microbiome Research. In Oral Microbiome. Methods in Molecular Biology; Adami, G., Ed.; Humana Press: New York, NY, USA, 2021; in press. [Google Scholar]
- Huybrechts, I.; Zouiouich, S.; Loobuyck, A.; Vandenbulcke, Z.; Vogtmann, E.; Pisanu, S.; Iguacel, I.; Scalbert, A.; Indave, I.; Smelov, V.; et al. The Human Microbiome in Relation to Cancer Risk: A Systematic Review of Epidemiologic Studies. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.K.; Gopinath, D. Eliminating bias and accelerating the clinical translation of oral microbiome research in oral oncology. Oral Oncol. 2018, 79, 84–85. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groeger, S.; Jarzina, F.; Domann, E.; Meyle, J. Porphyromonas gingivalis activates NFκB and MAPK pathways in human oral epithelial cells. BMC Immunol. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallimidi, A.B.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, L.; Jermanus, C.; Barbetta, B.; Choi, C.; Verbeke, P.; Ojcius, D.M.; Yilmaz, Ö. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Mol. Oral Microbiol. 2010, 25, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, O.; Jungas, T.; Verbeke, P.; Ojcius, D.M. Activation of the Phosphatidylinositol 3-Kinase/Akt Pathway Contributes to Survival of Primary Epithelial Cells Infected with the Periodontal Pathogen Porphyromonas gingivalis. Infect. Immun. 2004, 72, 3743–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asoudeh-Fard, A.; Barzegari, A.; Dehnad, A.; Bastani, S.; Golchin, A.; Omidi, Y. Lactobacillus plantarum induces apoptosis in oral cancer KB cells through upregulation of PTEN and downregulation of MAPK signalling pathways. BioImpacts 2017, 7, 193–198. [Google Scholar] [CrossRef]
- Abdulkareem, A.; Shelton, R.M.; Landini, G.; Cooper, P.; Milward, M.R. Periodontal pathogens promote epithelial-mesenchymal transition in oral squamous carcinoma cells in vitro. Cell Adhes. Migr. 2017, 12, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Liu, X.; Zhou, Y.; Acharya, A.; Savkovic, V.; Xu, C.; Wu, N.; Deng, Y.; Hu, X.; Li, H.; et al. Shared genetic and epigenetic mechanisms between chronic periodontitis and oral squamous cell carcinoma. Oral Oncol. 2018, 86, 216–224. [Google Scholar] [CrossRef] [Green Version]
- Kamarajan, P.; Ateia, I.; Shin, J.M.; Fenno, J.C.; Le, C.; Zhan, L.; Chang, A.; Darveau, R.; Kapila, Y.L. Periodontal pathogens promote cancer aggressivity via TLR/MyD88 triggered activation of Integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PLOS Pathog. 2020, 16, e1008881. [Google Scholar] [CrossRef] [PubMed]
- Duvallet, C.; Gibbons, S.M.; Gurry, T.; Irizarry, R.A.; Alm, E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017, 8, 1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulkareem, A.A.; Shelton, R.M.; Landini, G.; Cooper, P.R.; Milward, M.R. Potential role of periodontal pathogens in compromising epithelial barrier function by inducing epithelial-mesenchymal transition. J. Periodontal Res. 2018, 53, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez, C.; David, J.M.; Palena, C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin. Cancer Biol. 2017, 47, 177–184. [Google Scholar] [CrossRef]
- Stashenko, P.; Yost, S.; Choi, Y.; Danciu, T.; Chen, T.; Yoganathan, S.; Kressirer, C.; Ruiz-Tourrella, M.; Das, B.; Kokaras, A.; et al. The Oral Mouse Microbiome Promotes Tumorigenesis in Oral Squamous Cell Carcinoma. mSystems 2019, 4, e00323-19. [Google Scholar] [CrossRef] [Green Version]
| No. | Author, Year | Sample Type | Age (Mean/Median) | Nature of Control | Study Population Size (Case, Control) | Other Clinical Features Studied | Results: Diversity and Richness | Bacterial Taxa Associated with Tumors and Controls |
|---|---|---|---|---|---|---|---|---|
| 1 | Pushalkar et al. 2011 [12] | Saliva | Age: >50 | Healthy controls | Case: 3 Control: 2 | Smoking: at least one pack of cigarettes a day. Alcohol: more than five drinks a day | Increase in diversity in the control group | Streptococcus, Gemella, Rothia, Peptostreptococcus, Lactobacillus, Porphyromonas in OSCC group. Prevotella, Neisseria, Leptotrichia, Capnocytophaga, Actinobacillus and Oribacterium in the control group. |
| 2 | Schmidt et al. 2014 [13] | Oral swab | Cancer: 62 Control: 31 | Contralateral normal regions of the oral cavity | Case: 50 Control: 20 | N/A | N/A | Decreased relative abundance of Streptococcus and Rothia in the tumor group. Increased relative abundance of Fusobacterium in the tumor group. |
| 3 | Guerrero-Preston et al. 2016 [14] | Tissue and saliva | OSCC: 66 OPSCC: 62 Control: N/A | Healthy controls without smoking and drinking | Case: 19 Control: 25 | N/A | Decrease in richness and diversity in cases Significant increase of certain Lactobacillus and Weeksellaceae in HPV+ samples Significant abundance of Eikenella, Neisseria, and Leptotrichia in HPV– samples | Significant increase of Lactobacillus, Strepcoccus, Staphylococcus and Parvimonas in HNSCC group. Significant abundance of Haemophilus, Neisseria, Gemellaceae and Aggregatibacter in the control group. |
| 4 | Al-Hebshi et al. 2017 [30] | Tissue | Case: 53.6 ± 10.4 Controls: 52.3 ± 8.9 | Healthy, gender- and age-matched controls | Case: 20 Control: 20 | Smoking: shammah (smokeless tobacco) | Similar species richness and α-diversity in both groups (tissue biopsies) | Significant abundance of Fusobacterium in OSCC group. Significant abundance of Streptococcus and Rothia in the control group. |
| 5 | Banerjee et al. 2017 [16] | Tissue (FFPE) | N/A | Adjacent non-tumorous tissues, healthy controls | Cases: 100 Controls: 40 | N/A | N/A | Significant abundance of Proteobacteria Brevundimonas, Actinobacteria Mobiluncus, Frateuria, Caulobacter, Actinomyces and Aeromonas in OSCC group. Significant abundance of Actinomyces in the control group. |
| 6 | Bornigen et al. 2017 [17] | Oral rinse | 58 | Healthy controls | Case: 121 Control: 242 | N/A | Increase in diversity in smokers | Significant abundance of Dialister in the oral cancer group. Significant decrease of Actinomycetales and Lactobacillales in the oral cancer group. |
| 7 | Guerrero-Preston et al. 2017 [15] | Oral rinse | OSCC: 66 OPSCC: 62 Controls: N/A | Healthy controls | Case: 19 (HNSCC) Control: 25 | N/A | Lactobacillus gasseri/johnsonii, lactobacillus vaginalis in HPV+ patient | Significant increase of Lactobacillus spp, Streptococcus mutans, Fusobacterium nucleatum and Parvimonas micra in HNSCC group. Significant decrease of leptotrichia trevisanii, leptotrichia hofstadii and buccalis in HNSCC group |
| 8 | Lee et al. 2017 [26] | Saliva | Cancer Age: 53 ± 10 Control Age: 52 ± 14 | Healthy controls | Case: 125 Control: 127 | Betel nut chewing history Cigarette smoking history | N/A | Significant increase of Bacteroides, Escherichia, Cloacibacillus, Gemmiger, Oscillospira and Roseburia in cancer group. |
| 9 | Mok et al. 2017 [31] | Oral swab | Age: >20 | Healthy controls | Case: 9 Control: 9 | Smoking history Alcohol consumption history | Similar species richness between cancer and control group | Significant abundance of Streptococcus and Veillonella in the control group. Significant abundance of Neisseria, Gemella and Granulicatella in cancer group. |
| 10 | Shin et al. 2017 [19] | Tissue | Age: 59 ± 5.6 | Adjacent non-tumorous tissue | Case: 34 Control: 24 | N/A | Increase in α-diversity in the control group Decrease in β-diversity in tumor | Significant abundance of Firmicutes and Actinobacteria in the control group. Significant abundance of Fusobacteria in primary HNSCC group. |
| 11 | Zhao et al. 2017 [22] | Oral swabbing | Median age: 62 | Adjacent non-tumorous tissue | Case: 80 Control: 80 | N/A | Increase in diversity in cancer group | Significant increase of Spirochaetes, Fusobacteria, Bacteroidetes, Mycoplasma, Treponema, Campylobacter, Eikenella, Centipeda, Lachnospiraceae_G_7, Alloprevotella, Fusobacterium, Selenomonas, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, Parvimonas, Capnocytophaga and Peptostreptococcaceae_XI_G_7 in the cancer group. Significant increase of Firmicutes, Actinobacteria, Megasphaera, Stomatobaculum, Granulicatella, Lautropia, Veillonella, Streptococcus, Scardovia, Rothia, and Actinomyces in the control group. |
| 12 | Hayes et al. 2018 [18] | Oral rinse | Case: 60–70 Control: 60–70 | Healthy controls | Case: 129 Control: 254 | N/A | N/A | Significant increase of Actinobacteria in HNSCC group. Significant decrease of Parvimonas micra and Neisseria sicca in oral cancer group. Significant decrease of Genus Corynebacterium up to order Corynebacteriales, genus Kingella up to phylum Proteobacteria, Prevotella nanceiensis, Capnocytophaga leadbetteri and Selenomonas sputigena in HNSCC group. Significant increase of Actinomyces (oral taxon_170) in the oral cancer group. |
| 13 | Hsiao et al. 2018 [27] | Saliva | Cases: ≥20 Control: ≥20 | Healthy controls | Case: 138 Control: 151 | Betel nut chewing history Cigarette smoking history Alcohol consumption history Oral hygiene status | Significant increase of Prevotella intermedia in alcohol consumers and betel nut chewers Significant increase of F.nucleatum in smokers Significant increase of Prevotella tannerae and F.nucleatum in poor dental care group | Significant increase of Prevotella tannerae, F. nucleatum and Prevotella intermedia in the cancer group. |
| 14 | Lim et al. 2018 [32] | Oral rinse | Case: 65 Control: 20–60 | Healthy controls | Case: 63 Control: 20 | Smoking history Alcohol consumption history HPV status | Decrease in diversity in the cancer group | Significant increase of Oribacterium in OCC and OPC group. Significant decrease of Rothia, Haemophilus, Corynebacterium, Paludibacter, Porphyromonas, and Capnocytophaga in OCC and OPC group. Significant increase of Actinomyces, Parvimonas, Selenomonas, and Prevotella in OCC group. Significant increase of Haemophilus and Gemella in HPV+ group. |
| 15 | Perera et al. 2018 [35] | Tissue | Case: Age: 61.00 ± 9.5 Controls: Age: 50.58 ± 13.5 | Healthy controls | Case: 25 Control: 27 | Betel nut chewing history Smoking history Alcohol consumption history Oral hygiene and periodontal status Missing teeth | Decrease in diversity in cancer group | Significant increase of Capnocytophaga, Pseudomonas, Atopobium, Campylobacter concisus, Prevotella salivae, Prevotella loeschii, Fusobacterium oral taxon 204, F. nucleatum subsp. polymorphum, Streptococcus dysgalactiae, Citrobacter koseri, and Pseudomonas aeruginosa in OSCC group. Significant increase of Lautropia, Staphylococcus, Propionibacterium, Sphingomonas, Streptococcus parasanguinis, Streptococcus mitis, Streptococcus sp oral taxon 070, Streptococcus sp oral taxon 423, Streptococcus sp oral taxon 431, Streptococcus agalactiae, Rothia dentocariosa, Rothia mucilaginosa, Lautropia mirabilis, Leptotrichia oral taxon 225, and Staphylococcus epidermidis in control group. |
| 16 | Vesty et al. 2018 [36] | Saliva | Case: 49 to 81 Control: 20 to 35 | Healthy controls (non-smokers) | Case: 23 Control: 7 | Fungal communities concentrations of inflammatory cytokines | Increase in fungal diversity in dentally compromised group. IL-1 beta and Lachnoanaerobaculum as well as Actinomyces and IL-8 had negative correlations | Significant abundance of Treponema in cases. Significant abundance of Actinomyces and Fusobacterium in controls |
| 17 | Yang C. et al. 2018 [7] | Oral rinse | Case Age: 50–60 Control: Age: 35–35 | Healthy controls | Case: 197 Control: 51 | TNM stage Betel nut chewing history Alcohol consumption history | Significant increase of F. alocis in smokers | Significant abundance of Fusobacterium periodonticum, Parvimonas micra, Streptococcus constellatus, Haemophilus influenza and Filifactor alocis in OSCC (Stage 4) group. Significant abundance of Haemophilus parainfluenzae, Porphyromonas pasteri, Veillonella parvula and Actinomyces odontolyticus in control group. |
| 18 | Chang et al. 2019 [23] | Tissue (FFPE) | Case: 57.4 ± 10.4 Control: 55.4 ± 10.2 | Healthy controls | Case: 61 Control: 30 | Smoking history | Significant increase of P. gingivalis in clinical stage III-IV, low degree of tissue differentiation and lymph node metastasis group. | Significant increase of F. nucleatum and P. gingivalis in cancer group. |
| 19 | Ganly et al. 2019 [20] | Oral rinse | Mean age: 21 | Patients with benign or malignant thyroid nodules | Cases: 26 Controls: 12 | Smoking history Alcohol consumption history | N/A | Significant abundance of Alloprevotella, Fusobacterium and Prevotella in OSCC group. Significant abundance of Streptococcus in control group. |
| 20 | Hashimoto et al. 2019 [33] | Saliva | Case: 51 Control: 31 | Healthy controls | Case: 12 Control: 4 | N/A | N/A | Porphyromonas gingivalis in OSCC group. Significant abundance of Streptococcus anginosus in OSCC group. |
| 21 | Takahashi et al. 2019 [34] | Saliva | Case: 63.7 Control: 65.1 | Healthy controls (40 years of age) | Case: 60 Control: 80 | Age Sex Smoking history Alcohol consumption history Denture usage | Increase in diversity in OSCC group Decrease in diversity in OSCC Abundance of Peptostreptococcus in females Abundance of Haemophilus in males and alcohol consumers | Significant abundance of Peptostreptococcus, Fusobacterium, Alloprevotella, Capnocytophaga in OSCC group. Significant abundance of Rothia and Haemophilus in the control group. |
| 22 | Panda et al. 2020 [28] | Saliva | Case: 48–58 Control: 40 to 60 | Healthy controls | Case: 15 Control: 10 | Betel nut chewing history Smoking (smokeless tobacco) history | Increase in diversity in the control group | Significant abundance of Rothia mucilaginosa, Aggregatibacter segnis, Veillonella dispar, Prevotella nanceiensis, Rothia aeria, Capnocytophage ochracea, Neissseria bacilliformis, Prevotella nigrescens and Selenomonas noxia in the control group. Significant abundance of Haemophilus parainfluenzae, Haemophilus influenzae and Prevotella copri in the cancer group. Streptococcus anginosus was found only in oropharyngeal cancer tissues. |
| 23 | Sharma et al. 2020 [21] | Oral brushings | Cases: 58 Controls: 48 | Healthy controls | Case: 27 Control: 24 | Smoking | Increase in richness in OSCC group | Higher relative abundance of Stenotrophomonas ruminococcus and family Comamonadaceae in cases Tannerella, Capnocytophaga, Selenomonas, Veillonella, and Kingella, were higher in controls |
| 24 | Zhang et al. 2020 [24] | Tissue | Median age: 61 | Adjacent non-tumorous tissue | Case: 50 Control: 50 | Betel nut chewing history Smoking history Alcohol consumption history | Increase in diversity in OSCC group | Significant abundance of F.nucleatum, Prevotella intermedia, Aggregatibacter segnis, Campylobacter rectus, Capnocytophaga leadbetteri, Gemella morbillorum, Peptostreptococcus stomatis, Peptococcus sp. and Porphyromonas catoniae in OSCC group. Significant abundance of Corynebacterium matruchotii, Granulicatella elegans, Granulicatella adicens and Streptococcus oralis in control group. |
| 25 | Zhou et al. 2020 [25] | Tissue | 61.1 ± 12.4 | Adjacent paracancerous tissue 2 cm around edge of tumour | Case: 24 Control: 24 | N/A | N/A | Significant increase of Fusobacterium, Parvimonas, Peptostreptococcus and Streptococcus in cancer group. Significant decrease of Arthrobacter, Brevundimonas, Microbacterium, Mucispirillum, Paenibacillus and Streptophyta in cancer group. |
| 26 | Rai et al. 2020 [29] | Saliva | Case: 55.32 Control: 50.38 | Healthy controls | Case: 11 Control: 10 | Betel nut chewing history Tobacco chewing history Tobacco smoking history Alcohol consumption history Family history of cancer | N/A | Significant increase of Prevotella melaninogenica, Streptococcus anginosus, Veillonella parvula, Prevotella pallens, Porphyromonas endodontalis, Prevotella nanceiensis, Dialister sp., Campylobacter ureolyticus, Fusobacterium sp., Prevotella nigrescens, Neisseria bacilliformis, and Peptostreptococcus anaerobius in OSCC group. Significant increase of Neisseria subflava, Veillonella dispar, Rothia dentocariosa, and Rothia. Mucilaginosa in control group. Rare species of Ruminococcus gnavus, Lactobacillus plantarum, Bacteroides ovatus, Parabacteroides distasonis, Filifactor sp. and Dorea sp. found in saliva of OSCC group. |
| No. | Author, Year | Sample | Method of DNA Extraction | DNA Amplification | Sequencing | Reference Databases |
|---|---|---|---|---|---|---|
| 1 | Pushalkar et al. 2011 [12] | Saliva | DNA Purification Kit (MasterPure) | V4–V5 region. | 454 parallel DNA sequencing | RDP II |
| 2 | Schmidt et al. 2014 [13] | Oral swab | DNeasy Blood and Tissue Kit (Qiagen) | V4 region | 454 pyrosequencing | Greengenes |
| 3 | Guerrero-Preston et al. 2016 [14] | Tissue and saliva | Phenol-chloroform method | V3–V5 region | Roche/454 GS pyrosequencing | RDP |
| 4 | Al-Hebshi et al. 2017 [30] | Tissue | DDK DNA Isolation kit | V1–V3 region | Illumina MiSeq | HOMD |
| 5 | Banerjee et al. 2017 [16] | Tissue | All Prep DNA/RNA FFPE Kit | NA | Illumina MiSeq | RDP |
| 6 | Bornigen et al. 2017 [17] | Oral rinse | QIAsymphony virus/Bacteria Midi Kit | V4 variable region | Illumina MiSeq | Greengenes |
| 7 | Guerrero-Preston et al. 2017 [15] | Tissue and saliva | Phenol-chloroform method | V3–V5 region | Roche/454 GS pyrosequencing | RDP and Resphera Insight |
| 8 | Lee et al. 2017 [26] | Saliva | QIAamp DNA Blood Mini Kit | V4 region | Illumina MiSeq | SILVA |
| 9 | Mok et al. 2017 [31] | Oral swab | EURx commercial kit with modifications | V6–V9 region | NA | GenBank |
| 10 | Shin et al. 2017 [19] | Tissue | RNeasy Mini, RNA Isolation Kit (Qiagen) | V4 variable region | Ion Torrent Personal Genome Machine (PGM) | Greengenes |
| 11 | Zhao et al. 2017 [22] | Swab | QIAmp DNA Mini Kit | V4–V5 region | Illumina MiSeq | RDP |
| 12 | Hayes et al. 2018 [18] | Oral rinse | PowerSoil DNA Isolation Kit (MO BIO) | V3–V4 regions | 454 FLX Titanium pyrosequencing system (Roche) | HOMD |
| 13 | Hsiao et al. 2018 [27] | Saliva | QIAamp MinElute Virus Spin Kit | V3–V5 regions | Illumina MiSeq | RDP |
| 14 | Lim et al. 2018 [32] | Oral rinse | Maxwell® 16 LEV Blood DNA kit | V6–V8 region | Illumina MiSeq | Greengenes |
| 15 | Perera et al. 2018 [35] | Tissue | Gentra Puregene Tissue kit (Qiagen) | V1–V3 region | Illumina MiSeq | Species-level taxonomy assignment algorithm (BLASTN) |
| 16 | Vesty et al. 2018 [36] | Saliva | Phenol-chloroform based DNA extraction | V3–V4 region | Illumina MiSeq | Greengenes |
| 17 | Yang et al. 2018 [7] | Oral rinse | QIAamp DNA Microbiome Kit | V3–V4 region | Illumina MiSeq | Greengenes |
| 18 | Chang et al. 2019 [23] | Tissue | QIAampFast DNA Stool Mini Kit | V3-V4 region | Illumina MiSeq | NCBI |
| 19 | Ganly et al. 2019 [20] | Oral rinse | Modified QIAGEN DNA Extraction Method | V3 and V4 regions | 454 FLX platform | HOMD |
| 20 | Hashimoto et al. 2019 [33] | Saliva | NA | V4 region | Illumina MiSeq | Greengenes |
| 21 | Takahashi et al. 2019 [34] | Saliva | Gene Prep Star PI-80X device | V3–V4 region | Illumina MiSeq | SILVA 128 |
| 22 | Panda et al. 2020 [28] | Saliva | Qiagen DNeasy Blood and Tissue Kit | V3–V4 region | Illumina MiSeq | HOMD |
| 23 | Sharma et al. 2020 [21] | Oral brushings | DNA Purification Kit (Qiagen). | V4—region | Illumina MiSeq | Greengenes (v 13.8) |
| 24 | Zhang et al. 2020 [24] | Tissue | TIANamp Swab DNA Kit | V3–V4 region | Illumina MiSeq | RDP |
| 25 | Zhou et al. 2020 [25] | Tissue | NA | V3–V4 region | Illumina PE250 | Greengenes (v13.5) |
| 26 | Rai et al. 2020 [29] | Saliva | Qiagen DNeasy Blood and Tissue Kit | V3–V4 region | Illumina MiSeq | Greengenes (v 13.8) |
| No. | Author, Year | Sample | Microbial Functions Associated with Tumors and Controls |
|---|---|---|---|
| 1 | Zhao et al. 2017 [22] | Swabs | Translation, metabolism of cofactors and vitamins, metabolism of terpenoids and polyketides, replication and repair in cases |
| 2 | Al-Hebshi et al. 2017 [30] | Tissue | Bacterial mobility, flagellar assembly, bacterial chemotaxis, and LPS biosynthesis in cases DNA repair, glycolysis/gluconeogenesis, and biosynthesis of amino acids in controls |
| 3 | Perera et al. 2018 [35] | Tissue | Lipopolysaccharide biosynthesis, peptidases, carbon fixation in photosynthetic organisms in cases Base excision repair, glycolysis/gluconeogenesis, and biosynthesis of amino acids in controls |
| 4 | Yang et al. 2018 [7] | Oral rinse | Cytoskeleton proteins, methane metabolism, carbon fixation in photosynthetic organisms, restriction enzymes in cases. Amino acid synthesis and metabolism in controls |
| 5 | Zhang et al. 2020 [24] | Tissue | Proinflammatory bacterial component, such as lipopolysaccharide biosynthesis; metabolism of cofactors and vitamins, such as porphyrin and chlorophyll metabolism in cancer cases. |
| 6 | Zhou et al. 2020 [25] | Tissue | Methane metabolism, glucose-related metabolisms, such as phosphotransferase system (PTS) and glycolysis, were significantly enriched in cancer cases. |
| 7 | Sharma et al. 2020 [21] | Oral brushings | Xenobiotic and amine degradation in cases and sugar degradation pathways in controls |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su Mun, L.; Wye Lum, S.; Kong Yuiin Sze, G.; Hock Yoong, C.; Ching Yung, K.; Kah Lok, L.; Gopinath, D. Association of Microbiome with Oral Squamous Cell Carcinoma: A Systematic Review of the Metagenomic Studies. Int. J. Environ. Res. Public Health 2021, 18, 7224. https://doi.org/10.3390/ijerph18147224
Su Mun L, Wye Lum S, Kong Yuiin Sze G, Hock Yoong C, Ching Yung K, Kah Lok L, Gopinath D. Association of Microbiome with Oral Squamous Cell Carcinoma: A Systematic Review of the Metagenomic Studies. International Journal of Environmental Research and Public Health. 2021; 18(14):7224. https://doi.org/10.3390/ijerph18147224
Chicago/Turabian StyleSu Mun, Lee, See Wye Lum, Genevieve Kong Yuiin Sze, Cheong Hock Yoong, Kwek Ching Yung, Liong Kah Lok, and Divya Gopinath. 2021. "Association of Microbiome with Oral Squamous Cell Carcinoma: A Systematic Review of the Metagenomic Studies" International Journal of Environmental Research and Public Health 18, no. 14: 7224. https://doi.org/10.3390/ijerph18147224
APA StyleSu Mun, L., Wye Lum, S., Kong Yuiin Sze, G., Hock Yoong, C., Ching Yung, K., Kah Lok, L., & Gopinath, D. (2021). Association of Microbiome with Oral Squamous Cell Carcinoma: A Systematic Review of the Metagenomic Studies. International Journal of Environmental Research and Public Health, 18(14), 7224. https://doi.org/10.3390/ijerph18147224








