Abstract
Recent scientific trends have revealed that the collection and analysis of data on the occurrence and fate of SARS-CoV-2 in wastewater may serve as an early warning system for COVID-19. In South Africa, the first COVID-19 epicenter emerged in the Western Cape Province. The City of Cape Town, located in the Western Cape Province, has approximately 4 million inhabitants. This study reports on the monitoring of SARS-CoV-2 RNA in the wastewater of the City of Cape Town’s wastewater treatment plants (WWTPs) during the peak of the epidemic. During this period, the highest overall median viral RNA signal was observed in week 1 (9200 RNA copies/mL) and declined to 127 copies/mL in week 6. The overall decrease in the amount of detected viral SARS-CoV-2 RNA over the 6-week study period was associated with a declining number of newly identified COVID-19 cases in the city. The SARS-CoV-2 early warning system has now been established to detect future waves of COVID-19.
1. Introduction
Wastewater-based epidemiology has played an important role in the development of early warning systems (EWS) for various enteric viruses, including poliovirus, norovirus and hepatitis [1]. In the current COVID-19 pandemic, wastewater-based epidemiology has shown the potential to provide a platform for SARS-CoV-2 surveillance. SARS-CoV-2 RNA has been detected in the feces of both symptomatic and asymptomatic cases [2] and research in many countries, including France, the Netherlands and the United States of America (USA), has shown a correlation between SARS-CoV-2 RNA viral loads in wastewater and COVID-19 clinical case data [3,4]. For example, a study in the Netherlands detected SARS-CoV-2 RNA in wastewater even when the COVID-19 prevalence was low [4]. Such findings have provided the evidence-base for the Netherlands to incorporate sewage surveillance as an EWS within its national response to COVID-19 monitoring and other countries, including Australia, Germany, New Zealand and the USA, are taking similar steps. According to the World Health Organization (WHO) [5], it has been demonstrated that wastewater-based epidemiology has the potential to be used for monitoring COVID-19 prevalence and temporal trends, however it is necessary to pilot this approach in low- and middle-income (LMIC) settings to demonstrate its added value to support clinical surveillance. An effective wastewater-based epidemiology system may prove critical where health systems infrastructures, testing systems, personal protective equipment (PPE) and human resource capacities are constrained.
The City of Cape Town, in the Western Cape Province, is one of eight metropolitan municipalities in South Africa, with approximately 4 million inhabitants. It is one of the wealthiest cities in Africa but is also recognized as having one of the highest income inequalities globally. This inequality extends to health care access and living conditions, leading to variations in the transmission and spread of COVID-19 within the metropolis. As such, wastewater surveillance within the City of Cape Town can assist in unbiased tracking of COVID-19 at the sub-city scale to determine potential localized outbreaks. The first COVID-19 case in the Western Cape was reported on 11 March 2020 and the confirmed cases gradually increased after that point; from 1 May to 1 June 2020, the number increased from 226 cases to 1668 cases, respectively [6]. In early July 2020, the Western Cape emerged as the epicenter of COVID-19 in South Africa and at the time carried the highest proportion of COVID-19 cases (35%) as well as related deaths (64%) in the country [7]. To coincide with the COVID-19 peak in the Western Cape, this study undertook wastewater sampling in 23 WWTPs in the City of Cape Town and determined the spatial and temporal SARS-CoV-2 trends over a six-week period.
2. Materials and Methods
2.1. Wastewater Sampling
From 6 July 2020, one 500 mL untreated influent wastewater grab sample was collected at the raw inlet after coarse screening from each of the 23 wastewater treatment plants (WWTPs) in the City of Cape Town. Samples were collected once a week on a Monday with sample collection occurring at a similar time each week over a 6-week period. Two of the WWTPs (namely, Millers Point and Oudekraal) served as controls as they are situated on nature reserves and do not serve residential populations. A random computer-generated number was assigned to each WWTP and the laboratory staff were blinded to the incoming samples. All samples were transported to the laboratory on ice and processed immediately for the extraction of RNA, after which it was stored at −80 °C for subsequent quantitative real-time polymerase chain reaction (qRT-PCR).
2.2. Sample Concentration and RNA Extraction
A modified method described by Peccia et al. [8] and optimized by Johnson et al. [9] was used in order to extract the total RNA using the Qiagen RNeasy® PowerSoil® Kit, as per the manufacturer’s instructions (Qiagen, Hilden, Germany). Briefly, 100 mL of influent wastewater was spun down at 2500× g for 20 min, whereafter 2–5 mL of the resultant supernatant pellet was added to a 15 mL PowerBead® Tube containing a lysis buffer to inactivate the virus and stabilize the viral RNA. Thereafter, the sample was homogenized and phase-separated using an equal volume of phenol/chloroform and the upper aqueous phase was transferred to a new 15 mL tube and mixed with the required buffers as supplied within the RNeasy PowerSoil kit. The RNA isolation sample was then transferred to the RNeasy JetStar Mini Column to elute out the bound RNA before centrifugation at 13,000× g for 15 min. The resultant pellet was dried and dissolved in a final volume of 50 µL of ribonuclease-free water. The quantity and quality of the total RNA was measured by spectrophotometry using the NanoDrop® ND-1000 instrument (Nanodrop Technologies, Wilmington, NC, USA). In the absence of a surrogate, a clinical SARS-CoV-2 positive nasal swab sample with known viral copies was used to spike a wastewater sample in order to investigate the efficiency of the extraction method.
2.3. Quantitative Real-Time Polymerase Chain Reaction Analysis
The Centers for Disease Control and Prevention (CDC) approved quantitative real-time polymerase chain reaction (qRT-PCR) N1 and N2 primer/probe assays (Table 1) [10], which were purchased from Whitehead Scientific (Integrated DNA Technologies, Coralville, USA) and used for the detection of SARS-CoV-2 viral RNA in wastewater samples. Both N1 and N2 primer/probe sets aligned 100% with the N-protein of the SARS-CoV-2 strain. For qRT-PCR positive control and viral RNA copy number quantification, a 1:10 fold serial dilution was made with assays 2019-nCoV-N-Positive plasmid control, which was supplied at 200,000 copies/µL (Qauntabio, Beverley, MA, USA). For qualitative and quantitative analysis detection of SARS-CoV-2 viral RNA, a one-step qRT-PCR reaction was performed using the iTaqTM Universal Probes One-Step Reaction kit, according to the manufacturer’s instructions (Bio-Rad Laboratories, Richmond, CA, USA). Briefly, 1 µL of 0.2 µg/µL of total RNA was used for the qRT-PCR reaction in a final volume of 10 µL, using the QuantStudio™ 7 Flex Real-Time PCR System (ABI instrument, Life Technologies, Carlsbad, USA). All reactions were performed in technical duplicate and a template control was included for each experimental run. To minimize potential contamination, RNA extraction and qRT-PCR were performed in separate laboratories.

Table 1.
Thermal cycling protocol and primers and probes used in the study.
2.4. Statistical Analysis
The SARS-CoV-2 copies per milliliter of wastewater were described using summary statistics. The Shapiro–Wilk test was used to test the normality of SARS-CoV-2 copies per milliliter. Spearman’s rank correlation was used to determine the correlation between N1 and N2 primers. The signed rank test was used to test whether there was a significant change in SARS-Cov-2 RNA signals between weeks 1, 3 and 6. At week 1, quartiles estimated from the average of N1 and N2 were used to form the four RNA signal categories (category 1 with the lowest and category 4 with the highest RNA signal) which formed the basis for the maps presented. Additionally, a Spearman’s rho correlation analysis was performed on the wastewater and clinical cases.
2.5. Spatial Data
Suburb shapefiles were obtained from the City of Cape Town’s open data portal. Coordinates for each WWTP were collected using a handheld GPS and verified using Google Earth. All maps were produced using ArcGIS 10.6.1. (ESRI, Durban, South Africa). The WWTP catchment areas (comprising of 753 suburbs) were joined to the corresponding WWTPs.
3. Results
WWTP capacity ranged from 0.03 Ml/day to 200 Ml/day (Oudekraal and Cape Flats, respectively). The WWTP site characteristics are presented in Supplementary Table S1. A total of 138 grab samples, one per WWTP, were collected weekly over 6 weeks, of which 12 samples were collected from the two control sites. A 15% recovery efficiency of the extraction procedure was in line with previously published studies [11,12,13]. There was a strong correlation between viral RNA copies/mL for N1 and N2 primers (r = 0.897, p-value < 0.001). Samples from both control sites (Millers Point and Oudekraal WWTP) contained undetectable viral RNA loads over the 6-week period. Week 1 had the highest overall (median) viral RNA signal (9200 RNA copies/mL). The extremely high SARS-CoV-2 RNA signal recorded in week 1 at the Mitchells Plain WWTP (37,486,080 copies/mL) was flagged and re-analysis in the laboratory confirmed this result. There were a median decrease of 4779.5 RNA copies/mL between weeks 1 and 3 (p-value 0.001) and a median decrease of 3592 copies/mL between weeks 3 and 6 (p-value 0.0001). The median SARS-CoV-2 RNA for weeks 2, 4 and 5 were 3448, 4032 and 583 copies/mL, respectively.
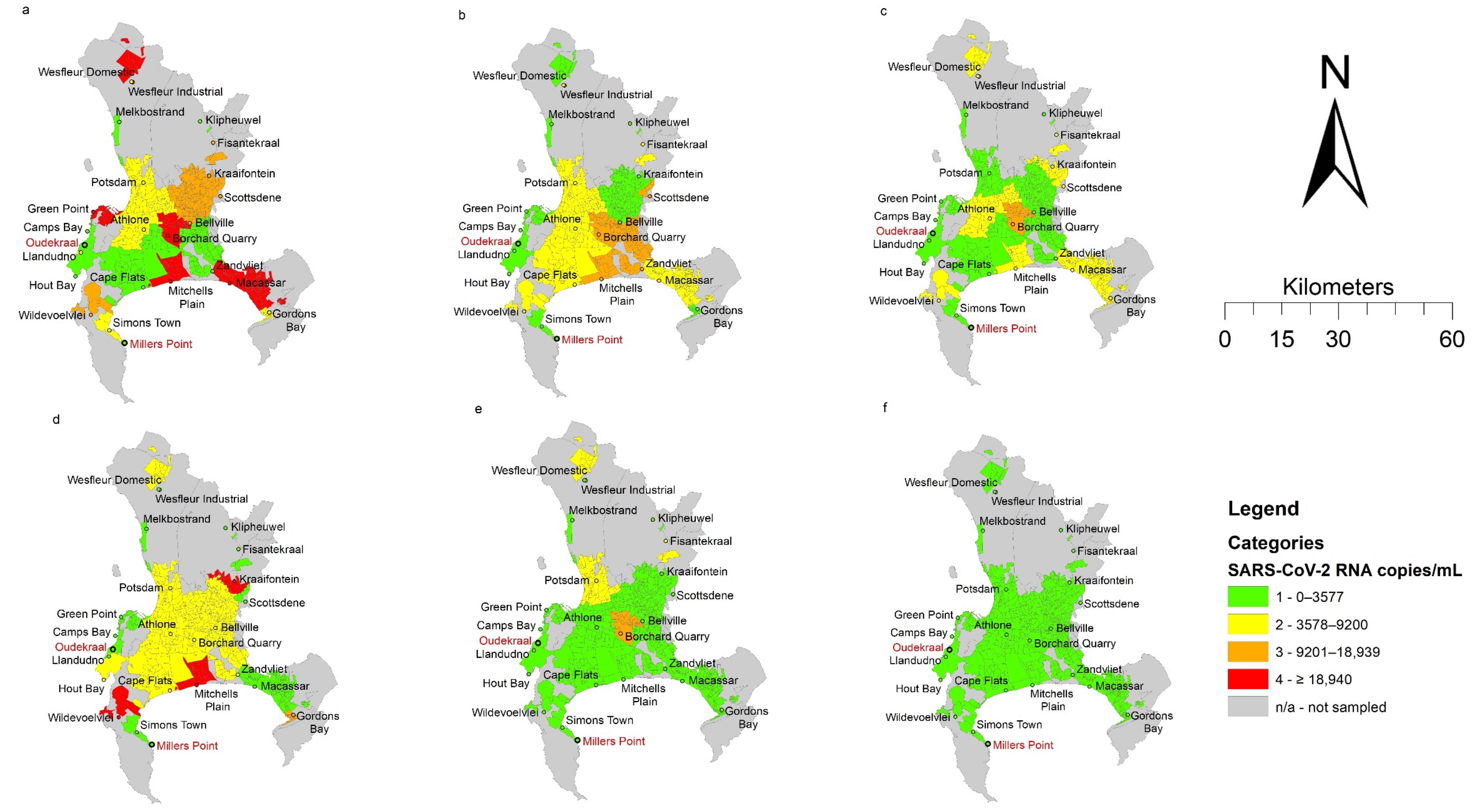
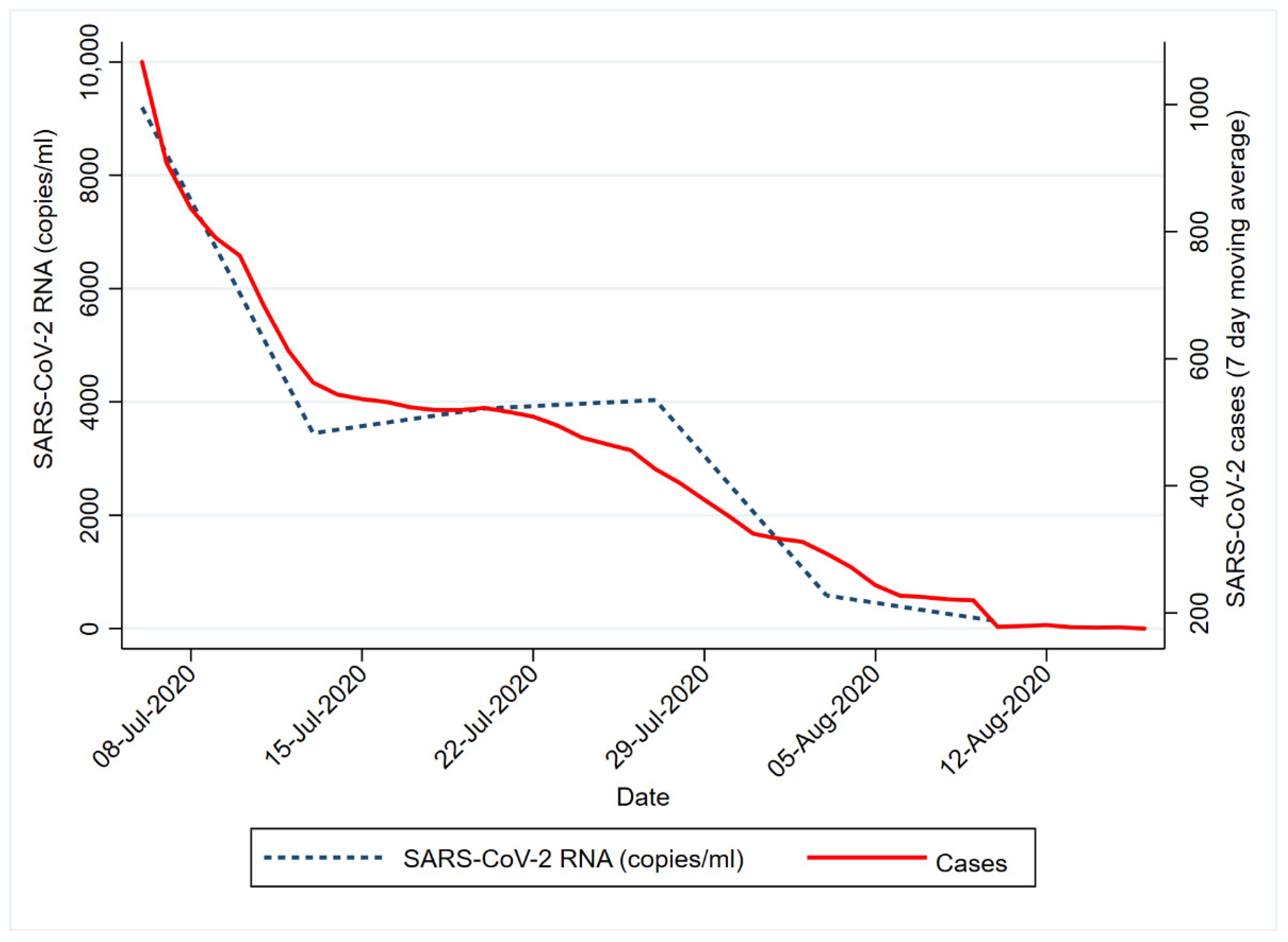
The spatial representation (Figure 1) shows the changing trend in SARS-CoV-2 RNA signals over time in relation to the starting time point (week 1). Based on the Inter Quartile Ranges (IQR), the RNA signals observed from the wastewater samples progressively decreased from week 1 to week 6. It must be noted that despite the decreased SARS-CoV-2 RNA load detected in most of the WWTPs, areas serviced by the Mitchells Plain and Borcherds Quarry WWTP had higher viral loads. Notably, both Mitchell’s Plain and Borcherds Quarry WWTPs were ‘hotspots’ (i.e., higher signal areas) in week 1. Additionally, the reduction in SARS-CoV-2 RNA corresponded with the decline in COVID-19 clinical cases over the same period (Figure 2). This was further confirmed by the Spearman’s rank test, which showed a significant positive correlation (r = 0.83; p = 0.0416) between the reported clinical cases and the SARS-CoV-2 viral RNA in the wastewater.
Figure 1.
Spatial representation of SARS-CoV-2 RNA/mL in wastewater from 23 wastewater treatments plants and related catchment areas (suburbs) in the City of Cape Town at 6 sampling timepoints: (a): 6 July 2020 (week 1); (b): 13 July 2020 (week 2); (c): 20 July 2020 (week 3); (d): 27 July 2020 (week 4); (e): 3 August 2020 (week 5); (f): 10 August 2020 (week 6).
Figure 2.
Number of confirmed new COVID-19 cases in the City of Cape Town from the period of 6 July 2020 to 10 August 2020. Data retrieved from the COVID-19 Dashboard from the Western Cape Provincial Government [6].
4. Discussion
The COVID-19 pandemic has had devastating societal and economic impacts on South Africa [14]. In this study, we determined spatial and temporal trends of SARS-CoV-2 in wastewater as the basis for a COVID-19 early warning system. The overall trends show the declining SARS-CoV-2 RNA signal that corresponds with the declining COVID-19 case numbers in Cape Town as reported by the Western Cape Provincial Government. Arora et al. [15] reported the presence of SARS-CoV-2 RNA in the wastewater over a period of three weeks in Jaipur City, India. Low SARS-CoV-2 RNA signal in wastewater corresponded to the low number of positive COVID-19 cases in the city. In Brazil, researchers monitored the SARS-CoV-2 RNA signal in wastewater for 41 weeks in the city of São Paulo and observed a positive correlation between the RNA signal in the wastewater and clinical cases [16]. Our results are consistent with these studies. In our study, untreated, influent wastewater samples were collected from the inlet of WWTPs, which other studies have demonstrated is the preferred sample type for investigating changes in SARS-CoV-2 RNA signals and correlations between clinical cases and hotspot identification [17,18].
The heterogeneous RNA signals within the City of Cape Town highlight the importance of temporal trends at a sub-city scale, especially for densely populated cities. A study in south-eastern Virginia (USA) suggested that as the COVID-19 pandemic wanes, it is likely that communities will see an increased incidence of small, localized outbreaks which can be detected by water-based epidemiology [19]. In South Africa, increased local transmission of the virus has been noted in the second and third waves, as well as following local super-spreader events [20,21]. The surveillance of suburbs allows for authorities to pin-point small outbreaks which for South Africa and other resource-constrained countries can guide targeted interventions and timeous public health responses [22].
In March of 2021, The European Commission issued a recommendation for nationwide wastewater monitoring system for SARS-CoV-2 surveillance as a complementary approach to monitor the spread of COVID-19 in all its member states [23]. Owing to reports of the detection and tracking of the virus in wastewater by several European countries in 2020, all EU member states were called to establish a wastewater surveillance system by October 2021. High-income countries with well-equipped sanitation systems, such as Finland [24], the Netherlands [25] and the USA [26], have established nationwide COVID-19 wastewater monitoring programs. Wastewater surveillance has shown to be a useful alternative and low-cost tool for population-level screening in these countries.
In LMICs, several studies have detected the presence of SARS-CoV-2 in wastewater including Argentina [27], Brazil [28] and India [29]. To our knowledge, this is the first study in an African setting that reports the spatial and temporal trends of the SARS-CoV-2 RNA signal in wastewater covering an entire Metro. This study demonstrates how wastewater monitoring of SARS-CoV-2 can be of benefit for and can support clinical testing, identifying hotspots, informing preparedness and monitoring the effectiveness of response measures brought about by public health officials.
The SARS-CoV-2 RNA signals in the study are reported as the virus concentration without normalization due to uncertainties in readily available information about flow rate and population size estimates. As highlighted by Hou et al. [30], census data is often outdated and does not accommodate for population mobility in cities, while the design capacity of a WWTW is not reflective of the real time load in a sewerage system. Moreover, Feng et al. [31] found that normalization to WWTP characteristics had a minimal impact when correlating SARS-CoV-2 RNA in wastewater to clinical cases. Given these uncertainties, the limited availability of such information in the study and the South African context, future research can explore normalization using a human fecal marker, such as pepper mild mottle virus.
At the time of sampling, South Africa was in lockdown level three, which indicated the most moderate of the five severity levels. Although the first wave of COVID-19 cases has since passed, there has been a resurgence of COVID-19 clusters and outbreaks, with the country currently experiencing a third wave [32]. A rapid public health response is therefore critical to prevent further morbidity and mortality, as health care systems in South Africa have come under strain. Moreover, limited and biased testing data may mask the estimates of COVID-19 infections, which are critical to informing public health responses [33,34]. The SARS-CoV-2 viral load in wastewater represents both asymptomatic and symptomatic shedding in a specific geographic location, thus providing a less biased dataset which can be used to assess infection prevalence [19]. Results from this study further support the need for prolonged wastewater surveillance as an additional strategy to current tracing methods employed to mitigate COVID-19 spread. Nonetheless, this study has a few key limitations. The wastewaters samples were collected using the grab sampling technique. Therefore, the results presented for each week are reflective of one time point. However, the routine sampling strategy adopted in this study allowed for weekly comparisons of concentrations to be made at each wastewater treatment plant. Furthermore, the SARS-CoV-2 variants of concern (VOC) have played an integral part in driving the pandemic. SARS-CoV-2 VOC testing was not undertaken in this study; however, our current and future wastewater surveillance studies now include VOC testing.
5. Conclusions
In conclusion, we report the detection of SARS-CoV-2 RNA in the wastewater of the City of Town’s WWTPs. We identified an overall decrease in the amount of detected viral RNA over the 6-week study period, associated with a declining number of newly identified COVID-19 cases. This is the first study on the African continent to determine spatial and temporal trends of SARS-CoV-2 spanning an entire metropolitan city. As South Africa has declining COVID-19 case numbers, ongoing wastewater surveillance is an economical early warning system that can guide public health responses. On the basis of these study results and the feasibility of trend detection, a pilot early warning system is underway in various cities across South Africa to identify potential COVID-19 resurgences at the sub-city scale.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijerph182212085/s1, S1: Wastewater treatment plant characteristics.
Author Contributions
Conceptualization, R.S., R.J., A.M., C.M., J.L.; Methodology, L.B., N.M. (Noluxabiso Mangwanaand), S.D., J.R.S., P.R., R.J.; T.R.; Software, T.R., N.M. (Noluxabiso Mangwanaand), C.W.; Validation, T.R.; Formal analysis, N.M. (Noluxabiso Mangwanaand), S.D., J.R.S., P.R., J.L., R.J., C.M.; Writing—original draft preparation, R.S., A.M., T.R., S.N., M.S.M., N.M. (Nomfundo Mahlangeni), C.W.; Writing—review and editing, All authors; Funding acquisition, R.S., A.M., M.M., C.M., J.L., R.J.; All authors have read and agreed to the published version of the manuscript.
Funding
The South African Medical Research Council grant number: 40118. The Michael & Susan Dell Foundation grant number: 96829.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available from author upon reasonable request.
Acknowledgments
The South African Medical Research Council and the Michael & Susan Dell Foundation are thanked for funding and support. We are grateful to the City of Cape Town for provision of wastewater samples.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The funders had no involvement in the study design, the collection, the analysis and the interpretation of data. The contents are solely the responsibility of the authors.
References
- Hovi, T.; Shulman, L.; Van Der Avoort, H.; Deshpande, J.; Roivainen, M.; De Gourville, E. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012, 140, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, F.; Deng, L.; Zhang, L.; Cai, Y.; Cheung, C.W.; Xia, Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J. Gen. Intern. Med. 2020, 35, 1545–1549. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–556. [Google Scholar] [CrossRef]
- WHO. Rapid expert consultation on environmental surveillance of SARS-CoV-2 in wastewater: Summary report: Virtual meeting. 23 July 2020. [Google Scholar]
- Government WC. COVID-19 Dashboard. Available online: https://coronavirus.westerncape.gov.za/covid-19-dashboard (accessed on 7 August 2021).
- NICD. Latest Confirmed Cases of Covid-19 in South Africa (6 July 2020). 2020. Available online: https://www.nicd.ac.za/latest-confirmed-cases-of-covid-19-in-south-africa-6-july-2020/ (accessed on 30 September 2021).
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M.; et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Muller, C.J.F.; Ghoor, S.; Louw, J.; Archer, E.; Surujlal-Naicker, S.; Berkowitz, N.; Volschenk, M.; Bröcker, L.H.L.; Wolfaardt, G.; et al. Qualitative and quantitative detection of SARS-CoV-2 RNA in untreated wastewater in Western Cape Province, South Africa. S. Afr. Med. J. 2021, 111, 5. [Google Scholar] [CrossRef]
- (CDC) CfDCaP. 2019-Novel Coronavirus (2019-nCoV) Real-time rRT-PCR Panel Primers and Probes; Department of Health and Human Services: Atlanta, Georgia. Available online: https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf (accessed on 6 October 2021).
- Randazzo, W.; Cuevas-Ferrando, E.; Sanjuan, R.; Domingo-Calap, P.; Sanchez, G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health 2020, 230, 113621. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simon, P.; Allende, A.; Sanchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Iaconelli, M.; Bonadonna, L.; Lucentini, L.; Suffredini, E. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021, 750, 141711. [Google Scholar] [CrossRef]
- Madhi, S.; Gray, E.G.; Ismail, N.; Izu, A.; Mendelson, M.; Cassim, N.; Stevens, W.; Venter, F. COVID-19 lockdowns in low-and middle-income countries: Success against COVID-19 at the price of greater costs. SAMJ S. Afr. Med. J. 2020, 110, 724–726. [Google Scholar] [CrossRef]
- Arora, S.; Nag, A.; Sethi, J.; Rajvanshi, J.; Saxena, S.; Shrivastava, S.K.; Gupta, A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 2020, 82, 2823–2836. [Google Scholar] [CrossRef]
- Claro, I.C.; Cabral, A.; Augusto, M.; Duran, A.; Graciosa, M.C.; Fonseca, F.L.; Speranca, M.A.; Bueno, R. Long term monitoring of SARA-CoV-2 RNA in wastewater in Brazil: A more responsive and economical approach. Water Res. 2021, 203, 117534. [Google Scholar] [CrossRef]
- Jawed, Z.; Krantzberg, G.; Voinson, S. Identifying Local Realities and Anticipating Challenges in Building Capacity of Ontario Municipal Wastewater Systems in Tracking for SARS-CoV-2. Int. J. World Policy Dev. Stud. 2021, 7, 13. [Google Scholar]
- Kumblathan, T.; Liu, Y.; Uppal, G.K.; Hrudey, S.E.; Li, X.F. Wastewater-Based Epidemiology for Community Monitoring of SARS-CoV-2: Progress and Challenges. ACS Environ. 2021. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Tompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef] [PubMed]
- Evans, J. Only 7 Active Covid-19 Cases Left after 113 Infections Linked to Cape Town ‘Superspreader’ Event. News24 2020. Available online: https://www.news24.com/news24/southafrica/news/only-7-active-covid-19-cases-left-after-113-infections-linked-to-cape-town-superspreader-event-20201030 (accessed on 6 October 2021).
- Meningitis CfRDa. An Update on COVID-19 Outbreak in South Africa The First and the Second Wave of COVID-19 Cases in South Africa. NICD-NHLS. Available online: https://www.nicd.ac.za/wp-content/uploads/2021/01/An-update-on-COVID-19-outbreak-in-South-Africa_The-first-and-second-wave.pdf (accessed on 18 January 2021).
- Richardson, H. How Waste Water is Helping South Africa Fight COVID-19. Nature 2021, 593, 616–617. Available online: https://www.nature.com/articles/d41586-021-01399-9 (accessed on 30 September 2021). [CrossRef] [PubMed]
- Commission Recommendations (EU) on a common approach to establish a systematic surveillance of SARS-CoV-2 and its variant in wastewater in the EU. Off. J. Eur. Union 2021, 472. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=uriserv:OJ.L_.2021.098.01.0003.01.ENG (accessed on 6 October 2021).
- Welfare FIfHa. Coronavirus Wastewater Monitoring. Available online: https://thl.fi/en/web/thlfi-en/research-and-development/research-and-projects/sars-cov-2-at-wastewater-treatment-plants/coronavirus-wastewater-monitoring (accessed on 30 September 2021).
- Rjiksoverheid. Coronavirus Dashboard. 2021. Available online: https://coronadashboard.government.nl/landelijk/rioolwater (accessed on 30 September 2021).
- (CDC) CfDCaP. National Wastewater Surveillance System. 2021. Available online: https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html (accessed on 30 September 2021).
- Barrios, M.E.D.S.; Torres, C.; Costamagna, D.M.; Fernández, M.D.B.; Mbayed, V.A. Dynamics of SARS-CoV-2 in wastewater in three districts of Buenos Aires metropolitan region, Argentina, throughout nine months of surveillance: A pilot study. Sci. Total Environ. 2021, 800, 149578. [Google Scholar] [CrossRef]
- Prado, T.; Fumian, T.M.; Mannarino, C.F.; Resende, P.C.; Motta, F.C.; Eppinghaus, A.L.F.; do Vale, V.H.C.; Braz, R.M.S.; da Silva Ribeiro de Andrade, J.; Maranhao, A.G. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021, 191, 116810. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef]
- Hou, C.; Chu, T.; Chen, M.; Hua, Z.; Xu, P.; Xu, H.; Wang, Y.; Liao, J.; Di, B. Application of multi-parameter population model based on endogenous population biomarkers and flow volume in wastewater epidemiology. Sci. Total Environ. 2021, 759, 143480. [Google Scholar] [CrossRef]
- Feng, S.; Roguet, A.; McClary-Gutierrez, J.S.; Newton, R.J.; Kloczko, N.; Meiman, J.G.; McLellan, S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. Acs EsT Water. 2021, 1, 1955–1965. [Google Scholar] [CrossRef]
- Blumberg, L.; Jassat, W.; Mendelson, M.; Cohen, C. The COVID-19 crisis in South Africa: Protecting the vulnerable. S. Afr. Med. J. 2020, 110, 825–826. [Google Scholar] [CrossRef] [PubMed]
- Pearce, N.; Vandenbroucke, J.P.; VanderWeele, T.J.; Greenland, S. Accurate statistics on COVID-19 are essential for policy guidance and decisions. Am. Public Health Assoc. 2020, 110, 949–951. [Google Scholar] [CrossRef]
- Wu, S.L.; Mertens, A.N.; Crider, Y.S.; Nguyen, A.; Pokpongkiat, N.N.; Djajadi, S.; Seth, A.; Hsiang, M.S.; Colford, J.M.; Reingold, A.; et al. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat. Commun. 2020, 11, 4507. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).