Biosignal-Based Digital Biomarkers for Prediction of Ventilator Weaning Success
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Study Population
2.3. Study Design
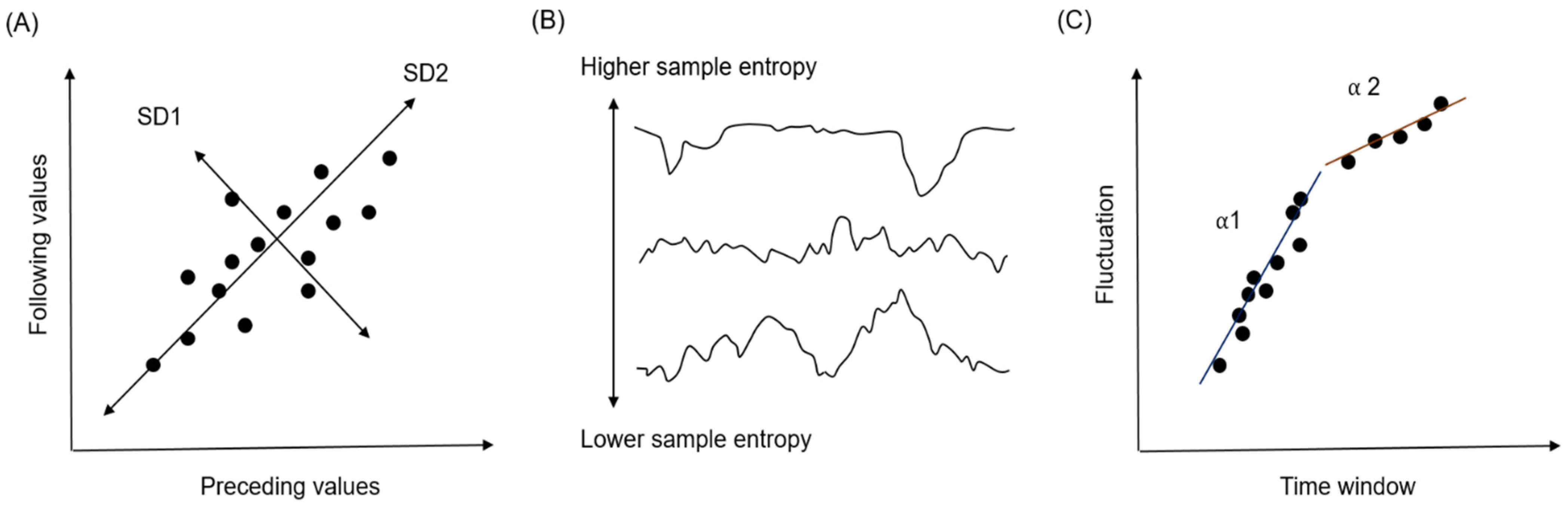
2.4. Feature Extraction
2.5. Statistical Analyses
2.6. Development of the Machine Learning Model
2.7. Software Used in the Study
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- MacIntyre, N.R.; Cook, D.J.; Ely, E.W., Jr.; Epstein, S.K.; Fink, J.B.; Heffner, J.E.; Hess, D.; Hubmayer, R.D.; Scheinhorn, D.J.; American College of Chest, P.; et al. Evidence-based guidelines for weaning and discontinuing ventilatory support: A collective task force facilitated by the American College of Chest Physicians; the American Association for Respiratory Care; and the American College of Critical Care Medicine. Chest 2001, 120, 375S–395S. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, B.; Bastuji-Garin, S.; Durand, M.C.; Malissin, I.; Rodrigues, P.; Cerf, C.; Outin, H.; Sharshar, T. Respiratory weakness is associated with limb weakness and delayed weaning in critical illness. Crit. Care Med. 2007, 35, 2007–2015. [Google Scholar] [CrossRef]
- Rello, J.; Ollendorf, D.A.; Oster, G.; Vera-Llonch, M.; Bellm, L.; Redman, R.; Kollef, M.H. Epidemiology and outcomes of ventilator-associated pneumonia in a large US database. Chest 2002, 122, 2115–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajakaruna, C.; Rogers, C.A.; Angelini, G.D.; Ascione, R. Risk factors for and economic implications of prolonged ventilation after cardiac surgery. J. Thorac. Cardiovasc. Surg. 2005, 130, 1270–1277. [Google Scholar] [CrossRef] [Green Version]
- Russotto, V.; Myatra, S.N.; Laffey, J.G.; Tassistro, E.; Antolini, L.; Bauer, P.; Lascarrou, J.B.; Szuldrzynski, K.; Camporota, L.; Pelosi, P.; et al. Intubation practices and adverse peri-intubation events in critically ill patients from 29 countries. JAMA 2021, 325, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.K.; Ciubotaru, R.L.; Wong, J.B. Effect of failed extubation on the outcome of mechanical ventilation. Chest 1997, 112, 186–192. [Google Scholar] [CrossRef]
- Esteban, A.; Alía, I.; Gordo, F.; Fernández, R.; Solsona, J.F.; Vallverdú, I.; Macías, S.; Allegue, J.M.; Blanco, J.; Carriedo, D.; et al. Extubation outcome after spontaneous breathing trials with T-tube or pressure support ventilation. The spanish lung failure collaborative group. Am. J. Respir. Crit. Care Med. 1997, 156, 459–465. [Google Scholar] [CrossRef]
- Capdevila, X.; Perrigault, P.F.; Ramonatxo, M.; Roustan, J.P.; Peray, P.; d’Athis, F.; Prefaut, C. Changes in breathing pattern and respiratory muscle performance parameters during difficult weaning. Crit. Care Med. 1998, 26, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Jubran, A.; Tobin, M.J. Pathophysiologic basis of acute respiratory distress in patients who fail a trial of weaning from mechanical ventilation. Am. J. Respir. Crit. Care Med. 1997, 155, 906–915. [Google Scholar] [CrossRef]
- Esteban, A.; Frutos, F.; Tobin, M.J.; Alía, I.; Solsona, J.F.; Valverdú, I.; Fernández, R.; de la Cal, M.A.; Benito, S.; Tomás, R.; et al. A comparison of four methods of weaning patients from mechanical ventilation. Spanish Lung Failure Collaborative Group. N. Engl. J. Med. 1995, 332, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.K. Predicting extubation failure: Is it in (on) the cards? Chest 2001, 120, 1061–1063. [Google Scholar] [CrossRef]
- Gershengorn, H.B. international variation in intubation and extubation practices and adverse events among critically ill patients receiving mechanical ventilation. JAMA 2021, 325, 1157–1159. [Google Scholar] [CrossRef] [PubMed]
- Vassilakopoulos, T.; Roussos, C.; Zakynthinos, S. Weaning from mechanical ventilation. J. Crit. Care 1999, 14, 39–62. [Google Scholar] [CrossRef]
- Boles, J.M.; Bion, J.; Connors, A.; Herridge, M.; Marsh, B.; Melot, C.; Pearl, R.; Silverman, H.; Stanchina, M.; Vieillard-Baron, A.; et al. Weaning from mechanical ventilation. Eur. Respir. J. 2007, 29, 1033–1056. [Google Scholar] [CrossRef]
- Burns, K.E.A.; Rizvi, L.; Cook, D.J.; Dodek, P.; Slutsky, A.S.; Jones, A.; Villar, J.; Kapadia, F.N.; Gattas, D.J.; Epstein, S.K.; et al. Variation in the practice of discontinuing mechanical ventilation in critically ill adults: Study protocol for an international prospective observational study. BMJ Open 2019, 9, e031775. [Google Scholar] [CrossRef] [Green Version]
- Burns, K.E.A.; Rizvi, L.; Cook, D.J.; Lebovic, G.; Dodek, P.; Villar, J.; Slutsky, A.S.; Jones, A.; Kapadia, F.N.; Gattas, D.J.; et al. Ventilator weaning and discontinuation practices for critically ill patients. JAMA 2021, 325, 1173–1184. [Google Scholar] [CrossRef]
- Perkins, G.D.; Mistry, D.; Lall, R.; Gao-Smith, F.; Snelson, C.; Hart, N.; Camporota, L.; Varley, J.; Carle, C.; Paramasivam, E.; et al. Protocolised non-invasive compared with invasive weaning from mechanical ventilation for adults in intensive care: The Breathe RCT. Health Technol. Assess. 2019, 23, 1–114. [Google Scholar] [CrossRef]
- Burns, K.E.A.; Rizvi, L.; Cook, D.J.; Seely, A.J.E.; Rochwerg, B.; Lamontagne, F.; Devlin, J.W.; Dodek, P.; Mayette, M.; Tanios, M.; et al. Frequency of screening and SBT technique trial—North American weaning collaboration (FAST-NAWC): A protocol for a multicenter, factorial randomized trial. Trials 2019, 20, 587. [Google Scholar] [CrossRef]
- Lewis, K.A.; Chaudhuri, D.; Guyatt, G.; Burns, K.E.A.; Bosma, K.; Ge, L.; Karachi, T.; Piraino, T.; Fernando, S.M.; Ranganath, N.; et al. Comparison of ventilatory modes to facilitate liberation from mechanical ventilation: Protocol for a systematic review and network meta-analysis. BMJ Open 2019, 9, e030407. [Google Scholar] [CrossRef] [Green Version]
- Vu, P.H.; Tran, V.D.; Duong, M.C.; Cong, Q.T.; Nguyen, T. Predictive value of the negative inspiratory force index as a predictor of weaning success: A crosssectional study. Acute Crit. Care 2020, 35, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Appavu, S.K. Prediction of extubation failure following mechanical ventilation: Where are we and where are we going? Crit. Care Med. 2020, 48, 1536–1538. [Google Scholar] [CrossRef] [PubMed]
- da Silva Neto, A.E.; de Souza, L.C.; Costa, H.L.; Guimarães, B.L.; de Azeredo, L.M.; Godoy, M.D.P.; Lugon, J.R. The timed inspiratory effort index as a weaning predictor: Analysis of intra- and interobserver reproducibility. Respir. Care 2020, 65, 636–642. [Google Scholar] [CrossRef]
- Frutos-Vivar, F.; Ferguson, N.D.; Esteban, A.; Epstein, S.K.; Arabi, Y.; Apezteguía, C.; González, M.; Hill, N.S.; Nava, S.; D’Empaire, G.; et al. Risk factors for extubation failure in patients following a successful spontaneous breathing trial. Chest 2006, 130, 1664–1671. [Google Scholar] [CrossRef] [Green Version]
- Epstein, S.K. Weaning from ventilatory support. Curr. Opin. Crit. Care 2009, 15, 36–43. [Google Scholar] [CrossRef]
- Yang, K.L.; Tobin, M.J. A prospective study of indexes predicting the outcome of trials of weaning from mechanical ventilation. N. Engl. J. Med. 1991, 324, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Casaseca-de-la-Higuera, P.; Martín-Fernández, M.; Alberola-López, C. Weaning from mechanical ventilation: A retrospective analysis leading to a multimodal perspective. IEEE Trans. Biomed. Eng. 2006, 53, 1330–1345. [Google Scholar] [CrossRef]
- El-Khatib, M.F.; Zeineldine, S.M.; Jamaleddine, G.W. Effect of pressure support ventilation and positive end expiratory pressure on the rapid shallow breathing index in intensive care unit patients. Intensive Care Med. 2008, 34, 505–510. [Google Scholar] [CrossRef]
- Krieger, B.P.; Isber, J.; Breitenbucher, A.; Throop, G.; Ershowsky, P. Serial measurements of the rapid-shallow-breathing index as a predictor of weaning outcome in elderly medical patients. Chest 1997, 112, 1029–1034. [Google Scholar] [CrossRef]
- Shang, P.; Zhu, M.; Baker, M.; Feng, J.; Zhou, C.; Zhang, H.L. Mechanical ventilation in Guillain-Barré syndrome. Expert Rev. Clin. Immunol. 2020, 16, 1053–1064. [Google Scholar] [CrossRef]
- Demiralp, B.; Koenig, L.; Xu, J.; Soltoff, S.; Votto, J. Time spent in prior hospital stay and outcomes for ventilator patients in long-term acute care hospitals. BMC Pulm. Med. 2021, 21, 104. [Google Scholar] [CrossRef]
- Villalba, D.; Gil Rossetti, G.; Scrigna, M.; Collins, J.; Rocco, A.; Matesa, A.; Areas, L.; Golfarini, N.; Pini, P.; Hannun, M.; et al. Prevalence of and risk factors for mechanical ventilation reinstitution in patients weaned from prolonged mechanical ventilation. Respir. Care 2020, 65, 210–216. [Google Scholar] [CrossRef]
- Meade, M.; Guyatt, G.; Cook, D.; Griffith, L.; Sinuff, T.; Kergl, C.; Mancebo, J.; Esteban, A.; Epstein, S. Predicting success in weaning from mechanical ventilation. Chest 2001, 120, 400S–424S. [Google Scholar] [CrossRef] [Green Version]
- Videtta, W.; Vallejos, J.; Roda, G.; Collazos, H.; Naccarelli, N.; Tamayo, A.; Calderón, N.; Bairaclioti, A.; Yoshida, M.; Vandaele, G.; et al. Predictors of successful extubation in neurocritical care patients. Acta Neurochir. Suppl. 2021, 131, 91–93. [Google Scholar] [CrossRef]
- Baptistella, A.R.; Mantelli, L.M.; Matte, L.; Carvalho, M.; Fortunatti, J.A.; Costa, I.Z.; Haro, F.G.; Turkot, V.L.O.; Baptistella, S.F.; de Carvalho, D.; et al. Prediction of extubation outcome in mechanically ventilated patients: Development and validation of the extubation predictive score (ExPreS). PLoS ONE 2021, 16, e0248868. [Google Scholar] [CrossRef]
- Leonov, Y.; Kisil, I.; Perlov, A.; Stoichev, V.; Ginzburg, Y.; Nazarenko, A.; Gimelfarb, Y. Predictors of successful weaning in patients requiring extremely prolonged mechanical ventilation. Adv. Respir. Med. 2020, 88, 477–484. [Google Scholar] [CrossRef]
- Wu, T.J.; Shiao, J.S.; Yu, H.L.; Lai, R.S. An integrative index for predicting extubation outcomes after successful completion of a spontaneous breathing trial in an adult medical intensive care unit. J. Intensive Care Med. 2019, 34, 640–645. [Google Scholar] [CrossRef]
- Liu, J.; Wang, C.J.; Ran, J.H.; Lin, S.H.; Deng, D.; Ma, Y.; Xu, F. The predictive value of brain natriuretic peptide or N-terminal pro-brain natriuretic peptide for weaning outcome in mechanical ventilation patients: Evidence from SROC. J. Renin Angiotensin Aldosterone Syst. 2021, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, J.; Webber, J.; Featherstone, R.; Sebastianski, M.; Vandermeer, B.; Senaratne, J.; Bagshaw, S.M. Brain natriuretic peptide to predict successful liberation from mechanical ventilation in critically ill patients: Protocol for a systematic review and meta-analysis. BMJ Open 2019, 9, e022600. [Google Scholar] [CrossRef] [PubMed]
- Fontela, P.C.; Glaeser, S.S.; Martins, L.F.; Condessa, R.L.; Prediger, D.T.; Forgiarini, S.G.; Forgiarini, L.A., Jr.; Lisboa, T.C.; Friedman, G. Medical research council scale predicts spontaneous breathing trial failure and difficult or prolonged weaning of critically ill individuals. Respir. Care 2021, 66, 733–741. [Google Scholar] [CrossRef]
- Mallat, J.; Baghdadi, F.A.; Mohammad, U.; Lemyze, M.; Temime, J.; Tronchon, L.; Thevenin, D.; Fischer, M.O. Central venous-to-arterial PCO2 difference and central venous oxygen saturation in the detection of extubation failure in critically ill patients. Crit. Care Med. 2020, 48, 1454–1461. [Google Scholar] [CrossRef]
- El-Khatib, M.; Jamaleddine, G.; Soubra, R.; Muallem, M. Pattern of spontaneous breathing: Potential marker for weaning outcome. Spontaneous breathing pattern and weaning from mechanical ventilation. Intensive Care Med. 2001, 27, 52–58. [Google Scholar] [CrossRef]
- Bien, M.Y.; Hseu, S.S.; Yien, H.W.; Kuo, B.I.; Lin, Y.T.; Wang, J.H.; Kou, Y.R. Breathing pattern variability: A weaning predictor in postoperative patients recovering from systemic inflammatory response syndrome. Intensive Care Med. 2004, 30, 241–247. [Google Scholar] [CrossRef]
- Fabregat, A.; Magret, M.; Ferré, J.A.; Vernet, A.; Guasch, N.; Rodríguez, A.; Gómez, J.; Bodí, M. A Machine Learning decision-making tool for extubation in intensive care unit patients. Comput. Methods Programs Biomed. 2021, 200, 105869. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujino, Y. Monitoring the patient for a safe-assisted ventilation. Curr. Opin. Crit. Care 2021, 27, 1–5. [Google Scholar] [CrossRef]
- Kennedy, H.L. Heart rate variability—A potential, noninvasive prognostic index in the critically ill patient. Crit. Care Med. 1998, 26, 213–214. [Google Scholar] [CrossRef]
- Camm, A.J.; Malik, M.; Bigger, J.T.; Breithardt, G.; Cerutti, S.; Cohen, R.J.; Coumel, P.; Fallen, E.L.; Kennedy, H.L.; Kleiger, R.E.; et al. Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Task force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation 1996, 93, 1043–1065. [Google Scholar]
- Kwon, J.M.; Cho, Y.; Jeon, K.H.; Cho, S.; Kim, K.H.; Baek, S.D.; Jeung, S.; Park, J.; Oh, B.H. A deep learning algorithm to detect anaemia with ECGs: A retrospective, multicentre study. Lancet Digit. Health 2020, 2, e358–e367. [Google Scholar] [CrossRef]
- Attia, Z.I.; Noseworthy, P.A.; Lopez-Jimenez, F.; Asirvatham, S.J.; Deshmukh, A.J.; Gersh, B.J.; Carter, R.E.; Yao, X.; Rabinstein, A.A.; Erickson, B.J.; et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction. Lancet 2019, 394, 861–867. [Google Scholar] [CrossRef]
- Raghunath, S.; Ulloa Cerna, A.E.; Jing, L.; van Maanen, D.P.; Stough, J.; Hartzel, D.N.; Leader, J.B.; Kirchner, H.L.; Stumpe, M.C.; Hafez, A.; et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat. Med. 2020, 26, 886–891. [Google Scholar] [CrossRef]
- He, J.; Tse, G.; Korantzopoulos, P.; Letsas, K.P.; Ali-Hasan-Al-Saegh, S.; Kamel, H.; Li, G.; Lip, G.Y.H.; Liu, T. P-wave indices and risk of ischemic stroke: A systematic review and meta-analysis. Stroke 2017, 48, 2066–2072. [Google Scholar] [CrossRef] [Green Version]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef]
- Zhu, H.; Cheng, C.; Yin, H.; Li, X.; Zuo, P.; Ding, J.; Lin, F.; Wang, J.; Zhou, B.; Li, Y.; et al. Automatic multilabel electrocardiogram diagnosis of heart rhythm or conduction abnormalities with deep learning: A cohort study. Lancet Digit. Health 2020, 2, e348–e357. [Google Scholar] [CrossRef]
- Kei Fong, M.W.; Ng, E.Y.K.; Er Zi Jian, K.; Hong, T.J. SVR ensemble-based continuous blood pressure prediction using multi-channel photoplethysmogram. Comput. Biol. Med. 2019, 113, 103392. [Google Scholar] [CrossRef]
- Lin, C.T.; Wang, C.Y.; Huang, K.C.; Horng, S.J.; Liao, L.D. Wearable, Multimodal, Biosignal Acquisition System for Potential Critical and Emergency Applications. Emerg Med. Int 2021, 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Elgendi, M.; Fletcher, R.; Liang, Y.; Howard, N.; Lovell, N.H.; Abbott, D.; Lim, K.; Ward, R. The use of photoplethysmography for assessing hypertension. NPJ Digit. Med. 2019, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Tomita, K.; Nakada, T.A.; Oshima, T.; Oami, T.; Aizimu, T.; Oda, S. Non-invasive monitoring using photoplethysmography technology. J. Clin. Monit. Comput. 2019, 33, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Buchman, T.G. The community of the self. Nature 2002, 420, 246–251. [Google Scholar] [CrossRef]
- Annane, D.; Trabold, F.; Sharshar, T.; Jarrin, I.; Blanc, A.S.; Raphael, J.C.; Gajdos, P. Inappropriate sympathetic activation at onset of septic shock: A spectral analysis approach. Am. J. Respir. Crit. Care Med. 1999, 160, 458–465. [Google Scholar] [CrossRef]
- Tobin, M.J.; Mador, M.J.; Guenther, S.M.; Lodato, R.F.; Sackner, M.A. Variability of resting respiratory drive and timing in healthy subjects. J. Appl. Physiol. (1985) 1988, 65, 309–317. [Google Scholar] [CrossRef]
- Tobin, M.J.; Yang, K.L.; Jubran, A.; Lodato, R.F. Interrelationship of breath components in neighboring breaths of normal eupneic subjects. Am. J. Respir. Crit. Care Med. 1995, 152, 1967–1976. [Google Scholar] [CrossRef]
- Jonkman, A.H.; Rauseo, M.; Carteaux, G.; Telias, I.; Sklar, M.C.; Heunks, L.; Brochard, L.J. Proportional modes of ventilation: Technology to assist physiology. Intensive Care Med. 2020, 46, 2301–2313. [Google Scholar] [CrossRef]
- Gertler, R. Respiratory Mechanics. Anesthesiol. Clin. 2021, 39, 415–440. [Google Scholar] [CrossRef]
- Brochard, L. Breathing: Does regular mean normal? Crit. Care Med. 1998, 26, 1773–1774. [Google Scholar] [CrossRef] [PubMed]
- Seely, A.J.; Bravi, A.; Herry, C.; Green, G.; Longtin, A.; Ramsay, T.; Fergusson, D.; McIntyre, L.; Kubelik, D.; Maziak, D.E.; et al. Do heart and respiratory rate variability improve prediction of extubation outcomes in critically ill patients? Crit. Care 2014, 18, R65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, M.; Watkins, W.J.; Tansey, K.; King, W.E.; Banerjee, S. Predicting extubation outcomes using the heart rate characteristics index in preterm infants: A cohort study. Eur. Respir. J. 2020, 56, 1–9. [Google Scholar] [CrossRef]
- Lilitsis, E.; Stamatopoulou, V.; Andrianakis, E.; Petraki, A.; Antonogiannaki, E.M.; Georgopoulos, D.; Vaporidi, K.; Kondili, E. Inspiratory effort and breathing pattern change in response to varying the assist level: A physiological study. Respir. Physiol. Neurobiol. 2020, 280, 103474. [Google Scholar] [CrossRef]
- Grasselli, G.; Brioni, M.; Zanella, A. Monitoring respiratory mechanics during assisted ventilation. Curr. Opin. Crit. Care 2020, 26, 11–17. [Google Scholar] [CrossRef]
- Yoon, D.; Lee, S.; Kim, T.Y.; Ko, J.; Chung, W.Y.; Park, R.W. System for collecting biosignal data from multiple patient monitoring systems. Healthc. Inform. Res. 2017, 23, 333–337. [Google Scholar] [CrossRef] [Green Version]
- Roh, J.H.; Synn, A.; Lim, C.M.; Suh, H.J.; Hong, S.B.; Huh, J.W.; Koh, Y. A weaning protocol administered by critical care nurses for the weaning of patients from mechanical ventilation. J. Crit. Care 2012, 27, 549–555. [Google Scholar] [CrossRef]
- Schmidt, G.A.; Girard, T.D.; Kress, J.P.; Morris, P.E.; Ouellette, D.R.; Alhazzani, W.; Burns, S.M.; Epstein, S.K.; Esteban, A.; Fan, E.; et al. Liberation from mechanical ventilation in critically ill adults: Executive summary of an official american college of chest physicians/American thoracic society clinical practice guideline. Chest 2017, 151, 160–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.N.; Lin, L.Y.; Chen, K.Y.; Kuo, P.H.; Yu, C.J.; Wu, H.D.; Yang, P.C. Changes of heart rate variability during ventilator weaning. Chest 2003, 123, 1222–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Rovere, M.T.; Pinna, G.D.; Maestri, R.; Mortara, A.; Capomolla, S.; Febo, O.; Ferrari, R.; Franchini, M.; Gnemmi, M.; Opasich, C.; et al. Short-term heart rate variability strongly predicts sudden cardiac death in chronic heart failure patients. Circulation 2003, 107, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Malpas, S.C. Neural influences on cardiovascular variability: Possibilities and pitfalls. Am. J. Physiol. Heart Circ. Physiol. 2002, 282, H6–H20. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.T.; Tsai, Y.J.; Lin, J.W.; Ruan, S.Y.; Wu, H.D.; Yu, C.J. Application of heart-rate variability in patients undergoing weaning from mechanical ventilation. Crit. Care 2014, 18, R21. [Google Scholar] [CrossRef] [Green Version]
- Engoren, M. Approximate entropy of respiratory rate and tidal volume during weaning from mechanical ventilation. Crit. Care Med. 1998, 26, 1817–1823. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, V.E.; Chouvarda, I.G.; Maglaveras, N.K.; Pneumatikos, I.A. Study of multiparameter respiratory pattern complexity in surgical critically ill patients during weaning trials. BMC Physiol. 2011, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- Tanios, M.A.; Nevins, M.L.; Hendra, K.P.; Cardinal, P.; Allan, J.E.; Naumova, E.N.; Epstein, S.K. A randomized, controlled trial of the role of weaning predictors in clinical decision making. Crit. Care Med. 2006, 34, 2530–2535. [Google Scholar] [CrossRef]
- Huang, C.T.; Yu, C.J. Conventional weaning parameters do not predict extubation outcome in intubated subjects requiring prolonged mechanical ventilation. Respir. Care 2013, 58, 1307–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otaguro, T.; Tanaka, H.; Igarashi, Y.; Tagami, T.; Masuno, T.; Yokobori, S.; Matsumoto, H.; Ohwada, H.; Yokota, H. Machine learning for the prediction of successful extubation among patients with mechanical ventilation in the intensive care unit: A retrospective observational study. J. Nippon Med. Sch. 2021. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.L.; McCann, E.M.; Lloyd, B.W.; Yup, G. Inspiratory time and pulmonary function in mechanically ventilated babies with chronic lung disease. Pediatr. Pulmonol. 1991, 11, 198–201. [Google Scholar] [CrossRef]
- Vassilakopoulos, T.; Zakynthinos, S.; Roussos, C. Respiratory muscles and weaning failure. Eur. Respir. J. 1996, 9, 2383–2400. [Google Scholar] [CrossRef] [Green Version]
- Vassilakopoulos, T.; Zakynthinos, S.; Roussos, C. The tension-time index and the frequency/tidal volume ratio are the major pathophysiologic determinants of weaning failure and success. Am. J. Respir. Crit. Care Med. 1998, 158, 378–385. [Google Scholar] [CrossRef]
- Yoon, D.; Jang, J.H.; Choi, B.J.; Kim, T.Y.; Han, C.H. Discovering hidden information in biosignals from patients using artificial intelligence. Korean J. Anesthesiol. 2020, 73, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, D.; Zou, J. Deep learning models to detect hidden clinical correlates. Lancet Digit. Health 2020, 2, e334–e335. [Google Scholar] [CrossRef]
- Chen, X.; Guo, W.; Zhao, L.; Huang, W.; Wang, L.; Sun, A.; Li, L.; Mo, F. Acute myocardial infarction detection using deep learning-enabled electrocardiograms. Front. Cardiovasc. Med. 2021, 8, 654515. [Google Scholar] [CrossRef]
- Jia, Y.; Kaul, C.; Lawton, T.; Murray-Smith, R.; Habli, I. Prediction of weaning from mechanical ventilation using convolutional neural networks. Artif. Intell. Med. 2021, 117, 102087. [Google Scholar] [CrossRef]
- Chang, Y.J.; Hung, K.C.; Wang, L.K.; Yu, C.H.; Chen, C.K.; Tay, H.T.; Wang, J.J.; Liu, C.F. A real-time artificial intelligence-assisted system to predict weaning from ventilator immediately after lung resection surgery. Int. J. Environ. Res. Public Health 2021, 18, 2713. [Google Scholar] [CrossRef]
- Hill, B.L.; Rakocz, N.; Rudas, Á.; Chiang, J.N.; Wang, S.; Hofer, I.; Cannesson, M.; Halperin, E. Imputation of the continuous arterial line blood pressure waveform from non-invasive measurements using deep learning. Sci. Rep. 2021, 11, 15755. [Google Scholar] [CrossRef]
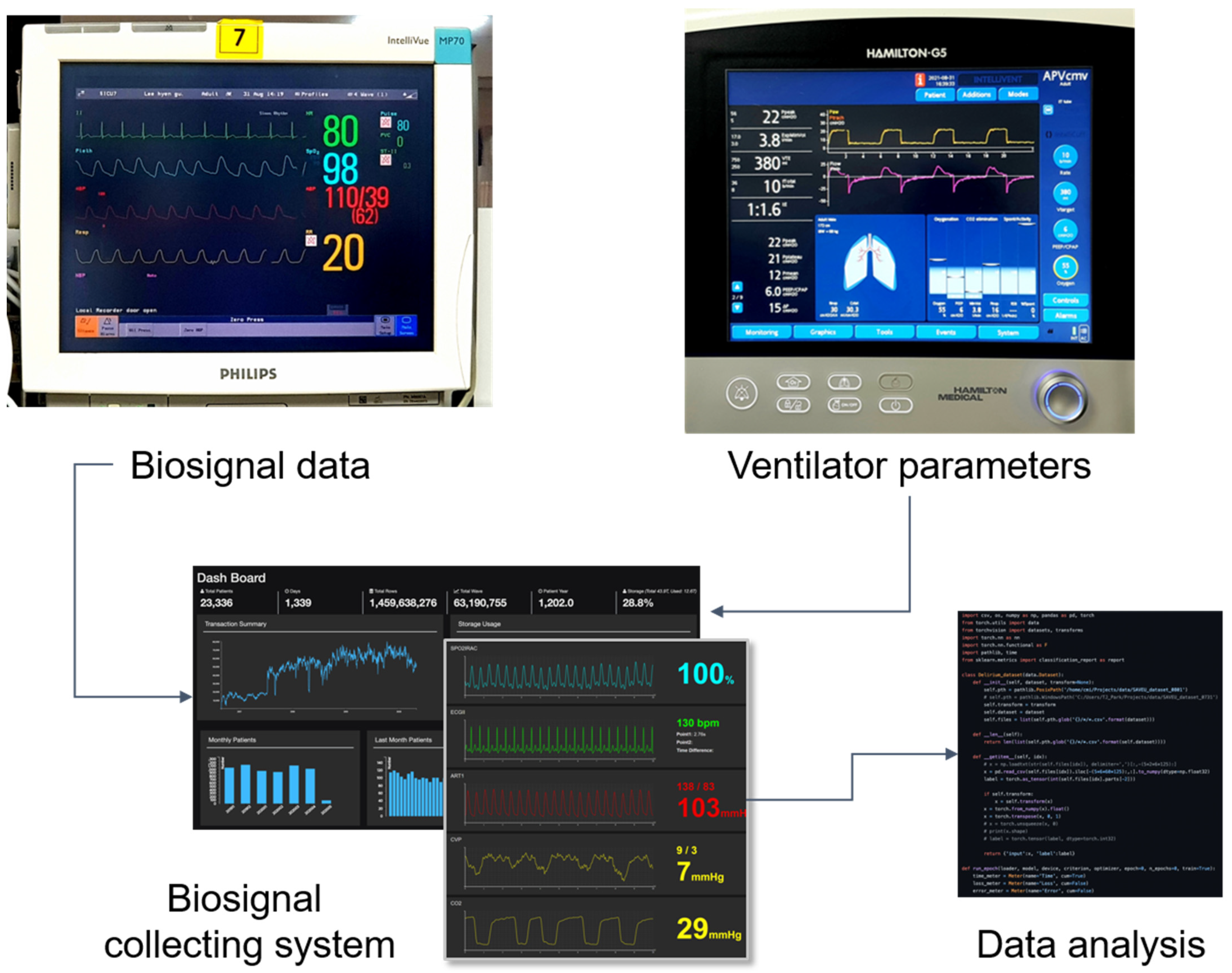

| Characteristics | Total (N = 89) | Success Group (N = 67) | Failure Group (N = 22) | p Value |
|---|---|---|---|---|
| Age, mean ± SD, year | 69.3 ± 14.3 | 69.8 ± 13.5 | 67.59 ± 16.5 | 0.533 |
| Sex (males/females), n | 54/35 | 40/27 | 14/8 | 0.743 |
| Body weight, mean ± SD, kg | 59.2 ± 11.7 | 59.6 ± 12.2 | 57.9 ± 10.3 | 0.568 |
| Height, mean ± SD, cm | 164.5 ± 9.6 | 163.8 ± 10.0 | 166.8 ± 8.0 | 0.193 |
| BMI, mean ± SD, kg/m2 | 21.9 ± 4.2 | 22.3 ± 4.3 | 20.8 ± 3.7 | 0.152 |
| Main cause of ICU admission, n(%) | 0.897 | |||
| Pneumonia | 59 (66.3) | 45 (67.2) | 14 (63.6) | |
| COPD/Asthma AE | 8 (9.0) | 6 (9.0) | 2 (9.1) | |
| Pulmonary hemorrhage | 4 (4.5) | 3 (4.5) | 1 (4.5) | |
| Sepsis | 3 (3.4) | 3 (4.5) | 0 (0) | |
| Gastrointestinal bleeding | 1 (1.1) | 1 (1.5) | 0 (0) | |
| Neurologic disease | 2 (2.2) | 1 (1.5) | 1 (4.5) | |
| Pulmonary edema | 7 (7.9) | 5 (7.5) | 2 (9.1) | |
| Others | 5 (5.6) | 3 (4.5) | 2 (9.1) | |
| Comorbidity, n(%) | ||||
| Cardiovascular disease | 52 (58.4) | 40 (59.7) | 12 (54.5) | 0.670 |
| Diabetes mellitus | 25 (28.1) | 20 (29.9) | 5 (22.7) | 0.519 |
| Chronic obstructive pulmonary disease | 16 (18.0) | 11 (16.4) | 5 (22.7) | 0.530 |
| Neurological disease | 24 (27.0) | 20 (29.9) | 4 (18.2) | 0.285 |
| Malignancy | 18 (20.2) | 14 (20.9) | 4 (18.2) | >0.99 |
| Renal disease | 10 (11.2) | 9 (13.4) | 1 (4.5) | 0.440 |
| Liver disease | 4 (4.5) | 4 (6.0) | 0 (0) | 0.568 |
| APACHE II score, mean ± SD | 21.8 ± 8.1 | 22.3 ± 8.3 | 20.2 ± 7.3 | 0.288 |
| Length of mechanical ventilation before SBT, mean ± SD, d | 7.3 ± 5.3 | 7.0 ± 5.5 | 8.1 ± 4.7 | 0.393 |
| Duration of MV ≥ 72 h, n(%) | 68 (76.4) | 50 (74.6) | 18 (81.8) | 0.491 |
| Use of neuromuscular blocker, n(%) | 18 (20.2) | 13 (19.4) | 5 (22.7) | 0.764 |
| Excess secretion, n(%) | 9 (10.1) | 6 (9.0) | 3 (13.6) | 0.684 |
| Arterial blood gas ananlysis, mean ± SD | ||||
| PaO2, mmHg | 107.8 ± 34.8 | 108.0 ± 31.4 | 106.9 ± 44.4 | 0.891 |
| PaCO2, mmHg | 38.7 ± 11.0 | 37.6 ± 10.4 | 41.9 ± 12.4 | 0.116 |
| PaO2/FiO2 ratio | 317.3 ± 102.3 | 320.5 ± 91.4 | 307.3 ± 132.1 | 0.666 |
| Upper airway disorder after extubation, n(%) | 2 (2.2) | 2 (3) | 0 (0) | >0.99 |
| Prior failed weaning attempt, n(%) | 15 (16.9) | 9 (13.4) | 6 (27.3) | 0.187 |
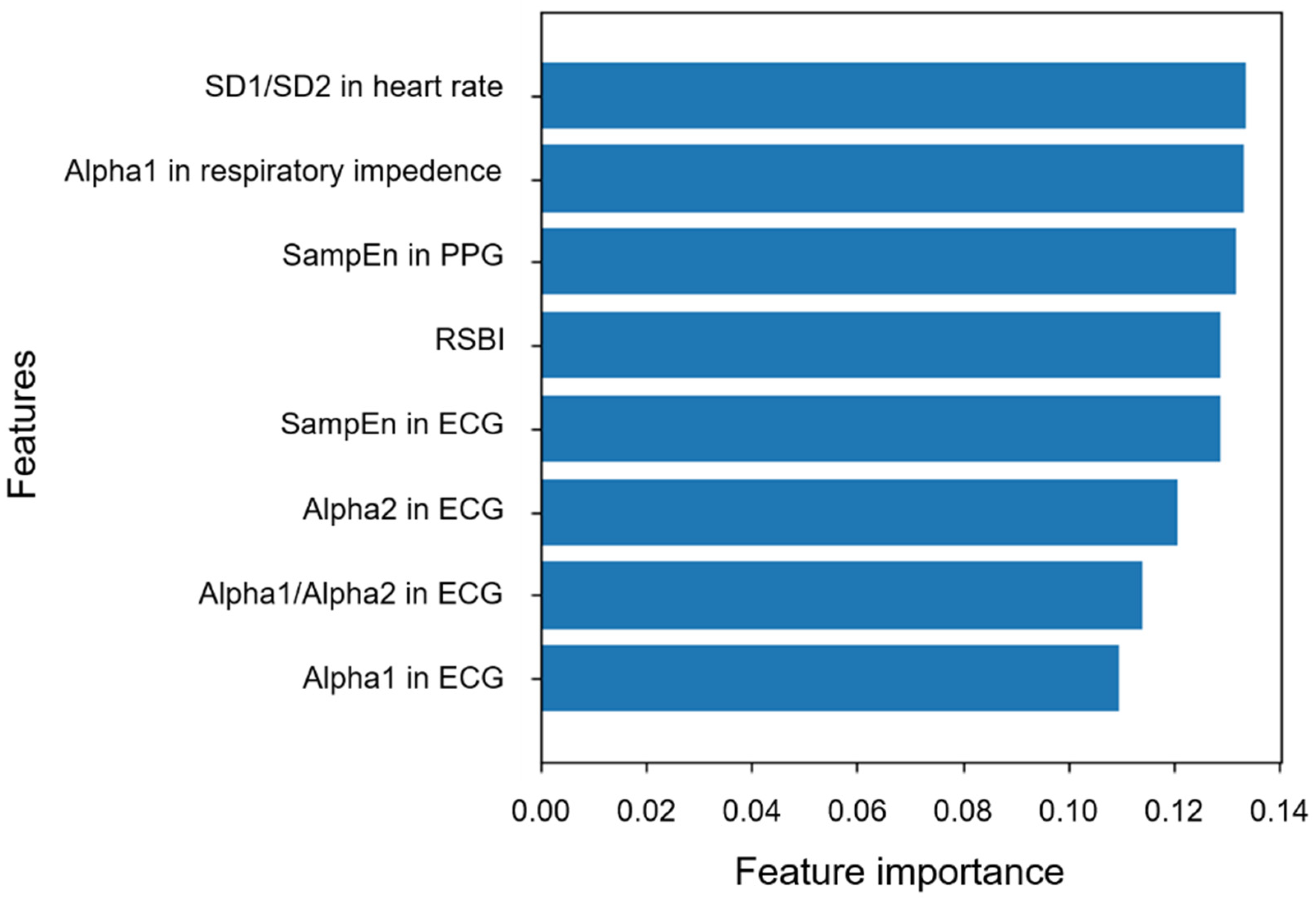
| Items | Variability Index | Success Group | Failure Group | p Value | αadj † |
|---|---|---|---|---|---|
| Heart rate | SD1 (mean ± SD) | 2.52 (1.45) | 2.17 (1.15) | 0.316 | 0.035 |
| SD2 (mean ± SD) | 6.74 (4.56) | 8.63 (6.98) | 0.741 | 0.076 | |
| SD1/SD2 (mean ± SD) | 0.43 (0.18) | 0.32 (0.13) | 0.015 * | 0.009 | |
| Respiratory rate | SD1 (mean ± SD) | 2.76 (1.22) | 1.77 (0.49) | 0.617 | 0.068 |
| SD2 (mean ± SD) | 2.94 (1.25) | 3.08 (1.25) | 0.561 | 0.059 | |
| SD1/SD2 (mean ± SD) | 0.62 (0.16) | 0.65 (0.24) | 0.592 | 0.065 | |
| Tidal volume | SD1 (mean ± SD) | 52.58 (32.96) | 27.64 (10.9) | 0.237 | 0.029 |
| SD2 (mean ± SD) | 72.90 (40.18) | 45.40 (9.86) | 0.747 | 0.079 | |
| SD1/SD2 (mean ± SD) | 0.72 (0.2) | 0.62 (0.19) | 0.496 | 0.053 | |
| IE ratio | SD1 (mean ± SD) | 61.23 (47.58) | 164.57 (297.75) | 0.882 | 0.094 |
| SD2 (mean ± SD) | 61.23 (47.58) | 189.32 (268.39) | 0.408 | 0.044 | |
| SD1/SD2 (mean ± SD) | 0.63 (0.22) | 0.55 (0.23) | 0.318 | 0.038 | |
| Inspiratory time | SD1 (mean ± SD) | 96.20 (68.04) | 79.98 (53.70) | 0.750 | 0.082 |
| SD2 (mean ± SD) | 146.79 (79.75) | 156.47 (117.67) | 0.567 | 0.062 | |
| SD1/SD2 (mean ± SD) | 0.66 (0.24) | 0.58 (0.28) | 0.511 | 0.056 | |
| Mean ABP | SD1 (mean ± SD) | 5.37 (4.58) | 5.99 (9.42) | 0.340 | 0.041 |
| SD2 (mean ± SD) | 10.80 (6.09) | 15.98 (23.12) | 0.832 | 0.088 | |
| SD1/SD2 (mean ± SD) | 0.5 (0.21) | 0.4 (0.17) | 0.087 | 0.026 | |
| ECG | SampEn (mean ± SD) | 2.04 (0.61) | 2.50 (0.46) | 0.005 ** | 0.006 |
| ɑ1 (mean ± SD) | 1.29 (0.15) | 1.22 (0.08) | 0.033 * | 0.021 | |
| ɑ2 (mean ± SD) | 0.57 (0.23) | 0.43 (0.20) | 0.016 * | 0.012 | |
| ɑ1/ɑ2 (mean ± SD) | 2.83 (2.46) | 3.47 (1.57) | 0.026 * | 0.018 | |
| Respiratory impedance | SampEn (mean ± SD) | 0.21 (0.05) | 0.22 (0.05) | 0.413 | 0.047 |
| ɑ1 (mean ± SD) | 2.03 (0.04) | 2.01 (0.03) | 0.018 * | 0.015 | |
| ɑ2 (mean ± SD) | 1.11 (0.27) | 1.07 (0.31) | 0.719 | 0.074 | |
| ɑ1/ɑ2 (mean ± SD) | 2.04 (1.11) | 2.70 (3.64) | 0.973 | 0.1 | |
| PPG | SampEn (mean ± SD) | 0.14 (0.05) | 0.18 (0.11) | 0.002 ** | 0.003 |
| ɑ1 (mean ± SD) | 1.96 (0.12) | 1.91 (0.19) | 0.062 | 0.024 | |
| ɑ2 (mean ± SD) | 1.96 (0.12) | 0.78 (0.49) | 0.429 | 0.05 | |
| ɑ1/ɑ2 (mean ± SD) | 5.61 (10.72) | −2.15 (29.37) | 0.947 | 0.097 | |
| ABP | SampEn (mean ± SD) | 0.36 (0.39) | 0.43 (0.46) | 0.247 | 0.032 |
| ɑ1 (mean ± SD) | 2.09 (0.01) | 2.08 (0.02) | 0.627 | 0.071 | |
| ɑ2 (mean ± SD) | 1.83 (0.10) | 1.82 (0.12) | 0.775 | 0.085 | |
| ɑ1/ɑ2 (mean ± SD) | 1.14 (0.07) | 1.15 (0.08) | 0.853 | 0.091 |
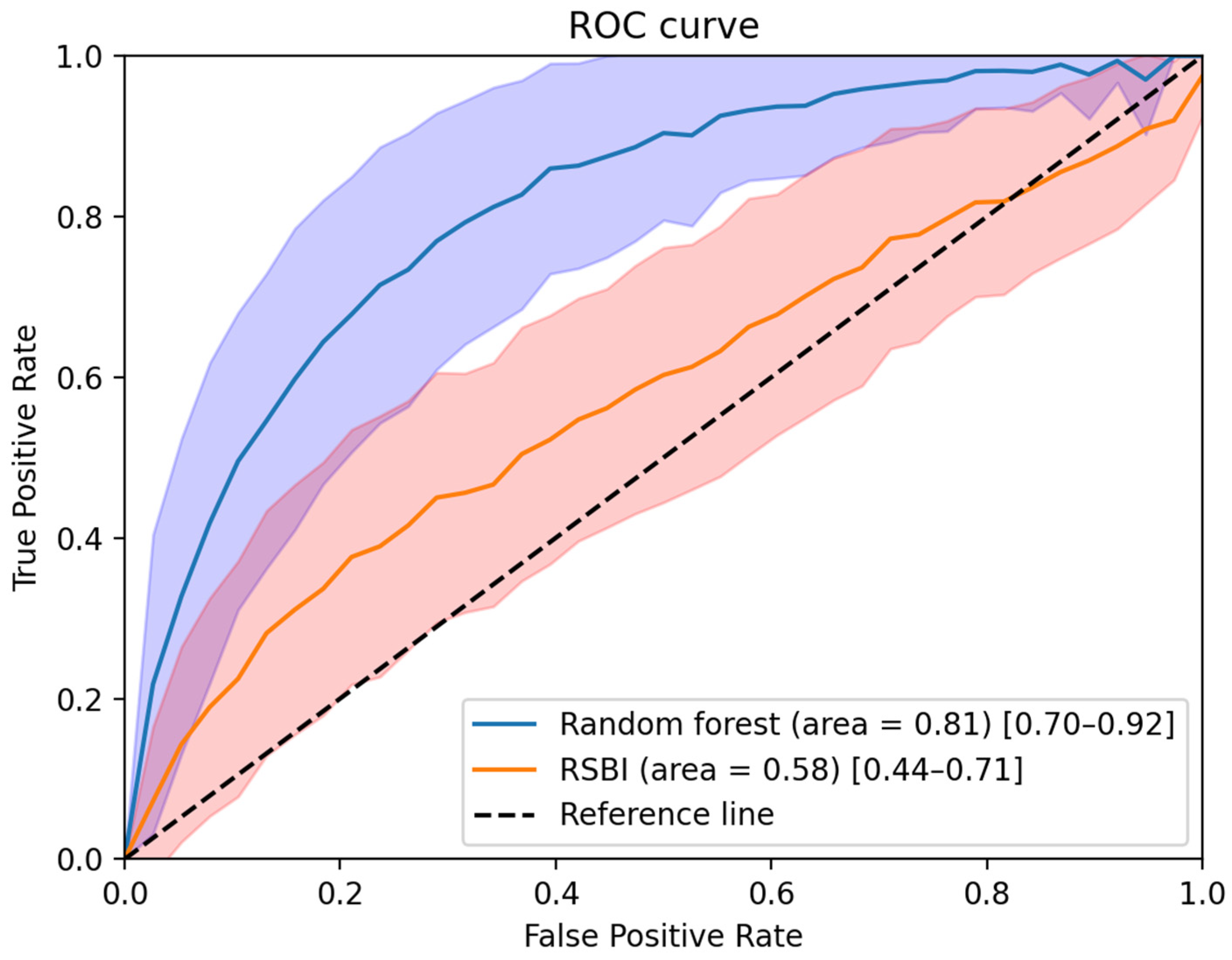
| Sensitivity | Specificity | Accuracy | PPV | NPV | F-1 Score | |
|---|---|---|---|---|---|---|
| RSBI (≥105) | 0.91 (0.87–0.96) | 0.26 (0.13–0.38) | 0.80 (0.75–0.84) | 0.85 (0.83–0.88) | 0.40 (0.20–0.61) | 0.30 (0.17–0.44) |
| RSBI + biosignal (Random Forest) | 0.91 (0.85–0.97) | 0.52 (0.36–0.69) | 0.84 (0.79–0.89) | 0.90 (0.87–0.93) | 0.58 (0.40–0.76) | 0.53 (0.40–0.66) |
| RSBI + biosignal (Multiple regression) | 0.91 (0.86–0.97) | 0.41 (0.25–0.57) | 0.82 (0.78–0.87) | 0.88 (0.85–0.91) | 0.53 (0.33–0.73) | 0.44 (0.30–0.58) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.E.; Kim, T.Y.; Jung, Y.J.; Han, C.; Park, C.M.; Park, J.H.; Park, K.J.; Yoon, D.; Chung, W.Y. Biosignal-Based Digital Biomarkers for Prediction of Ventilator Weaning Success. Int. J. Environ. Res. Public Health 2021, 18, 9229. https://doi.org/10.3390/ijerph18179229
Park JE, Kim TY, Jung YJ, Han C, Park CM, Park JH, Park KJ, Yoon D, Chung WY. Biosignal-Based Digital Biomarkers for Prediction of Ventilator Weaning Success. International Journal of Environmental Research and Public Health. 2021; 18(17):9229. https://doi.org/10.3390/ijerph18179229
Chicago/Turabian StylePark, Ji Eun, Tae Young Kim, Yun Jung Jung, Changho Han, Chan Min Park, Joo Hun Park, Kwang Joo Park, Dukyong Yoon, and Wou Young Chung. 2021. "Biosignal-Based Digital Biomarkers for Prediction of Ventilator Weaning Success" International Journal of Environmental Research and Public Health 18, no. 17: 9229. https://doi.org/10.3390/ijerph18179229
APA StylePark, J. E., Kim, T. Y., Jung, Y. J., Han, C., Park, C. M., Park, J. H., Park, K. J., Yoon, D., & Chung, W. Y. (2021). Biosignal-Based Digital Biomarkers for Prediction of Ventilator Weaning Success. International Journal of Environmental Research and Public Health, 18(17), 9229. https://doi.org/10.3390/ijerph18179229





