Characterisation of Environmental Biofilms Colonising Wall Paintings of the Fornelle Cave in the Archaeological Site of Cales
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling Points
2.3. Light and Scanning Electron Microscopy Analyses
2.4. X-ray Diffraction (XRD) Analysis
2.5. Molecular Analyses
2.5.1. Culture-Dependent Characterisation of Biological Community
2.5.2. Automated Ribosomal Intergenic Spacer Analysis (ARISA) Capillary Electrophoresis and Community Analyses
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barton, H.A. Introduction to cave microbiology: A review for the non-specialist. J. Cave Karst Stud. 2006, 68, 43–54. [Google Scholar]
- Kováč, L. Caves as Oligotrophic Ecosystems. In Cave Ecology; Moldovan, O.T., Kováč, L., Halse, S., Eds.; Springer: Cham, Switzerland, 2018; pp. 297–307. ISBN 978-3-319-98852-8. [Google Scholar]
- Braack, L.E.O. Arthropod inhabitants of a tropical cave “Island” environment provisioned by bats. Biol. Conserv. 1989, 48, 77–84. [Google Scholar] [CrossRef]
- Falasco, E.; Ector, L.; Isaia, M.; Wetzel, C.; Hoffmann, L.; Bona, F. Diatom flora in subterranean ecosystems: A review. Int. J. Speleol. 2014, 43, 231–251. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, L. Caves and Other Low-Light Environments: Aerophitic Photoautotrophic Microorganisms. In Encyclopedia of Environmental Microbiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003. [Google Scholar] [CrossRef]
- Ghosh, S.; Kuisiene, N.; Cheeptham, N. The cave microbiome as a source for drug discovery: Reality or pipe dream? Biochem. Pharmacol. 2017, 134, 18–34. [Google Scholar] [CrossRef]
- Grobbelaar, J.U. Lithophytic algae: A major threat to the karst formation of show caves. J. Appl. Phycol. 2000, 12, 309–315. [Google Scholar] [CrossRef]
- Summers Engel, A. Microbial Diversity of Cave Ecosystems. Geomicrobiol. Mol. Environ. Perspect. 2010, 219–238. [Google Scholar] [CrossRef]
- Albertano, P. Epilithic algal communities in hypogean environment. Giorn. Bot. Ital. 1993, 127, 385–392. [Google Scholar] [CrossRef]
- Stal, L.J. Cyanobacterial mats and stromatolites. In The Ecology of the Cyanobacteria: Their Diversity in Space and Time; Whitton, B.A., Potts, M., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 61–120. [Google Scholar]
- Guillitte, O. Bioreceptivity: A new concept for building ecology studies. Sci. Total Environ. 1995, 167, 215–220. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Piñar, G.; Lubitz, W.; Rölleke, S. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 2002, 211, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Cuezva, S.; Sanchez-Moral, S.; Saiz-Jimenez, C.; Cañaveras, J.C. Microbial Communities and Associated Mineral Fabrics in Altamira Cave, Spain. Int. J. Speleol. 2009, 38, 83–92. [Google Scholar]
- Groth, I.; Vettermann, R.; Schuetze, B.; Schumann, P.; Saiz-Jimenez, C. Actinomycetes in Karstic caves of northern Spain (Altamira and Tito Bustillo). Microbiol. Methods 1999, 36, 115–122. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Piñar, G.; Lubitz, W.; Rolleke, S. Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ. Microbiol. 2002, 4, 392–400. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Piñar, G.; Lubitz, W.; Rölleke, S. Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonín and La Garma). FEMS Microbiol. Ecol. 2004, 47, 235–247. [Google Scholar] [CrossRef]
- Groth, L.; Laiz, S.; Sanchez-Moral, J.C.; Cañveras, C.; Saiz-Jimenez, I.P.S. Geomicrobiological Study of the Grotta dei Cervi, Porto Badisco, Italy. Geomicrobiol. J. 2001, 18, 241–258. [Google Scholar] [CrossRef]
- Zona, M. Calvi Antica e Moderna; Miranda: Napoli, Italy, 1820. [Google Scholar]
- Carcaiso, E. Storia Dell’Antica Cales; Museo Provinciale Campano: Capua, Italy, 1980. [Google Scholar]
- Piazza, S. Pittura Rupestre Medievale Lazio e Campania Settentrionale (Secoli VI–XIII); Publications de l’École française de Rome, Ed.; Collection de l’École française de Rome: Rome, Italy, 2006. [Google Scholar]
- Bonacci, C. Cales: Un’Area Archeologica da Riscoprire; Vertigo: Roma, Italy, 2013. [Google Scholar]
- Passaro, C. Cales: Dalla Cittadella Medievale Alla Città antica: Recenti Scavi e Nuove Acquisizioni; Grafiche Mincione Sparanise: Sparanise, Italy, 2009. [Google Scholar]
- Calvi Risorta (CE), Grotta delle Fornelle. Available online: https://care.huma-num.fr/it/index.php?title=CALVI_RISORTA_(CE),_Grotta_delle_Fornelle&oldid=5284 (accessed on 29 July 2021).
- Carotti, A. Gli Affreschi Della Grotta Delle Fornelle a Calvi Vecchia; Luca, C.-D., Ed.; Consiglio Nazionale delle Ricerche: Rome, Italy, 1974. [Google Scholar]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Pröschold, T.; Leliaert, F. Systematics of the green algae: Conflict of classic and modern approaches. In Unravelling the Algae: The Past, Present and Future of Algal Systematics; Brodie, J., Lewis, J., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 123–153. [Google Scholar]
- Starr, R.C.; Zeikus, J.A. UTEX: The culture collection of algae at the University of Texas at Austin. J. Phycol. 1987, 23, 1–47. [Google Scholar]
- Rippka, R.; Herdman, H. Pasteur Culture Collection of Cyanobacterial Strains in Axenic Culture. In Catalogue & Taxonomic Handbook, Catalogue of Strains; Institut Pasteur: Paris, France, 1992; Volume 1. [Google Scholar]
- Castenholz, R.W. Culturing Methods for Cyanobacteria. Methods Enzymol. 1988, 167, 68–93. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- Cennamo, P.; Caputo, P.; Giorgio, A.; Moretti, A.; Pasquino, N. Biofilms on tuff stones at historical sites: Identification and removal by nonthermal effects of radiofrequencies. Microb. Ecol. 2013, 66, 659–668. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Fisher, M.M.; Triplett, E.W. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 1999, 65, 4630–4636. [Google Scholar] [CrossRef] [Green Version]
- Ranjard, L.; Poly, F.; Lata, J.C.; Mougel, C.; Thioulouse, J.; Nazaret, S. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: Biological and methodological variability. Appl. Environ. Microbiol. 2001, 67, 4479–4487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinker, P.; Weig, A.; Rambold, G.; Feldhaar, H.; Tragust, S. Microbial community composition of nest-carton and adjoining soil of the ant Lasius fuliginosus and the role of host secretions in structuring microbial communities. Fungal Ecol. 2019, 38, 44–53. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.r-project.org/index.html (accessed on 29 April 2021).
- Oksanen, J.; Guillaume Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 29 April 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. la Sociètè vaudoise des Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Cennamo, P.; Montuori, N.; Trojsi, G.; Fatigati, G.; Moretti, A. Biofilms in churches built in grottoes. Sci. Total Environ. 2016, 543, 727–738. [Google Scholar] [CrossRef]
- Popović, S.; Krizmanić, J.; Vidaković, D.; Karadžić, V.; Milovanović, Ž.; Pećić, M.; Simić, G.S. Biofilms in caves: Easy method for the assessment of dominant phototrophic groups/taxa in situ. Environ. Monit. Assess. 2020, 192, 1–17. [Google Scholar] [CrossRef]
- Zhou, J.P.; Gu, Y.Q.; Zou, C.S.; Mo, M.H. Phylogenetic diversity of bacteria in an earth-cave in Guizhou Province, Southwest of China. J. Microbiol. 2007, 45, 105–112. [Google Scholar]
- Karbowska-Berent, J. Microbiodeterioration of mural paintings: A review. In Art, Biology and Conservation: Biodeterioration of Works of Art; Koestler, R.J., Koestler, V.H., Charola, A.E., Nieto-Fernandez, F.E., Eds.; The Metropolitan Museum of Art: New York, NY, USA, 2003; pp. 266–301. [Google Scholar]
- Radaelli, A.; Paganini, M.; Basavecchia, V.; Elli, V.; Neri, M.; Zanotto, C.; Pontieri, E.; De Giuli Morghen, C. Identification, molecular biotyping and ultrastructural studies of bacterial communities isolated from two damaged frescoes of St Damian’s Monastery in Assisi. Lett. Appl. Microbiol. 2004, 38, 447–453. [Google Scholar] [CrossRef]
- Karpovich-Tate, N.; Rebrikova, N.L. Microbial communities on damaged frescoes and building materials in the Cathedral of the Nativity of the Virgin in the Pafnutii-Borovskii monastery, Russia. Int. Biodeterior. 1991, 27, 281–296. [Google Scholar] [CrossRef]
- Saiz-Jimenez, C.; Samson, R.A. Biodegradacion de obras de arte. Hongos implicados en la degradacion de los frescos del monasterio de la Rabida (Huelva). Bot. Macaronesica 1981, 8–9, 255–264. [Google Scholar]
- Pfendler, S.; Karimi, B.; Maron, P.A.; Ciadamidaro, L.; Valot, B.; Bousta, F.; Alaoui-Sosse, L.; Alaoui-Sosse, B.; Aleya, L. Biofilm biodiversity in French and Swiss show caves using the metabarcoding approach: First data. Sci. Total Environ. 2018, 615, 1207–1217. [Google Scholar] [CrossRef]
- Kozlova, E.V.; Mazina, S.E.; Pešić, V. Biodiversity of phototrophs in illuminated entrance zones of seven caves in Montenegro. Ecol. Montenegrina 2019, 20, 24–39. [Google Scholar] [CrossRef]
- Lee, N.M.; Meisinger, D.B.; Aubrecht, R.; Kovačik, L.; Saiz-Jimenez, C.; Baskar, S.; Liebl, W.; Porter, M.L.; Summers Engel, A. Caves and karst environments. In Life at Extremes: Environments, Organisms and Strategies For Survival; Bell, E.M., Ed.; CAB International: Egham, UK, 2012; pp. 320–344. [Google Scholar]
- Roldán, M.; Hernández-Mariné, M. Exploring the secrets of the three-dimensional architecture of phototrophic biofilms in caves. Int. J. Speleol. 2009, 38, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Martinez, A.; Asencio, A.D. Distribution of cyanobac-teria at the Gelada Cave (Spain) by physical parameters. J. Cave Karst Stud. 2010, 72, 11–20. [Google Scholar] [CrossRef]
- Urzì, C.; de Leo, F.; Bruno, L.; Albertano, P. Microbial diversity in paleolithic caves: A study case on the phototrophic biofilms of the cave of bats (Zuheros, Spain). Microb. Ecol. 2010, 60, 116–129. [Google Scholar] [CrossRef]
- Vinogradova, O.N.; Kovalenko, O.V.; Wasser, S.P.; Nevo, E.; Weinstein-Evron, M. Species diversity gradient to darkness stress in blue-green algae/cyanobacteria: A microscale test in a prehistoric cave, Mount Carmel, Israel. Isr. J. Plant Sci. 1998, 46, 229–238. [Google Scholar] [CrossRef]
- Czerwik-Marcinkowska, J.; Mrozińska, T. Algae and cyanobacteria in caves of the Polish Jura. Polish Bot. J. 2011, 56, 203–243. [Google Scholar]
- Lamprinou, V.; Danielidis, D.B.; Economou-Amilli, A.; Pantazidou, A. Distribution survey of Cyanobacteria in three Greek caves of Peloponnese. Int. J. Speleol. 2012, 41, 267–272. [Google Scholar] [CrossRef] [Green Version]
- Del Mondo, A.; Pinto, G.; Carbone, D.A.; Pollio, A.; De Natale, A. Biofilm architecture on different substrates of an Oculatella subterranea (Cyanobacteria) strain isolated from Pompeii archaeological site (Italy). Environ. Sci. Pollut. Res. 2018, 25, 26079–26089. [Google Scholar] [CrossRef]
- Vázquez-Martínez, J.; Gutierrez-Villagomez, J.M.; Fonsecagarcía, C.; Ramírez-Chávez, E.; Mondragón-Sánchez, M.L.; Partidamartínez, L.; Johansen, J.R.; Molina-Torres, J. Nodosilinea chupicuarensis sp. nov. (Leptolyngbyaceae, Synechococcales) a subaerial cyanobacterium isolated from a stone monument in central Mexico. Phytotaxa 2018, 334, 167–182. [Google Scholar] [CrossRef] [Green Version]
- Cuzman, O.A.; Ventura, S.; Sili, C.; Mascalchi, C.; Turchetti, T.; D’Acqui, L.P.; Tiano, P. Biodiversity of phototrophic biofilms dwelling on monumental fountains. Microb. Ecol. 2010, 60, 81–95. [Google Scholar] [CrossRef]
- Ogórek, R.; Lejman, A.; Matkowski, K. Fungi isolated from Niedźwiedzia Cave in Kletno (Lower Silesia, Poland). Int. J. Speleol. 2013, 42, 161–166. [Google Scholar] [CrossRef] [Green Version]
- Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F.; Forbes, G.J. A world review of fungi, yeasts, and slime molds in caves. Int. J. Speleol. 2013, 42, 77–96. [Google Scholar] [CrossRef]
- Pusz, W.; Ogórek, R.; Knapik, R.; Kozak, B.; Bujak, H. The occurrence of fungi in the recently discovered Jarkowicka cave in the Karkonosze Mts. (Poland). Geomicrobiol. J. 2015, 32, 59–67. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Du, Y.; Tian, T.; Xiang, T.; Liu, X.; Wu, F.; An, L.; Wang, W.; Gu, J.D.; et al. The community distribution of bacteria and fungi on ancient wall paintings of the Mogao Grottoes. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mitova, M.M.; Iliev, M.; Nováková, A.; Gorbushina, A.A.; Groudeva, V.I.; Martin-Sanchez, P.M. Diversity and biocide susceptibility of fungal assemblages dwelling in the Art Gallery of Magura Cave, Bulgaria. Int. J. Speleol. 2017, 46, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
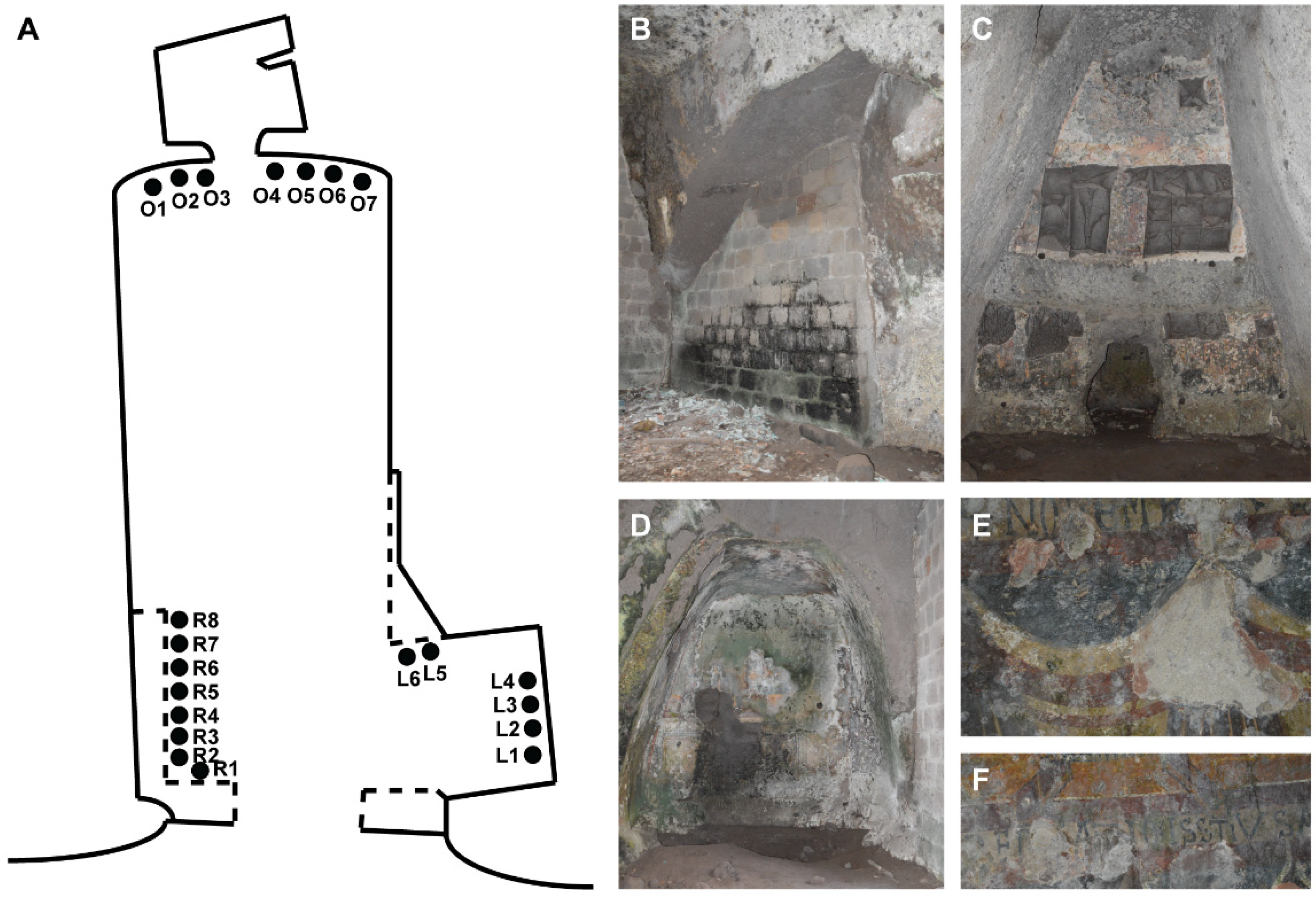
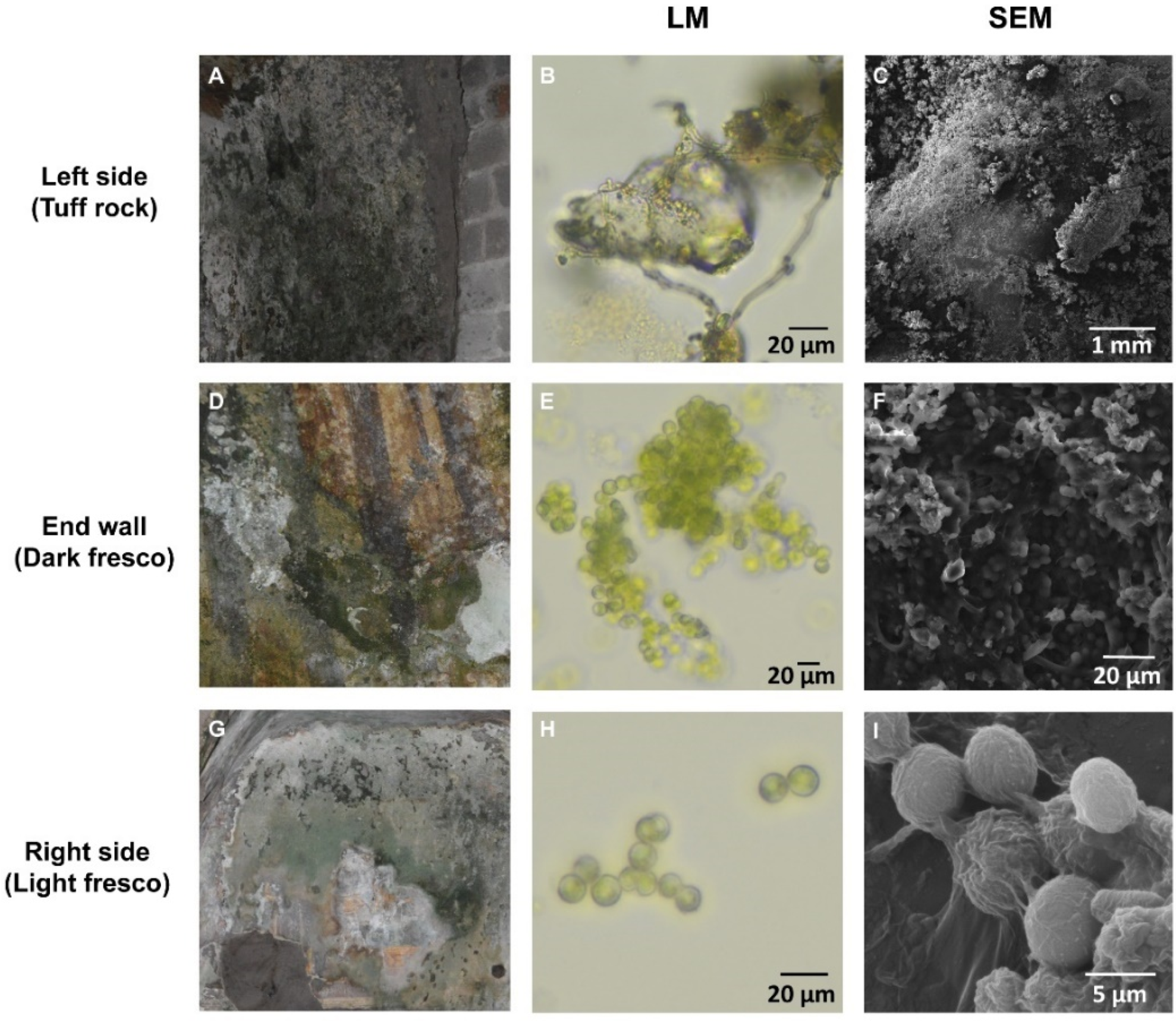

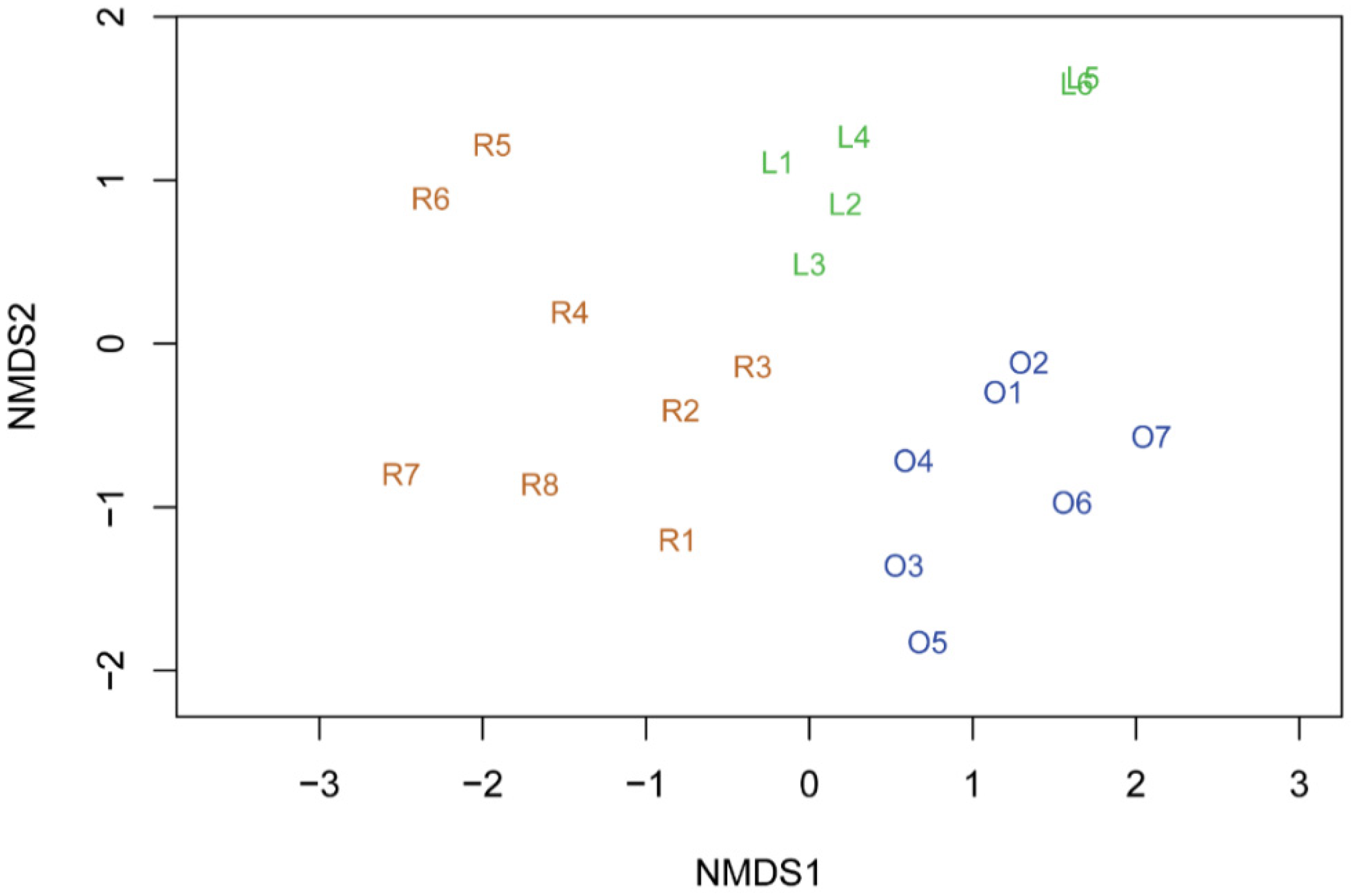

| Sample | Cave Side | Substratum | Temperature (°C) | Light Intensity (lx) | Relative Humidity, RH (%) |
|---|---|---|---|---|---|
| L1 | right | Fresco (light) | 27 ± 0.2 | 499 ± 0.1 | 70 ± 1 |
| L2 | right | Fresco (light) | 27 ± 0.2 | 500 ± 0.2 | 71 ± 1 |
| L3 | right | Fresco (light) | 27 ± 0.1 | 494 ± 0.3 | 70 ± 2 |
| L4 | right | Fresco (light) | 27 ± 0.1 | 497 ± 0.1 | 68 ± 1 |
| L5 | right | Fresco (light) | 28 ± 0.1 | 680 ± 0.1 | 68 ± 1 |
| L6 | right | Fresco (light) | 28 ± 0.2 | 678 ± 0.1 | 69 ± 1 |
| O1 | end wall | Fresco (shadow) | 25 ± 0.1 | 68 ± 0.1 | 73 ± 1 |
| O2 | end wall | Fresco (shadow) | 25 ± 0.1 | 68 ± 0.2 | 73 ± 1 |
| O3 | end wall | Fresco (shadow) | 25 ± 0.3 | 70 ± 0.2 | 73 ± 1 |
| O4 | end wall | Fresco (shadow) | 25 ± 0.2 | 69 ± 0.1 | 72 ± 1 |
| O5 | end wall | Fresco (shadow) | 25 ± 0.1 | 67 ± 0.3 | 73 ± 1 |
| O6 | end wall | Fresco (shadow) | 25 ± 0.2 | 70 ± 0.2 | 73 ± 1 |
| O7 | end wall | Fresco (shadow) | 25 ± 0.1 | 70 ± 0.1 | 73 ± 1 |
| R1 | left | Tuff rock | 26 ± 0.1 | 280 ± 0.3 | 75 ± 1 |
| R2 | left | Tuff rock | 26 ± 0.1 | 278 ± 0.3 | 75 ± 2 |
| R3 | left | Tuff rock | 26 ± 0.2 | 280 ± 0.2 | 75 ± 1 |
| R4 | left | Tuff rock | 26 ± 0.1 | 200 ± 0.2 | 75 ± 1 |
| R5 | left | Tuff rock | 26 ± 0.2 | 200 ± 0.2 | 75 ± 1 |
| R6 | left | Tuff rock | 26 ± 0.1 | 198 ± 0.1 | 75 ± 1 |
| R7 | left | Tuff rock | 26.1 ± 0.1 | 200 ± 0.1 | 75 ± 1 |
| R8 | left | Tuff rock | 26.2 ± 0.1 | 200 ± 0.1 | 75 ± 1 |
| Light Fresco | Dark Fresco | Tuff (Bare Rock) | |
|---|---|---|---|
| Calcite | ++ | +++ | +++ |
| Feldspars | + | + | ++ |
| Iron oxides | + | + | + |
| Mica | + | + | + |
| Plaster | +++ | ++ | + |
| Pyroxenes | + | + | + |
| Quartz | + | + | + |
| Taxa | Dark Fresco | Light Fresco | Tuff Rock |
|---|---|---|---|
| Bacteria | |||
| Bacillus megaterum | - | + | + |
| Bacillus mycoides | - | + | + |
| Bacillus sp. | - | + | + |
| Bacteroides sp. | - | - | + |
| Microbacterium sp. | + | - | - |
| Micrococcus sp. | - | - | + |
| Pseudomonas sp. | - | + | - |
| Staphylococcus sp. | - | + | - |
| Cyanobacteria | |||
| Aphanothece naegelii | - | + | - |
| Halospirulina tapeticola | - | - | + |
| Jaaginema sp. | - | + | - |
| Leptolyngbya africana | + | - | - |
| Leptolyngbya faveolarum | + | + | - |
| Leptolyngbya norvegica | + | + | - |
| Microcoleus sp. | - | - | + |
| Nodosilinea bijugata | + | - | - |
| Nodosilinea cf. nodulosa | + | - | - |
| Nodosilinea sp. | + | - | - |
| Oculatella subterranean | - | + | - |
| Oculatella ucrainica | - | + | - |
| Oscillatoria angusta | - | + | - |
| Phormidium sp. | - | + | - |
| Prochlorococcus sp. | - | + | - |
| Pseudanabaena limnetica | - | - | - |
| Spirulina sp. | - | - | + |
| Synechococcus sp. | - | + | - |
| Algae | |||
| Auxenochlorella protothecoides | - | + | - |
| Bracteacoccus xerophilus | - | + | - |
| Chlamydomonas chlamydogama | - | + | - |
| Chlorella sorokiniana | + | - | + |
| Chlorella sp. | - | + | - |
| Chlorella thermophile | - | + | - |
| Chlorella vulgaris | - | + | + |
| Chloroidium saccharophilum | - | + | + |
| Dictyosphaerium ehrenbergianum | - | + | - |
| Didymogenes sphaerica | - | + | + |
| Eremochloris sphaerica | - | - | + |
| Marvania coccoides | - | + | - |
| Micractinium reisseri | - | + | - |
| Neochloris aquatica | - | + | - |
| Fungi | |||
| Alternaria sp. | + | - | + |
| Aspergillus sp. | + | - | + |
| Cladosporium sp. | - | + | - |
| Colletotrichum sp. | - | - | + |
| Fusarium verticilloides | - | + | - |
| Penicillium sp. | + | - | + |
| Pleosporales sp. | - | - | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Luca, D.; Caputo, P.; Perfetto, T.; Cennamo, P. Characterisation of Environmental Biofilms Colonising Wall Paintings of the Fornelle Cave in the Archaeological Site of Cales. Int. J. Environ. Res. Public Health 2021, 18, 8048. https://doi.org/10.3390/ijerph18158048
De Luca D, Caputo P, Perfetto T, Cennamo P. Characterisation of Environmental Biofilms Colonising Wall Paintings of the Fornelle Cave in the Archaeological Site of Cales. International Journal of Environmental Research and Public Health. 2021; 18(15):8048. https://doi.org/10.3390/ijerph18158048
Chicago/Turabian StyleDe Luca, Daniele, Paolo Caputo, Teresa Perfetto, and Paola Cennamo. 2021. "Characterisation of Environmental Biofilms Colonising Wall Paintings of the Fornelle Cave in the Archaeological Site of Cales" International Journal of Environmental Research and Public Health 18, no. 15: 8048. https://doi.org/10.3390/ijerph18158048






