Exposomes to Exosomes: Exosomes as Tools to Study Epigenetic Adaptive Mechanisms in High-Altitude Humans
Abstract
:1. Introduction
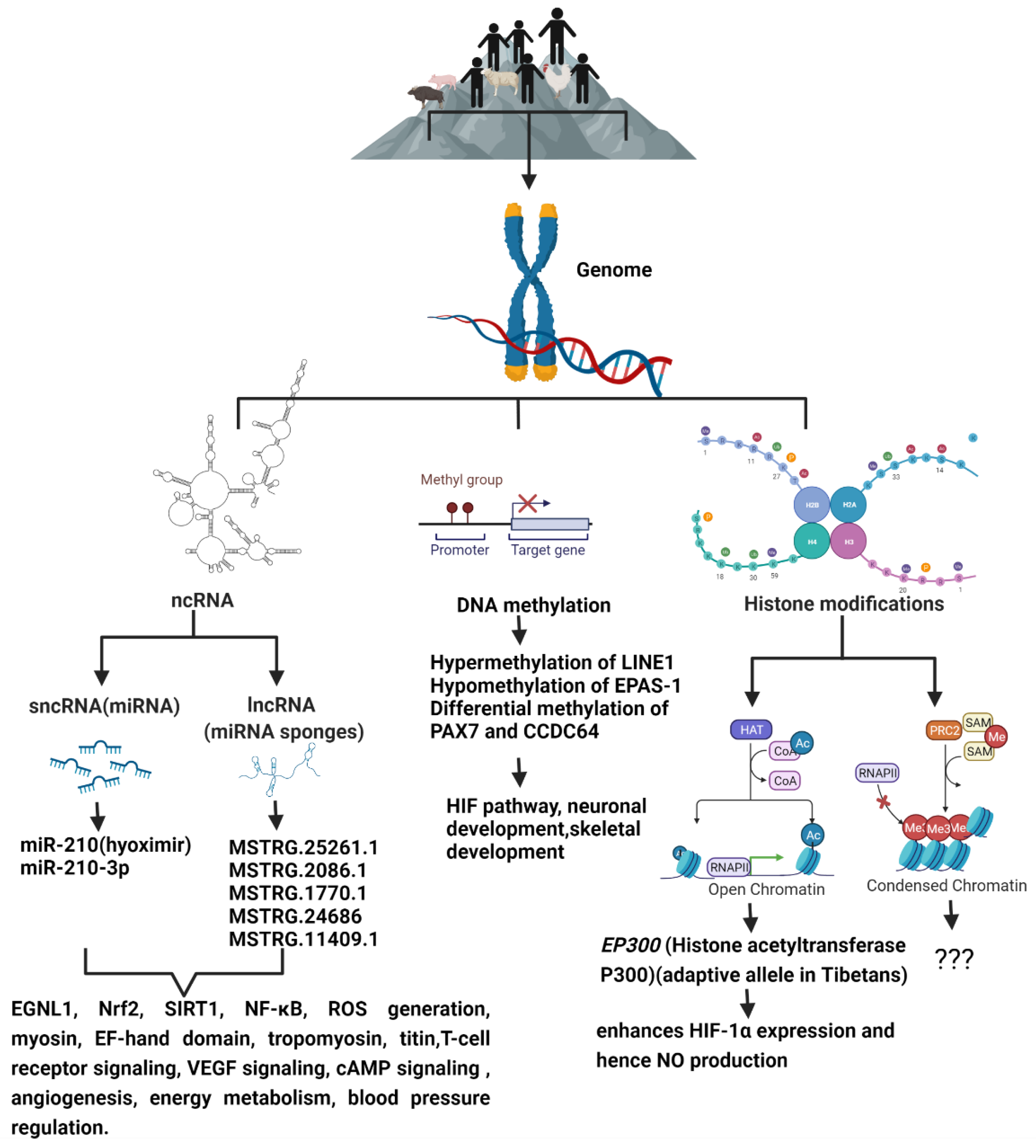
2. Epigenetics of Hypoxia
3. Epigenetic Adaptation of Animals to High Altitude
4. Epigenetics of High-Altitude-Induced (Hypobaric Hypoxia-Induced) Pulmonary Hypertension
5. Exosomes and Hypoxia
6. Exosome for High-Altitude Epigenetic Research
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wild, C.P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.J.; Rupert, J.L. Hypoxia and environmental epigenetics. High Alt. Med. Biol. 2014, 15, 323–330. [Google Scholar] [CrossRef]
- Petersson, J.; Glenny, R.W. Gas exchange and ventilation-perfusion relationships in the lung. Eur. Respir. J. 2014, 44, 1023–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imray, C.; Wright, A.; Subudhi, A.; Roach, R. Acute mountain sickness: Pathophysiology, prevention, and treatment. Prog. Cardiovasc. Dis. 2010, 52, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Beall, C.M. Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr. Comp. Biol. 2006, 46, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petousi, N.; Robbins, P.A. Human adaptation to the hypoxia of high altitude: The Tibetan paradigm from the pregenomic to the postgenomic era. J. Appl. Physiol. (1985) 2014, 116, 875–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheviron, Z.A.; Brumfield, R.T. Genomic insights into adaptation to high-altitude environments. Heredity 2012, 108, 354–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisancho, A.R. Developmental functional adaptation to high altitude: Review. Am. J. Hum. Biol. 2013, 25, 151–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sydykov, A.; Mamazhakypov, A.; Maripov, A.; Kosanovic, D.; Weissmann, N.; Ghofrani, H.A.; Sarybaev, A.S.; Schermuly, R.T. Pulmonary Hypertension in Acute and Chronic High Altitude Maladaptation Disorders. Int. J. Environ. Res. Public Health 2021, 18, 1692. [Google Scholar] [CrossRef]
- Arias-Stella, J.; Kruger, H.; Recavarren, S. Pathology of chronic mountain sickness. Thorax 1973, 28, 701–708. [Google Scholar] [CrossRef] [Green Version]
- Garrido, E.; Botella de Maglia, J.; Castillo, O. Acute, subacute and chronic mountain sickness. Rev. Clin. Esp. 2020. [Google Scholar] [CrossRef]
- Brito, J.; Siques, P.; Pena, E. Long-term chronic intermittent hypoxia: A particular form of chronic high-altitude pulmonary hypertension. Pulm. Circ. 2020, 10, 5–12. [Google Scholar] [CrossRef]
- Newman, J.H.; Holt, T.N.; Hedges, L.K.; Womack, B.; Memon, S.S.; Willers, E.D.; Wheeler, L.; Phillips, J.A., 3rd; Hamid, R. High-altitude pulmonary hypertension in cattle (brisket disease): Candidate genes and gene expression profiling of peripheral blood mononuclear cells. Pulm. Circ. 2011, 1, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Shirley, K.L.; Beckman, D.W.; Garrick, D.J. Inheritance of pulmonary arterial pressure in Angus cattle and its correlation with growth. J. Anim. Sci. 2008, 86, 815–819. [Google Scholar] [CrossRef]
- Jungmann, H. Acclimatization and adaptation to high altitude. Prog. Biometeorol. 1974, 1 Pt 1B, 474–481, 695–698. [Google Scholar]
- Cogo, A. The lung at high altitude. Multidiscip. Respir. Med. 2011, 6, 14–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremona, G.; Asnaghi, R.; Baderna, P.; Brunetto, A.; Brutsaert, T.; Cavallaro, C.; Clark, T.M.; Cogo, A.; Donis, R.; Lanfranchi, P.; et al. Pulmonary extravascular fluid accumulation in recreational climbers: A prospective study. Lancet 2002, 359, 303–309. [Google Scholar] [CrossRef]
- Aaronson, P.I.; Robertson, T.P.; Knock, G.A.; Becker, S.; Lewis, T.H.; Snetkov, V.; Ward, J.P. Hypoxic pulmonary vasoconstriction: Mechanisms and controversies. J. Physiol. 2006, 570, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Hakim, T.S.; Michel, R.P.; Minami, H.; Chang, H.K. Site of pulmonary hypoxic vasoconstriction studied with arterial and venous occlusion. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1983, 54, 1298–1302. [Google Scholar] [CrossRef] [PubMed]
- Stream, J.O.; Luks, A.M.; Grissom, C.K. Lung disease at high altitude. Expert Rev. Respir. Med. 2009, 3, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Naeije, R. Pulmonary circulation at high altitude. Respiration 1997, 64, 429–434. [Google Scholar] [CrossRef]
- Bigham, A.W.; Mao, X.; Mei, R.; Brutsaert, T.; Wilson, M.J.; Julian, C.G.; Parra, E.J.; Akey, J.M.; Moore, L.G.; Shriver, M.D. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum. Genom. 2009, 4, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonson, T.S.; Yang, Y.; Huff, C.D.; Yun, H.; Qin, G.; Witherspoon, D.J.; Bai, Z.; Lorenzo, F.R.; Xing, J.; Jorde, L.B.; et al. Genetic evidence for high-altitude adaptation in Tibet. Science 2010, 329, 72–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beall, C.M.; Cavalleri, G.L.; Deng, L.; Elston, R.C.; Gao, Y.; Knight, J.; Li, C.; Li, J.C.; Liang, Y.; McCormack, M.; et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA 2010, 107, 11459–11464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigham, A.; Bauchet, M.; Pinto, D.; Mao, X.; Akey, J.M.; Mei, R.; Scherer, S.W.; Julian, C.G.; Wilson, M.J.; Lopez Herraez, D.; et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010, 6, e1001116. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Jin, Z.B.; Chen, J.; Huang, X.F.; Li, X.M.; Liang, Y.B.; Mao, J.Y.; Chen, X.; Zheng, Z.; Bakshi, A.; et al. Genetic signatures of high-altitude adaptation in Tibetans. Proc. Natl. Acad. Sci. USA 2017, 114, 4189–4194. [Google Scholar] [CrossRef] [Green Version]
- Childebayeva, A.; Harman, T.; Weinstein, J.; Goodrich, J.M.; Dolinoy, D.C.; Day, T.A.; Bigham, A.W.; Brutsaert, T.D. DNA Methylation Changes Are Associated With an Incremental Ascent to High Altitude. Front. Genet. 2019, 10, 1062. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [Green Version]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- John, R.M.; Rougeulle, C. Developmental Epigenetics: Phenotype and the Flexible Epigenome. Front. Cell Dev. Biol. 2018, 6, 130. [Google Scholar] [CrossRef]
- Oey, H.; Whitelaw, E. On the meaning of the word ‘epimutation’. Trends Genet. 2014, 30, 519–520. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.A.; Alda-Catalinas, C.; Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018, 19, 436–450. [Google Scholar] [CrossRef]
- Galan, C.; Krykbaeva, M.; Rando, O.J. Early life lessons: The lasting effects of germline epigenetic information on organismal development. Mol. Metab. 2020, 38, 100924. [Google Scholar] [CrossRef]
- Lai, K.P.; Wang, S.Y.; Li, J.W.; Tong, Y.; Chan, T.F.; Jin, N.; Tse, A.; Zhang, J.W.; Wan, M.T.; Tam, N.; et al. Hypoxia Causes Transgenerational Impairment of Ovarian Development and Hatching Success in Fish. Environ. Sci. Technol. 2019, 53, 3917–3928. [Google Scholar] [CrossRef]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquiere, B.; Van Dyck, L.; Boeckx, B.; Schoonjans, L.; et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 2016, 537, 63–68. [Google Scholar] [CrossRef]
- Xin, J.; Zhang, H.; He, Y.; Duren, Z.; Bai, C.; Chen, L.; Luo, X.; Yan, D.S.; Zhang, C.; Zhu, X.; et al. Chromatin accessibility landscape and regulatory network of high-altitude hypoxia adaptation. Nat. Commun. 2020, 11, 4928. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Semenza, G.L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995, 270, 1230–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L.; Wang, G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell Biol. 1992, 12, 5447–5454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L. Hydroxylation of HIF-1: Oxygen sensing at the molecular level. Physiology 2004, 19, 176–182. [Google Scholar] [CrossRef] [Green Version]
- Guillemin, K.; Krasnow, M.A. The hypoxic response: Huffing and HIFing. Cell 1997, 89, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Weidemann, A.; Johnson, R.S. Biology of HIF-1alpha. Cell Death Differ. 2008, 15, 621–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, J.A.; Watson, C.J.; McCann, A.; Baugh, J. Epigenetics, the epicenter of the hypoxic response. Epigenetics 2010, 5, 293–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, J.A.; Watson, C.J.; McCrohan, A.M.; Woodfine, K.; Tosetto, M.; McDaid, J.; Gallagher, E.; Betts, D.; Baugh, J.; O’Sullivan, J.; et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum. Mol. Genet. 2009, 18, 3594–3604. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.B.; Denko, N.; Barton, M.C. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res. 2008, 640, 174–179. [Google Scholar] [CrossRef] [Green Version]
- Choudhry, H.; Harris, A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018, 27, 281–298. [Google Scholar] [CrossRef]
- Place, T.L.; Fitzgerald, M.P.; Venkataraman, S.; Vorrink, S.U.; Case, A.J.; Teoh, M.L.; Domann, F.E. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS ONE 2011, 6, e14617. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, Z.; Zhao, S.; Chen, C.; Cong, X.; Li, Z.; Ren, M. The Clinicopathological Significance of Epigenetic Silencing of VHL Promoter and Renal Cell Carcinoma: A Meta-Analysis. Cell Physiol. Biochem. 2016, 40, 1465–1472. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, M.; Song, K.; Wuren, T.; Yan, J.; Ge, R.L.; Ji, L.; Cui, S. VHL gene methylation contributes to excessive erythrocytosis in chronic mountain sickness rat model by upregulating the HIF-2alpha/EPO pathway. Life Sci. 2021, 266, 118873. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Kong, X.; Sang, N. Effects of histone deacetylase inhibitors on HIF-1. Cell Cycle 2006, 5, 2430–2435. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Dioum, E.M.; Hogg, R.T.; Gerard, R.D.; Garcia, J.A. Hypoxia increases sirtuin 1 expression in a hypoxia-inducible factor-dependent manner. J. Biol. Chem. 2011, 286, 13869–13878. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.H.; Lee, Y.M.; Chun, Y.S.; Chen, J.; Kim, J.E.; Park, J.W. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol. Cell 2010, 38, 864–878. [Google Scholar] [CrossRef]
- Dioum, E.M.; Chen, R.; Alexander, M.S.; Zhang, Q.; Hogg, R.T.; Gerard, R.D.; Garcia, J.A. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 2009, 324, 1289–1293. [Google Scholar] [CrossRef]
- Mizuno, S.; Bogaard, H.J.; Kraskauskas, D.; Alhussaini, A.; Gomez-Arroyo, J.; Voelkel, N.F.; Ishizaki, T. p53 Gene deficiency promotes hypoxia-induced pulmonary hypertension and vascular remodeling in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 300, L753–L761. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Choi, K.; Oh, H.; Park, Y.K.; Park, H. HIF-1-dependent induction of Jumonji domain-containing protein (JMJD) 3 under hypoxic conditions. Mol. Cells 2014, 37, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Yao, L.; Shen, Y.; Guo, X.; Duan, X. Histone H3K9 demethylase JMJD1A is a co-activator of erythropoietin expression under hypoxia. Int. J. Biochem. Cell Biol. 2019, 109, 33–39. [Google Scholar] [CrossRef]
- Steinmann, K.; Richter, A.M.; Dammann, R.H. Epigenetic silencing of erythropoietin in human cancers. Genes Cancer 2011, 2, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 24R–29R. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Hu, Z.; Lin, Y.; Zhang, C.; Perez-Polo, J.R. Downregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc. Res. 2010, 87, 535–544. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Le, Q.T.; Giaccia, A.J. MiR-210–micromanager of the hypoxia pathway. Trends Mol. Med. 2010, 16, 230–237. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Zuo, J. Emerging roles of miR-210 and other non-coding RNAs in the hypoxic response. Acta Biochim. Biophys. Sin. 2014, 46, 220–232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Ban, D.; Gou, X.; Zhang, Y.; Yang, L.; Chamba, Y.; Zhang, H. Genome-wide DNA methylation profiles in Tibetan and Yorkshire pigs under high-altitude hypoxia. J. Anim. Sci. Biotechnol. 2019, 10, 25. [Google Scholar] [CrossRef] [Green Version]
- Barth, D.A.; Prinz, F.; Teppan, J.; Jonas, K.; Klec, C.; Pichler, M. Long-Noncoding RNA (lncRNA) in the Regulation of Hypoxia-Inducible Factor (HIF) in Cancer. Noncoding RNA 2020, 6, 27. [Google Scholar] [CrossRef]
- Hancock, A.M.; Alkorta-Aranburu, G.; Witonsky, D.B.; Di Rienzo, A. Adaptations to new environments in humans: The role of subtle allele frequency shifts. Philos. Trans. R. Soc. Lond. B Biol Sci. 2010, 365, 2459–2468. [Google Scholar] [CrossRef]
- Audergon, P.N.; Catania, S.; Kagansky, A.; Tong, P.; Shukla, M.; Pidoux, A.L.; Allshire, R.C. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 2015, 348, 132–135. [Google Scholar] [CrossRef] [Green Version]
- Gluckman, P.D.; Hanson, M.A.; Spencer, H.G. Predictive adaptive responses and human evolution. Trends Ecol. Evol. 2005, 20, 527–533. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zheng, X.; Zhang, Y.; Zhang, H.; Zhang, X.; Zhang, H. Comparative transcriptomic and proteomic analyses provide insights into functional genes for hypoxic adaptation in embryos of Tibetan chickens. Sci. Rep. 2020, 10, 11213. [Google Scholar] [CrossRef]
- Zheng, W.S.; He, Y.X.; Cui, C.Y.; Ouzhu, L.; Deji, Q.; Peng, Y.; Bai, C.J.; Duoji, Z.; Gongga, L.; Bian, B.; et al. EP300 contributes to high-altitude adaptation in Tibetans by regulating nitric oxide production. Zool. Res. 2017, 38, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuyama, M.; WuWong, D.J.; Horvath, S.; Matsuyama, S. Epigenetic clock analysis of human fibroblasts in vitro: Effects of hypoxia, donor age, and expression of hTERT and SV40 largeT. Aging 2019, 11, 3012–3022. [Google Scholar] [CrossRef] [PubMed]
- Childebayeva, A.; Goodrich, J.M.; Leon-Velarde, F.; Rivera-Chira, M.; Kiyamu, M.; Brutsaert, T.D.; Dolinoy, D.C.; Bigham, A.W. Genome-Wide Epigenetic Signatures of Adaptive Developmental Plasticity in the Andes. Genome Biol. Evol. 2021, 13, evaa239. [Google Scholar] [CrossRef]
- Medina, D.; Morrison, D.G. Current ideas on selenium as a chemopreventive agent. Pathol. Immunopathol. Res. 1988, 7, 187–199. [Google Scholar] [CrossRef]
- Julian, C.G.; Moore, L.G. Human Genetic Adaptation to High Altitude: Evidence from the Andes. Genes 2019, 10, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beall, C.M. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. USA 2007, 104 (Suppl. 1), 8655–8660. [Google Scholar] [CrossRef] [Green Version]
- Adkins, R.M.; Krushkal, J.; Tylavsky, F.A.; Thomas, F. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 728–736. [Google Scholar] [CrossRef] [Green Version]
- Childebayeva, A.; Jones, T.R.; Goodrich, J.M.; Leon-Velarde, F.; Rivera-Chira, M.; Kiyamu, M.; Brutsaert, T.D.; Dolinoy, D.C.; Bigham, A.W. LINE-1 and EPAS1 DNA methylation associations with high-altitude exposure. Epigenetics 2019, 14, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Foll, M.; Gaggiotti, O.E.; Daub, J.T.; Vatsiou, A.; Excoffier, L. Widespread signals of convergent adaptation to high altitude in Asia and america. Am. J. Hum. Genet. 2014, 95, 394–407. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Song, K.; Qi, G.; Yan, R.; Yang, Y.; Li, Y.; Wang, S.; Bai, Z.; Ge, R.L. Adipose-derived exosomal miR-210/92a cluster inhibits adipose browning via the FGFR-1 signaling pathway in high-altitude hypoxia. Sci. Rep. 2020, 10, 14390. [Google Scholar] [CrossRef]
- Bell, A.; Bell, D.; Chakravarti, N.; Ma, J.; Henton, N.; Prieto, V.G. Detection of a MicroRNA molecular signature of ultraviolet radiation in the superficial regions of melanocytic nevi on sun-exposed skin. Mod. Pathol. 2018, 31, 1744–1755. [Google Scholar] [CrossRef] [PubMed]
- Long, K.; Feng, S.; Ma, J.; Zhang, J.; Jin, L.; Tang, Q.; Wang, X.; Mai, M.; Xiao, W.; Liu, L.; et al. Small non-coding RNA transcriptome of four high-altitude vertebrates and their low-altitude relatives. Sci. Data 2019, 6, 192. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, C.; Zhou, W.; Shi, Y.; Guo, P.; Liu, Y.; Wang, J.; Zhang, C.Y.; Zhang, C. Elevation of Circulating miR-210-3p in High-Altitude Hypoxic Environment. Front. Physiol. 2016, 7, 84. [Google Scholar] [CrossRef] [Green Version]
- Kayser, B.; Hoppeler, H.; Claassen, H.; Cerretelli, P. Muscle structure and performance capacity of Himalayan Sherpas. J. Appl. Physiol. (1985) 1991, 70, 1938–1942. [Google Scholar] [CrossRef]
- Carbonell, T.; Gomes, A.V. MicroRNAs in the regulation of cellular redox status and its implications in myocardial ischemia-reperfusion injury. Redox Biol. 2020, 36, 101607. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Gao, L.; Zimmerman, M.C.; Zucker, I.H. Myocardial infarction-induced microRNA-enriched exosomes contribute to cardiac Nrf2 dysregulation in chronic heart failure. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H928–H939. [Google Scholar] [CrossRef] [PubMed]
- Climent, M.; Viggiani, G.; Chen, Y.W.; Coulis, G.; Castaldi, A. MicroRNA and ROS Crosstalk in Cardiac and Pulmonary Diseases. Int. J. Mol. Sci. 2020, 21, 4370. [Google Scholar] [CrossRef]
- R Babu, K.; Tay, Y. The Yin-Yang Regulation of Reactive Oxygen Species and MicroRNAs in Cancer. Int. J. Mol. Sci. 2019, 20, 5335. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Wang, R.J.; Li, G.Z.; Zhang, Y.; Yu, S.; Liu, Y.F.; Chen, X.Y.; Hou, S.K. miRNA array analysis of plasma miRNA alterations in rats exposed to a high altitude hypoxic environment. Mol. Med. Rep. 2018, 18, 5502–5510. [Google Scholar] [CrossRef]
- Sutton, J.R.; Reeves, J.T.; Wagner, P.D.; Groves, B.M.; Cymerman, A.; Malconian, M.K.; Rock, P.B.; Young, P.M.; Walter, S.D.; Houston, C.S. Operation Everest II: Oxygen transport during exercise at extreme simulated altitude. J. Appl. Physiol. (1985) 1988, 64, 1309–1321. [Google Scholar] [CrossRef]
- Xin, J.W.; Chai, Z.X.; Zhang, C.F.; Yang, Y.M.; Zhang, Q.; Zhu, Y.; Cao, H.W.; Yang Ji, C.; Zhong, J.C.; Ji, Q.M. Transcriptome analysis identified long non-coding RNAs involved in the adaption of yak to high-altitude environments. R. Soc. Open Sci. 2020, 7, 200625. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, J.; Wang, J.; Chai, Z.; Zhang, C.; Xin, J.; Wang, J.; Cai, X.; Wu, Z.; Ji, Q. Whole-Transcriptome Analysis of Yak and Cattle Heart Tissues Reveals Regulatory Pathways Associated with High-Altitude Adaptation. Front. Genet. 2021, 12, 579800. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, W.; Zhang, B.; Ling, Y.; Kim, W.K.; Zhang, H. Comprehensive analysis of coding and non-coding RNA transcriptomes related to hypoxic adaptation in Tibetan chickens. J. Anim. Sci. Biotechnol. 2021, 12, 60. [Google Scholar] [CrossRef]
- Wang, J.; Chai, Z.; Deng, L.; Wang, J.; Wang, H.; Tang, Y.; Zhong, J.; Ji, Q. Detection and integrated analysis of lncRNA and mRNA relevant to plateau adaptation of Yak. Reprod. Domest. Anim. 2020, 55, 1461–1469. [Google Scholar] [CrossRef]
- Shimoda, L.A.; Laurie, S.S. Vascular remodeling in pulmonary hypertension. J. Mol. Med. 2013, 91, 297–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merklinger, S.L.; Jones, P.L.; Martinez, E.C.; Rabinovitch, M. Epidermal growth factor receptor blockade mediates smooth muscle cell apoptosis and improves survival in rats with pulmonary hypertension. Circulation 2005, 112, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakao, S.; Tatsumi, K.; Voelkel, N.F. Endothelial cells and pulmonary arterial hypertension: Apoptosis, proliferation, interaction and transdifferentiation. Respir. Res. 2009, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Mirrakhimov, A.E.; Strohl, K.P. High-altitude Pulmonary Hypertension: An Update on Disease Pathogenesis and Management. Open Cardiovasc. Med. J. 2016, 10, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Aldashev, A.A.; Sarybaev, A.S.; Sydykov, A.S.; Kalmyrzaev, B.B.; Kim, E.V.; Mamanova, L.B.; Maripov, R.; Kojonazarov, B.K.; Mirrakhimov, M.M.; Wilkins, M.R.; et al. Characterization of high-altitude pulmonary hypertension in the Kyrgyz: Association with angiotensin-converting enzyme genotype. Am. J. Respir. Crit. Care Med. 2002, 166, 1396–1402. [Google Scholar] [CrossRef] [PubMed]
- Sime, F.; Penaloza, D.; Ruiz, L. Bradycardia, increased cardiac output, and reversal of pulmonary hypertension in altitude natives living at sea level. Br. Heart J. 1971, 33, 647–657. [Google Scholar] [CrossRef] [Green Version]
- Pugliese, S.C.; Yeager, M.E.; Stenmark, K.R. Hypoxic Pulmonary Hypertension. In PanVascular Medicine; Springer: Berlin/Heidelberg, Germany, 2015; pp. 4169–4209. [Google Scholar]
- Stenmark, K.R.; Frid, M.G.; Yeager, M.; Li, M.; Riddle, S.; McKinsey, T.; El Kasmi, K.C. Targeting the adventitial microenvironment in pulmonary hypertension: A potential approach to therapy that considers epigenetic change. Pulm. Circ. 2012, 2, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Sun, M.; Ramchandran, R.; Raj, J.U. IGF-1 signaling in neonatal hypoxia-induced pulmonary hypertension: Role of epigenetic regulation. Vascul. Pharmacol. 2015, 73, 20–31. [Google Scholar] [CrossRef] [Green Version]
- Caruso, P.; MacLean, M.R.; Khanin, R.; McClure, J.; Soon, E.; Southgate, M.; MacDonald, R.A.; Greig, J.A.; Robertson, K.E.; Masson, R.; et al. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 716–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parikh, V.N.; Jin, R.C.; Rabello, S.; Gulbahce, N.; White, K.; Hale, A.; Cottrill, K.A.; Shaik, R.S.; Waxman, A.B.; Zhang, Y.Y.; et al. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: Results of a network bioinformatics approach. Circulation 2012, 125, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Blissenbach, B.; Nakas, C.T.; Kronke, M.; Geiser, T.; Merz, T.M.; Pichler Hefti, J. Hypoxia-induced changes in plasma micro-RNAs correlate with pulmonary artery pressure at high altitude. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 314, L157–L164. [Google Scholar] [CrossRef] [PubMed]
- Grunig, G.; Eichstaedt, C.A.; Verweyen, J.; Durmus, N.; Saxer, S.; Krafsur, G.; Stenmark, K.; Ulrich, S.; Grunig, E.; Pylawka, S. Circulating MicroRNA Markers for Pulmonary Hypertension in Supervised Exercise Intervention and Nightly Oxygen Intervention. Front. Physiol. 2018, 9, 955. [Google Scholar] [CrossRef]
- Yang, Q.; Lu, Z.; Ramchandran, R.; Longo, L.D.; Raj, J.U. Pulmonary artery smooth muscle cell proliferation and migration in fetal lambs acclimatized to high-altitude long-term hypoxia: Role of histone acetylation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 303, L1001–L1010. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, H.; Li, M.; Frid, M.G.; Flockton, A.R.; McKeon, B.A.; Yeager, M.E.; Fini, M.A.; Morrell, N.W.; Pullamsetti, S.S.; et al. MicroRNA-124 controls the proliferative, migratory, and inflammatory phenotype of pulmonary vascular fibroblasts. Circ. Res. 2014, 114, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; He, Y.Y.; Jiang, X.; Wang, Y.; Chen, J.W.; Zhao, J.H.; Ye, J.; Lian, T.Y.; Zhang, X.; Zhang, R.J.; et al. DNA methyltransferase 3B deficiency unveils a new pathological mechanism of pulmonary hypertension. Sci. Adv. 2020, 6, eaba2470. [Google Scholar] [CrossRef]
- Mishra, A.; Kohli, S.; Dua, S.; Thinlas, T.; Mohammad, G.; Pasha, M.A. Genetic differences and aberrant methylation in the apelin system predict the risk of high-altitude pulmonary edema. Proc. Natl. Acad. Sci. USA 2015, 112, 6134–6139. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef]
- Das, A.; Mohan, V.; Krishnaswamy, V.R.; Solomonov, I.; Sagi, I. Exosomes as a storehouse of tissue remodeling proteases and mediators of cancer progression. Cancer Metastasis Rev. 2019, 38, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Barberis, E.; Vanella, V.V.; Falasca, M.; Caneapero, V.; Cappellano, G.; Raineri, D.; Ghirimoldi, M.; De Giorgis, V.; Puricelli, C.; Vaschetto, R.; et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 2021, 8, 632290. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Kosaka, N.; Araya, J.; Kuwano, K.; Ochiya, T. Extracellular vesicles in lung microenvironment and pathogenesis. Trends Mol. Med. 2015, 21, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Lin, Z.X.; Jiang, X.Y.; Yu, X.Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Ekstrom, K.; Valadi, H.; Sjostrand, M.; Malmhall, C.; Bossios, A.; Eldh, M.; Lotvall, J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles 2012, 1, 18389. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Barbagallo, C.; Cirnigliaro, M.; Battaglia, R.; Brex, D.; Caponnetto, A.; Barbagallo, D.; Di Pietro, C.; Purrello, M. Asymmetric RNA Distribution among Cells and Their Secreted Exosomes: Biomedical Meaning and Considerations on Diagnostic Applications. Front. Mol. Biosci. 2017, 4, 66. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A. Bioinformatic analysis revealing association of exosomal mRNAs and proteins in epigenetic inheritance. J. Theor. Biol. 2014, 357, 143–149. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [Green Version]
- Shao, C.; Yang, F.; Miao, S.; Liu, W.; Wang, C.; Shu, Y.; Shen, H. Role of hypoxia-induced exosomes in tumor biology. Mol. Cancer 2018, 17, 120. [Google Scholar] [CrossRef]
- Kore, R.A.; Edmondson, J.L.; Jenkins, S.V.; Jamshidi-Parsian, A.; Dings, R.P.M.; Reyna, N.S.; Griffin, R.J. Hypoxia-derived exosomes induce putative altered pathways in biosynthesis and ion regulatory channels in glioblastoma cells. Biochem. Biophys. Rep. 2018, 14, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Kenneweg, F.; Bang, C.; Xiao, K.; Boulanger, C.M.; Loyer, X.; Mazlan, S.; Schroen, B.; Hermans-Beijnsberger, S.; Foinquinos, A.; Hirt, M.N.; et al. Long Noncoding RNA-Enriched Vesicles Secreted by Hypoxic Cardiomyocytes Drive Cardiac Fibrosis. Mol. Ther. Nucleic Acids 2019, 18, 363–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Han, Y.; Jo, H.A.; Lee, J.; Song, Y.S. Non-coding RNAs shuttled via exosomes reshape the hypoxic tumor microenvironment. J. Hematol. Oncol. 2020, 13, 67. [Google Scholar] [CrossRef]
- Chan, Y.C.; Banerjee, J.; Choi, S.Y.; Sen, C.K. miR-210: The master hypoxamir. Microcirculation 2012, 19, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Caby, M.P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlotogorski-Hurvitz, A.; Dayan, D.; Chaushu, G.; Korvala, J.; Salo, T.; Sormunen, R.; Vered, M. Human saliva-derived exosomes: Comparing methods of isolation. J. Histochem. Cytochem. 2015, 63, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, Y.; Sun, W.; Zhang, Q.; Gu, T.; Li, G. Plasma exosomes as novel biomarker for the early diagnosis of gastric cancer. Cancer Biomark. 2018, 21, 805–812. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019, 10, 3854. [Google Scholar] [CrossRef] [PubMed]
- van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130502. [Google Scholar] [CrossRef]
- Vautrot, V.; Chanteloup, G.; Elmallah, M.; Cordonnier, M.; Aubin, F.; Garrido, C.; Gobbo, J. Exosomal miRNA: Small Molecules, Big Impact in Colorectal Cancer. J. Oncol. 2019, 2019, 8585276. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Quek, C.Y.; Sun, X.; Bellingham, S.A.; Hill, A.F. The detection of microRNA associated with Alzheimer’s disease in biological fluids using next-generation sequencing technologies. Front. Genet. 2013, 4, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, G.K.; Khan, M.A.; Zubair, H.; Srivastava, S.K.; Khushman, M.; Singh, S.; Singh, A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019, 9, 5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padmasekar, M.; Savai, R.; Seeger, W.; Pullamsetti, S.S. Exposomes to Exosomes: Exosomes as Tools to Study Epigenetic Adaptive Mechanisms in High-Altitude Humans. Int. J. Environ. Res. Public Health 2021, 18, 8280. https://doi.org/10.3390/ijerph18168280
Padmasekar M, Savai R, Seeger W, Pullamsetti SS. Exposomes to Exosomes: Exosomes as Tools to Study Epigenetic Adaptive Mechanisms in High-Altitude Humans. International Journal of Environmental Research and Public Health. 2021; 18(16):8280. https://doi.org/10.3390/ijerph18168280
Chicago/Turabian StylePadmasekar, Manju, Rajkumar Savai, Werner Seeger, and Soni Savai Pullamsetti. 2021. "Exposomes to Exosomes: Exosomes as Tools to Study Epigenetic Adaptive Mechanisms in High-Altitude Humans" International Journal of Environmental Research and Public Health 18, no. 16: 8280. https://doi.org/10.3390/ijerph18168280






