Using Social Network Analysis to Identify Spatiotemporal Spread Patterns of COVID-19 around the World: Online Dashboard Development
Abstract
:1. Introduction
1.1. Social Network Analysis for Identifying Spread Patterns Based on DCC
1.2. Item Response Theory and the Infection Point on an Ogive Curve
1.3. Two Phenomena Observed in the COVID-19 Pandemic
1.4. The Aims of This Study
2. Materials and Methods
2.1. Data Source
2.2. Spread Routes of COVID-19 across Continents
2.3. The Three Steps Below Were to Identify the Spread Patterns of COVID-19
- Step 1: Using log (CNIC) to define the correlation coefficients (CCs) in countries/regions
- Step 2: Applying SNA to classify the spread clusters of COVID-19
- Step 3: Plotting the SNA to classify the spread clusters of COVID-19
2.4. Building the Model-Based on IRT
2.4.1. Percentage-Type Observed Data (OPi)
2.4.2. The IRT Probability Model
2.4.3. Ogive Curved in a Model
2.4.4. Transforming Epi into the Number of Expected CNIC
2.5. Model Parameter Estimation
2.5.1. The Attributes of the Ogive Curve
2.5.2. Parameter Estimation
- To minimize the total residuals, we used the Microsoft function as shown below.
- Estimated parametersIn Equation (2), a and b were estimated.
- Constrained termsWe set a and b in a range between (0, 4) and (−5, 5), respectively. In addition, the correlation coefficient (CC) between OPi and Epi was set beyond 0.9.
- Perform the Solver add-inThe Microsoft Solver add-in was performed for each country/region to estimate the model parameters (see Appendix A for more details). The ogive curve can be plotted to predict the future CNIC and determine IP days as explained in the next section.
2.6. Determining IP Using a Search Scheme
2.7. Statistical Tools and Data Analysis
3. Results
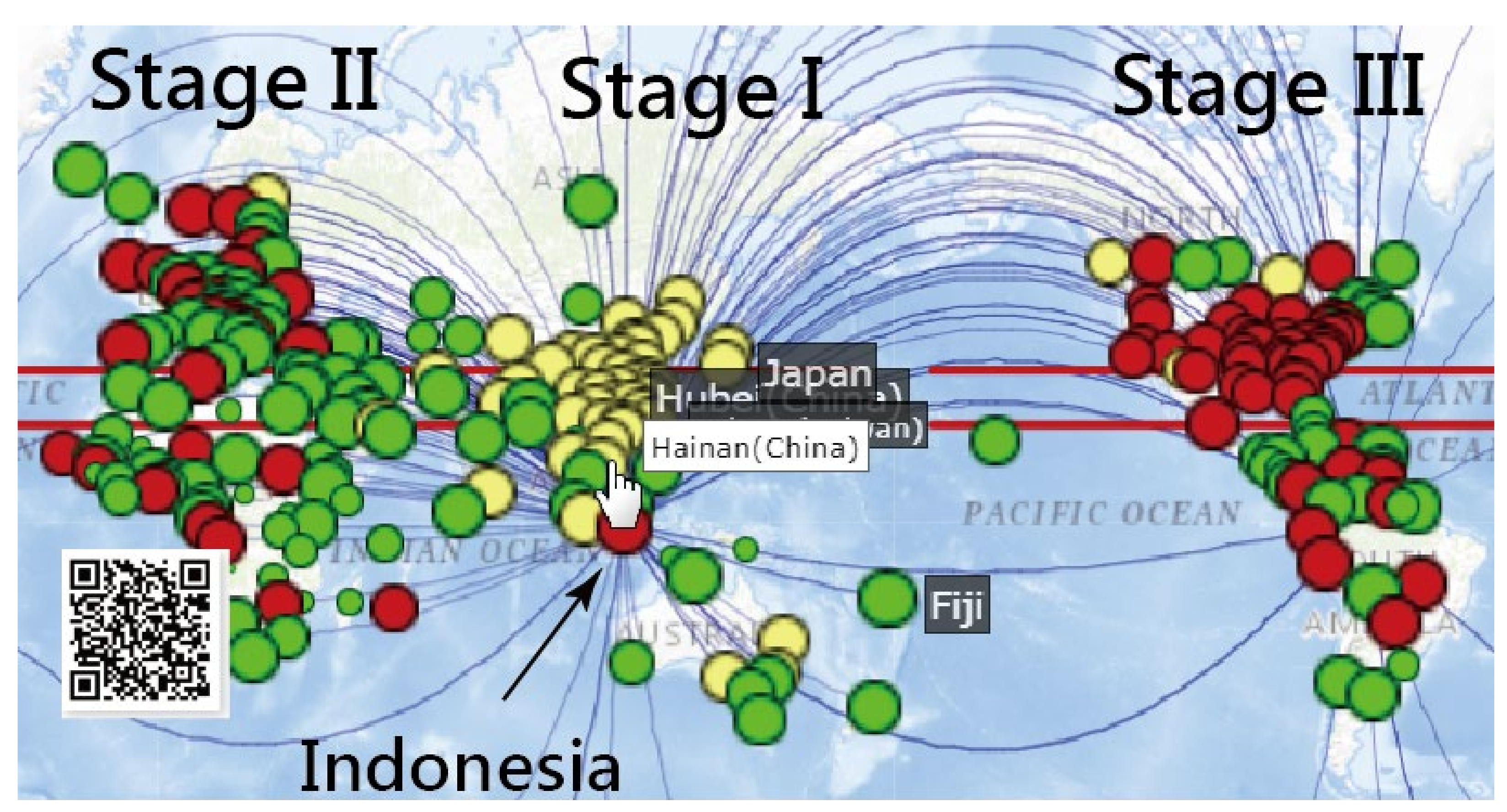
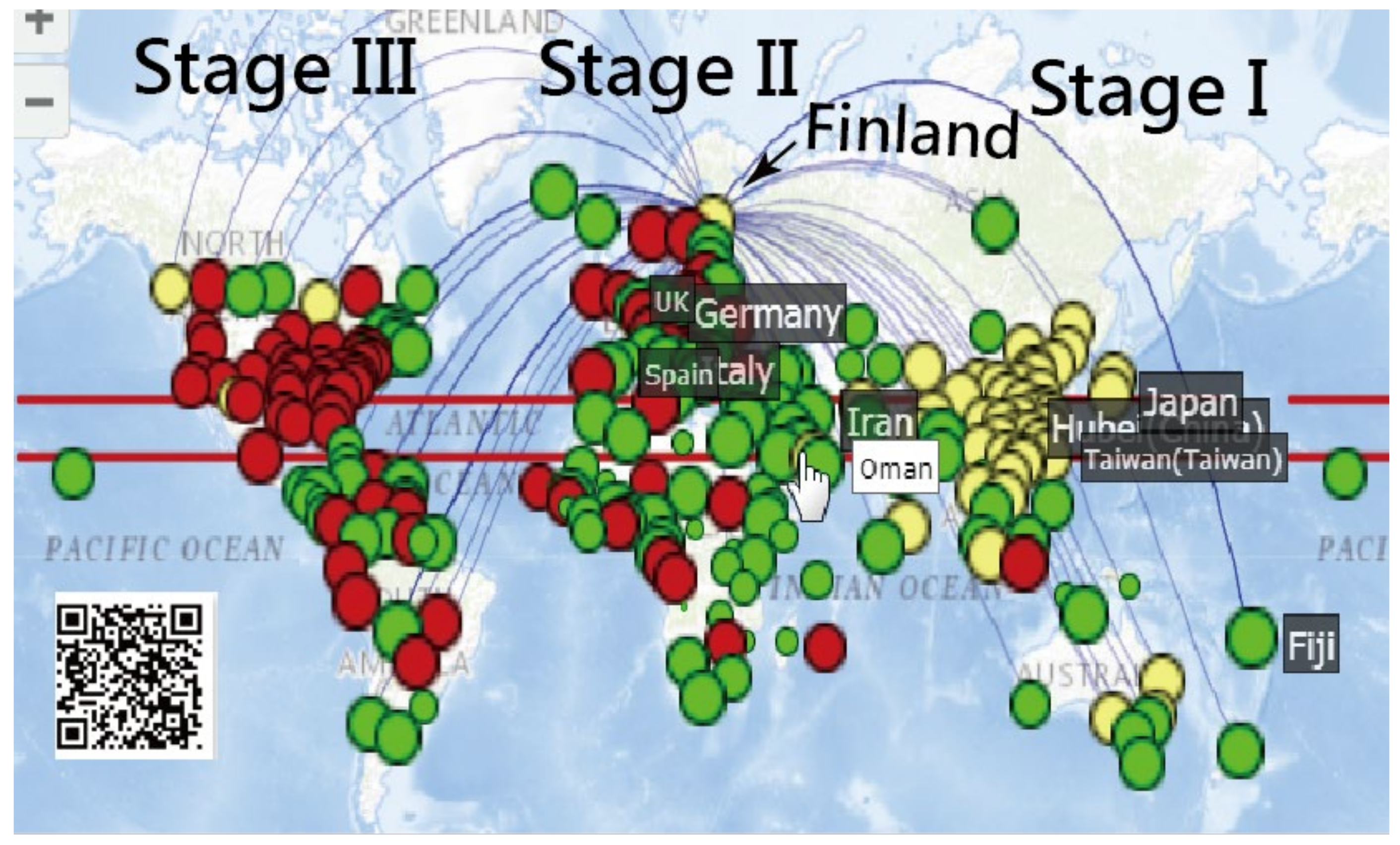
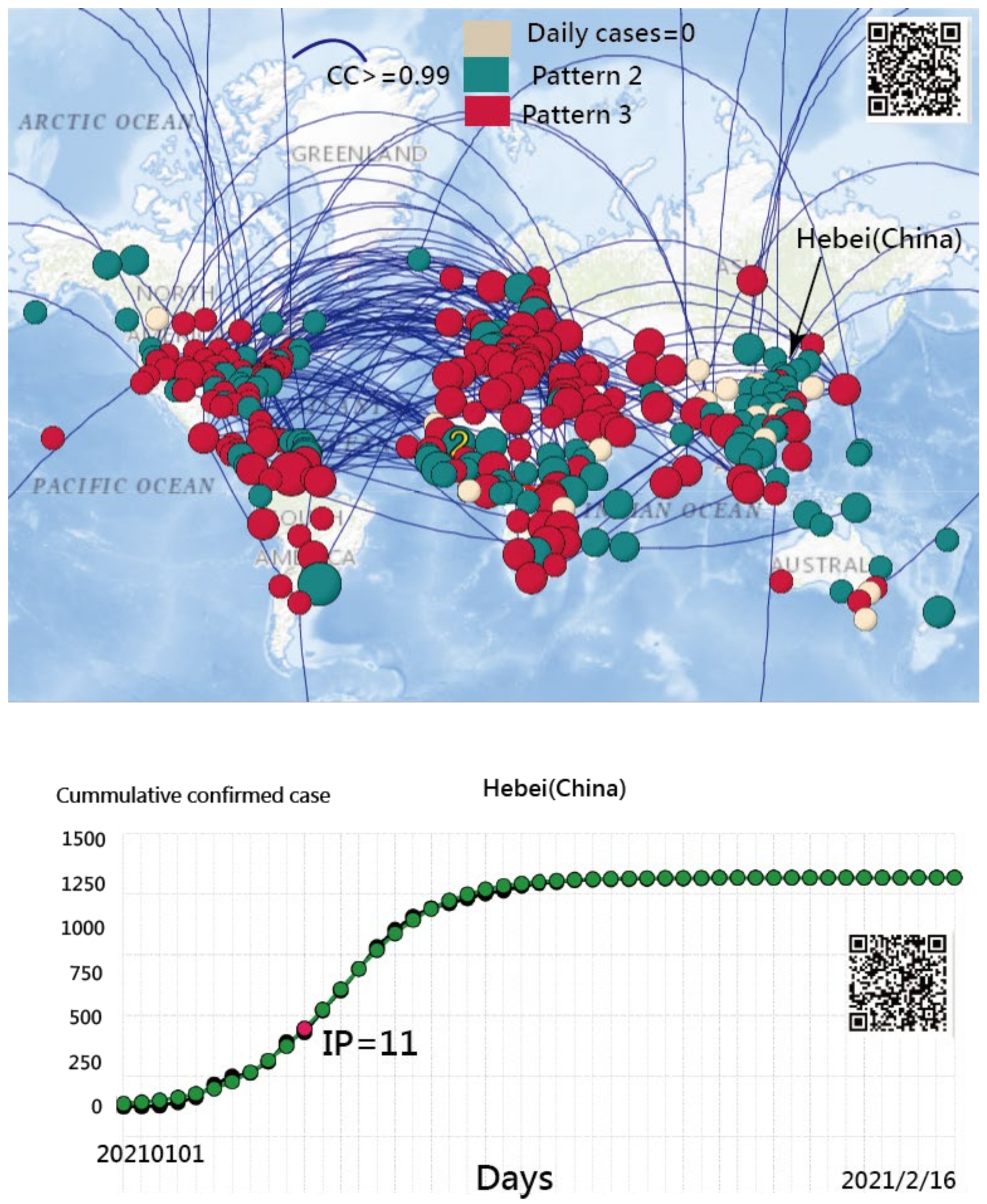
3.1. Spread Clusters in Color on Google Maps
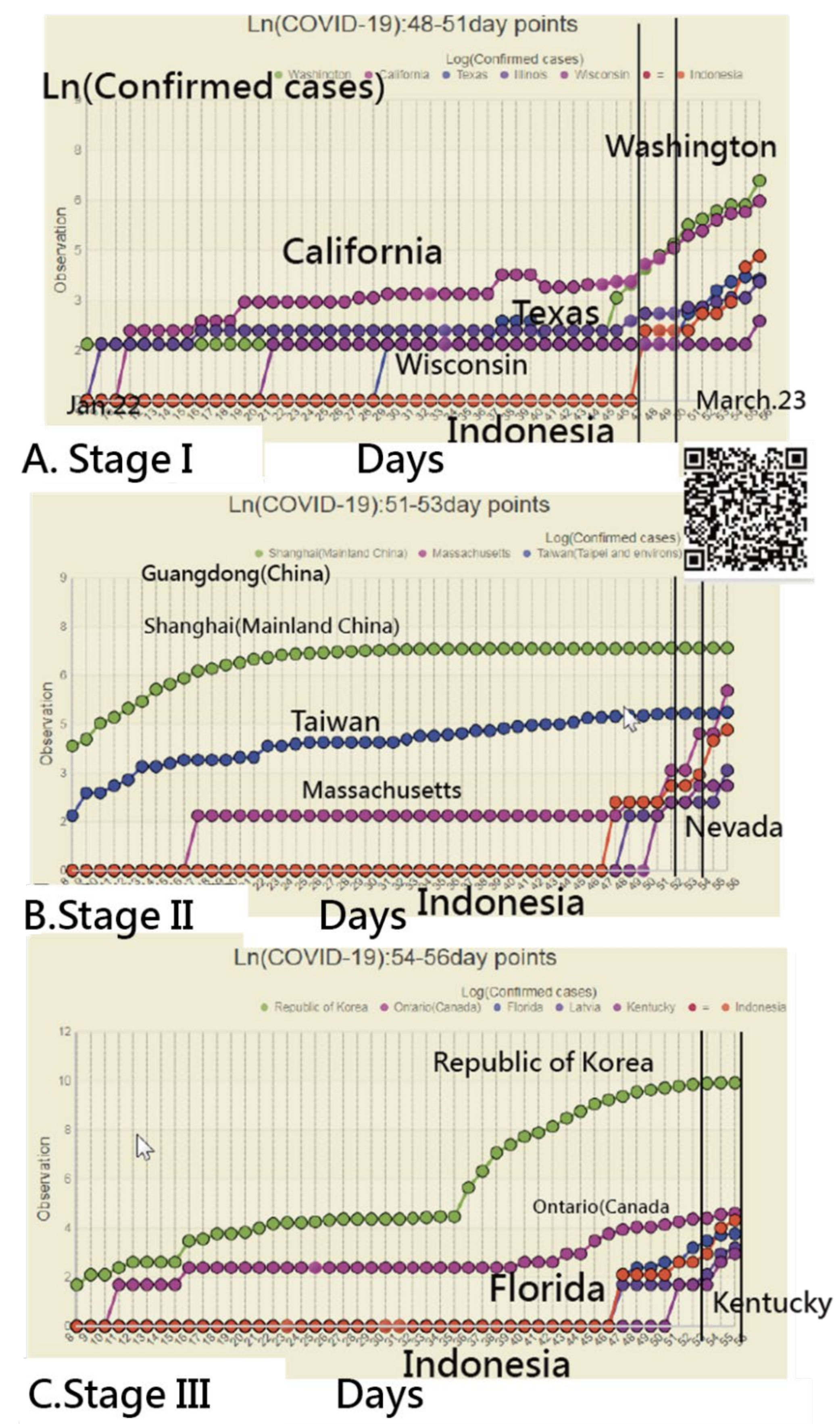
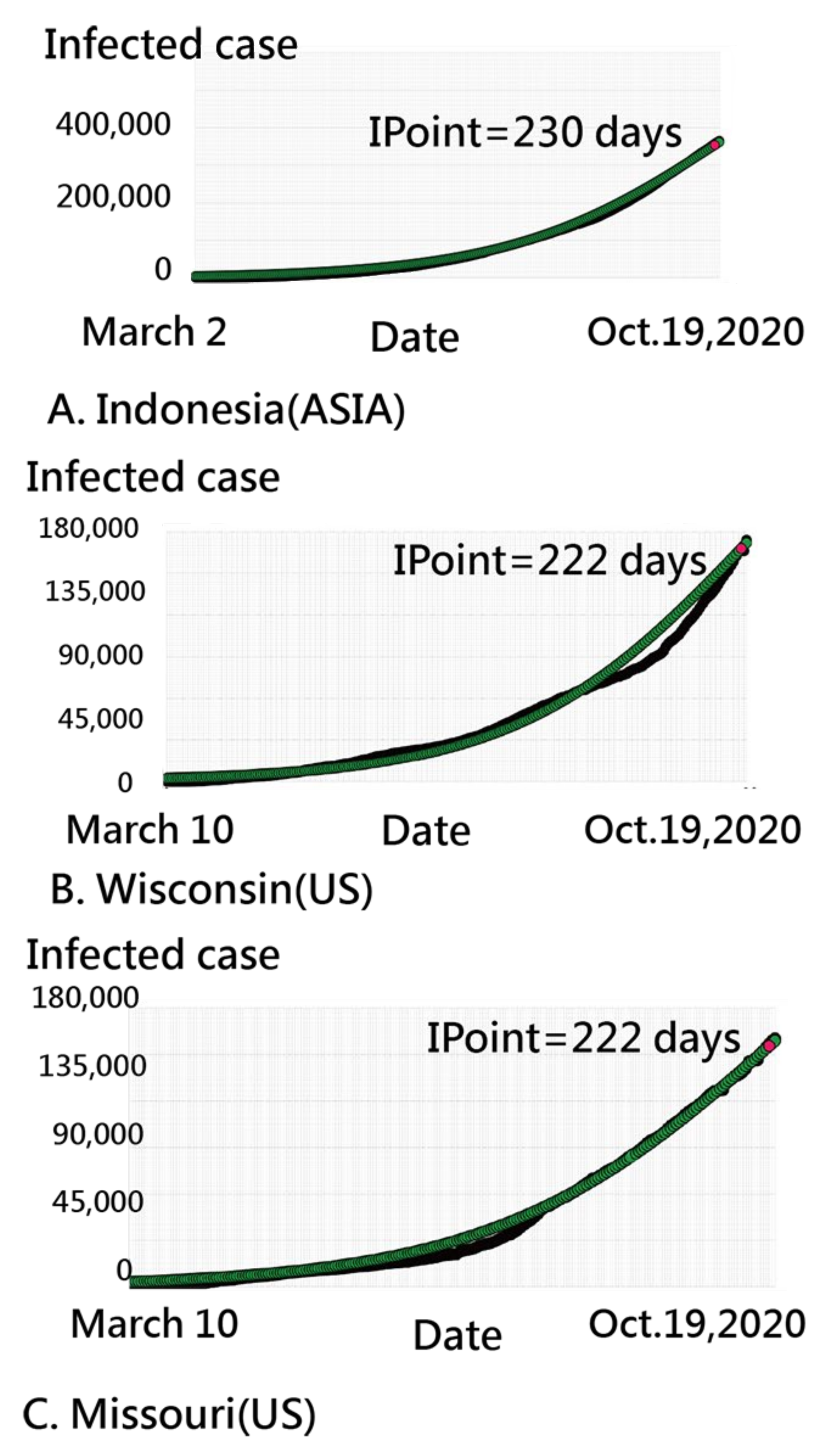
3.2. Details about Indonesia in the CNIC Pattern of COVID-19
3.3. Finland’s CNIC Pattern of COVID-19
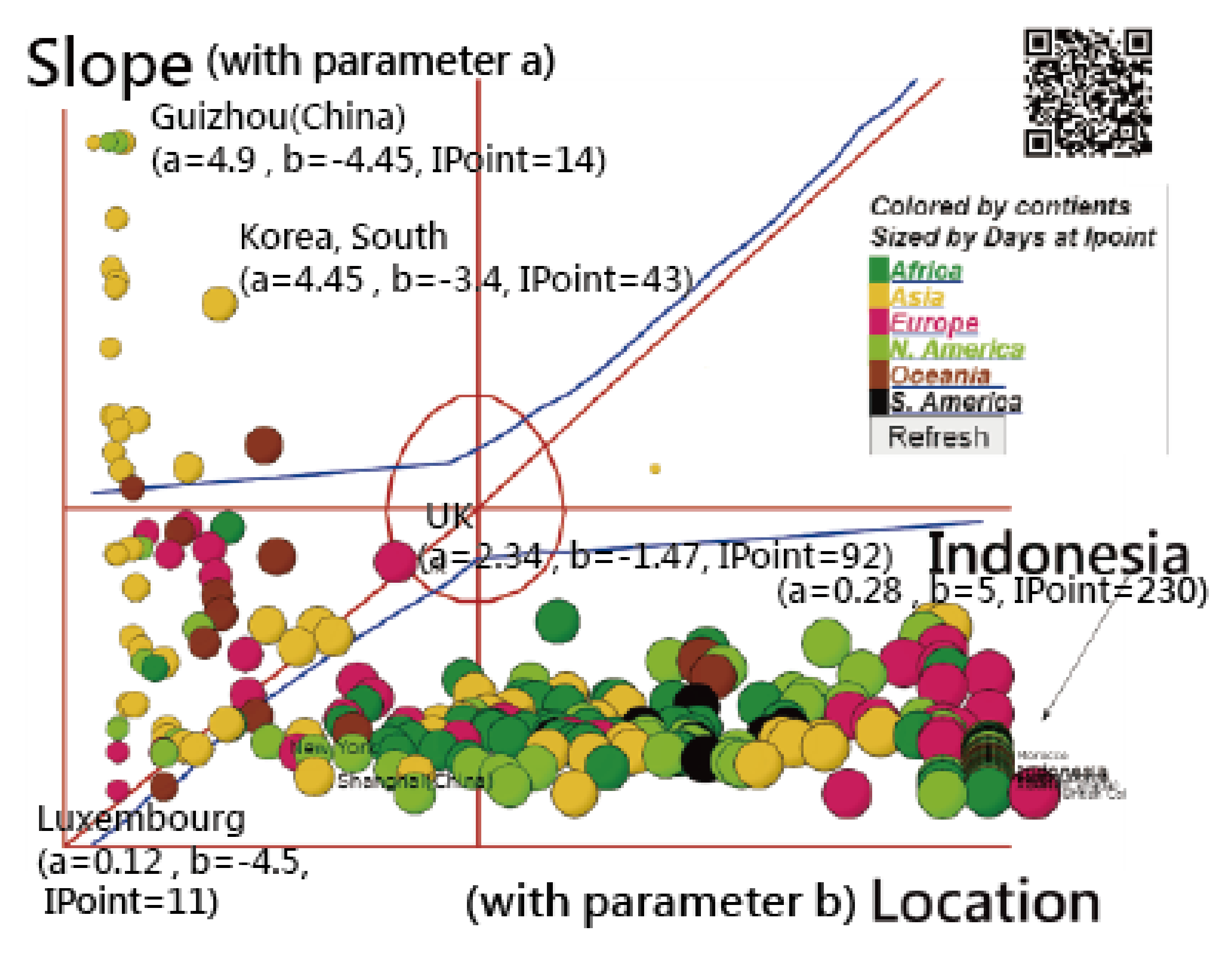
3.4. Using an IRT-Based Model to Examine Spread Patterns
3.5. The Spread Patterns of COVID-19 in January 2021
3.6. Online Dashboards Shown on Google Maps
4. Discussion
4.1. Findings and Implications
4.2. What This Finding Adds to What We Already Knew
4.3. What Is Implied and What Should Be Changed
4.4. Strengths of This Study
4.5. Limitations and Future Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Feng, Y.; Li, Q.; Tong, X.; Wang, R.; Zhai, S.; Gao, C.; Lei, Z.; Chen, S.; Zhou, Y.; Wang, J.; et al. Spatiotemporal spread pattern of the COVID-19 cases in China. PLoS ONE 2020, 15, e0244351. [Google Scholar] [CrossRef]
- Boulos, M.N.K.; Geraghty, E.M. Geographical tracking and mapping of coronavirus disease COVID-19/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic and associated events around the world: How 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. Int. J. Health Geogr. 2020, 19, 8. [Google Scholar] [CrossRef] [Green Version]
- JHU. Dashboard Online for COVID-19 in Near Real Time. Available online: https://coronavirus.jhu.edu/map.html (accessed on 20 February 2021).
- Leszkiewicz, A. Dashboard Online for COVID-19 in Near Real Time. Available online: https://avatorl.org/covid-19/ (accessed on 20 February 2021).
- World Health Organization. Novel Coronavirus (COVID-19) Situation (Public Dashboard). Available online: https://covid19.who.int/ (accessed on 20 February 2021).
- HealthMap. Novel Coronavirus 2019-nCoV (Interactive Map). Available online: https://healthmap.org/wuhan/ (accessed on 20 February 2021).
- Schiffmann, A. A Creator of One COVID-19 Dashboard. Available online: https://ncov2019.live/data (accessed on 20 February 2021).
- Google Team. 2019 Novel Coronavirus (nCoV) Data Repository. Available online: https://github.com/CSSEGISandData/2019-nCoV (accessed on 20 February 2021).
- World Health Organization (WHO). Novel Coronavirus (2019-nCoV) Outbreak. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 20 February 2021).
- Centers for Disease Control and Prevention (CDC). CDC Tests for 2019-nCoV. Available online: https://www.cdc.gov/ (accessed on 20 February 2021).
- European Centre for Disease Prevention and Control (ECDC). Novel Coronavirus. Available online: https://www.ecdc.europa.eu/en/home (accessed on 20 February 2021).
- DXY. Novel Coronavirus (2019-nCoV). Available online: https://ncov.dxy.cn/ncovh5/view/pneumonia (accessed on 20 February 2021).
- Duan, Q.; Wu, J.; Wu, G.; Wang, Y.G. Predication of Inflection Point and Outbreak Size of COVID-19 in New Epicentres. Nonlinear Dyn. 2020, 101, 1561–1581. [Google Scholar]
- Chatham, W.W. Treating Covid-19 at the Inflection Point. J. Rheumatol. 2020, 47, 1–10. [Google Scholar]
- Gu, C.; Zhu, J.; Sun, Y.; Ahou, K.; Gu, J. The inflection point about COVID-19 may have passed. Sci. Bull. 2020, 65, 865–867. [Google Scholar] [CrossRef]
- Fan, R.G.; Wang, Y.B.; Luo, M.; Zhang, Y.Q.; Zhu, C.P. SEIR-Based COVID-19 Transmission Model and Inflection Point Prediction Analysis. J. Univ. Electron. Sci. Technol. China 2020, 49. [Google Scholar] [CrossRef]
- Evelien, O.; Ronald, R. Social network analysis: A powerful strategy, also for the information sciences. J. Inf. Sci. 2002, 28, 441–453. [Google Scholar]
- Martin, G. A social network analysis of Twitter: Mapping the digital humanities community. Cogent Arts Humanit. 2016, 3, 1171458. [Google Scholar] [CrossRef]
- Hagen, L.; Neely, S.; Robert-Cooperman, C.; Keller, T.; DePaula, N. Crisis Communications in the Age of Social Media: A Network Analysis of Zika-Related Tweets. Soc. Sci. Comput. Rev. Soc. Sci. Comput. Rev. 2018, 36, 523–541. [Google Scholar] [CrossRef]
- Hsieh, W.T.; Chien, T.W.; Kuo, S.C.; Lin, H.J. Whether productive authors using the national health insurance database also achieve higher individual research metrics: A bibliometric study. Medicine 2020, 99, e18631. [Google Scholar] [CrossRef]
- Lin, C.H.; Chou, P.H.; Chou, W.; Chien, T.W. Using the Kano model to display the most cited authors and affiliated countries in schizophrenia research. Schizophr Res. 2019, 216, 422–428. [Google Scholar] [CrossRef]
- Chien, T.W.; Wang, H.Y.; Chang, Y.; Kan, W.C. Using Google Maps to display the pattern of coauthor collaborations on the topic of schizophrenia: A systematic review between 1937 and 2017. Schizophr Res. 2019, 204, 206–213. [Google Scholar] [CrossRef]
- Chien, T.W.; Chang, Y.; Wang, H.Y. Understanding the productive author who published papers in medicine using National Health Insurance Database: A systematic review and meta-analysis. Medicine 2018, 97, e9967. [Google Scholar] [CrossRef]
- Perc, M.; Miksić, N.G.; Slavinec, M.; Stožer, A. Forecasting COVID-19. Front. Phys. 2020, 8, 127. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Nie, Y.; Penny, M. Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: A data-driven analysis. J. Med. Virol. 2020, 92, 645–659. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Anastassopoulou, C.; Russo, L.; Tsakris, A.; Siettos, C. Data-based analysis, modeling and forecasting of the COVID-19 outbreak. PLoS ONE 2020, 15, e0230405. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Chen, H. Modeling the epidemic dynamics and control of COVID-19 outbreak in China. Quant. Biol. 2020, 8, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Rong, X.; Yang, L.; Chu, H.; Fan, M. Effect of delay in diagnosis on transmission of COVID-19. Math. Biosci. Eng. 2020, 17, 2725–2740. [Google Scholar] [CrossRef]
- Mandal, M.; Jana, S.; Nandi, S.K.; Khatua, A.; Adak, S.; Kar, T.K. A model based study on the dynamics of COVID-19: Prediction and control. Chaos Soliton Fract. 2020, 136, 109889. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Qi, G. Effects of control measures on the dynamics of COVID-19 and double-peak behavior in Spain. Nonlinear Dyn. 2020. [Google Scholar] [CrossRef]
- Wang, L.-Y.; Chien, T.-W.; Chou, W. Using the Ipcase-Index with Inflection Points and the Corresponding Case Numbers to Identify the Impact Hit by COVID-19 in China: An Observation Study. Int. J. Environ. Res. Public Health 2021, 18, 1994. [Google Scholar] [CrossRef]
- Lord, F.M. Practical applications of item characteristic curve theory. J. Educ. Meas. 1977, 14, 117–138. [Google Scholar] [CrossRef]
- Lord, F.M. Applications of Item Response Theory to Practical Testing Problems; Lawrence Eribaum Associates: Hillsdale, NJ, USA, 1980. [Google Scholar]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 95, 497–506. [Google Scholar] [CrossRef] [Green Version]
- CGTN News. COVID-19 in China: What’s Happened in Hebei and Heilongjiang. Available online: https://news.cgtn.com/news/2021-01-12/COVID-19-in-China-What-s-happened-in-Hebei-and-Heilongjiang-WZDgaNQeys/index.html (accessed on 12 January 2021).
- CDC. Search COVID-19 Risk Assessment by Country. Available online: https://www.cdc.gov/coronavirus/2019-ncov/travelers/index.html (accessed on 1 March 2020).
- Batagelj, V.; Mrvar, A. Pajek—Analysis, and Visualization of Large Networks. In Graph Drawing Software; Jünger, M., Mutzel, P., Eds.; Springer: Berlin, Germany, 2003; pp. 77–103. [Google Scholar]
- Chien, T.W. Cronbach’s Alpha with the Dimension Coefficient to Jointly Assess a Scale’s Quality. Rasch Meas. Trans. 2012, 26, 1379. [Google Scholar]
- Lee, C.J.; Chou, W.; Chien, T.W.; Yeh, Y.T.; Jen, T.H. Using the separation index for identifying the dominant role in an organization: A case of publications in organization innovation. Int. J. Organ. Innov. 2020, 12, 135–145. [Google Scholar]
- Chang, C.S.; Yeh, Y.T.; Chien, T.W.; Lin, J.C.J.; Cheng, B.W.; Lai, F.J. Using the separation index to identify the most dominant role: A case of application on COVID-19 outbreak. Int. J. Organ. Innov. 2020, 12, 10–20. [Google Scholar]
- Kano, N.; Seraku, N.; Takahashi, F.; Tsuji, S. Attractive Quality and Must-Be Quality. J. Jpn. Soc. Qual. Control 1984, 41, 39–48. [Google Scholar]
- Chou, P.H.; Yeh, Y.T.; Kan, W.C.; Chien, T.W.; Kuo, S.C. Using Kano diagrams to display the most cited article types, affiliated countries, authors and MeSH terms on spinal surgery in recent 12 years. Eur. J. Med. Res. 2021, 26, 22. [Google Scholar] [CrossRef] [PubMed]
- Kan, W.C.; Chou, W.; Chien, T.W.; Yeh, Y.T.; Chou, P.H. The Most-Cited Authors Who Published Papers in JMIR mHealth and uHealth Using the Authorship-Weighted Scheme: Bibliometric Analysis. JMIR Mhealth Uhealth. 2020, 8, e11567. [Google Scholar] [CrossRef]
- Chien, T.W. Comparison of Parameters in IRT Model for COVID-19 on the Kano Diagram. Available online: http://www.healthup.org.tw/gps/snapatternipoint.htm (accessed on 6 February 2021).
- Park, J.; Kim, S. Machine Learning-Based Activity Pattern Classification Using Personal PM2.5 Exposure Information. Int. J. Environ. Res. Public Health 2020, 17, 6573. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, L. Support Vector Machine Classification of Drunk Driving Behaviour. Int. J. Environ. Res. Public Health 2017, 14, 108. [Google Scholar] [CrossRef]
- Quan, J. Multi-Temporal Effects of Urban Forms and Functions on Urban Heat Islands Based on Local Climate Zone Classification. Int. J. Environ. Res. Public Health 2019, 16, 2140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florez, H.; Singh, S. Online dashboard and data analysis approach for assessing COVID-19 case and death data. F1000Research 2020, 9, 570. [Google Scholar] [CrossRef] [PubMed]
- Wissel, B.D.; Van Camp, P.J.; Kouril, M.; Weis, C.; Glauser, T.A.; White, P.S.; Kohane, I.S.; Dexheimer, J.W. An interactive online dashboard for tracking COVID-19 in U.S. counties, cities, and states in real time. J. Am. Med. Inform. Assoc. 2020, 27, 1121–1125. [Google Scholar] [CrossRef]
- Alwan, N.A.; Bhopal, R.; Burgess, R.A.; Colburn, T.; Cuevas, L.E.; Smith, G.D.; Egger, M.; Eldridge, S.; Gallo, V.; Gilthorpe, M.S.; et al. Evidence informing the UK’s COVID-19 public health response must be transparent. Lancet 2020, 395, 1036–1037. [Google Scholar] [CrossRef]
- THead, M.G. A real-time policy dashboard can aid global transparency in the response to coronavirus disease 2019. Int. Health 2020, 12, 373–374. [Google Scholar] [CrossRef]
- Pubmed. Articles Related to COVID-19 and Social Network Analysis. Available online: https://pubmed.ncbi.nlm.nih.gov/?term=%28%28Social+network+analysis%29+%29+AND+%28COVID-19%5BMeSH+Major+T (accessed on 17 February 2021).
- Gio, H.R. The Exploration of Author Network. Available online: https://researchoutput.ncku.edu.tw/en/persons/how-ran-guo/network/ (accessed on 16 February 2021).
- Chang, C.S.; Yeh, Y.T.; Chien, T.W.; Lin, J.J.; Cheng, B.W.; Kuo, S.C. The computation of case fatality rate for novel coronavirus (COVID-19) based on Bayes theorem: An observational study. Medicine 2020, 99, e19925. [Google Scholar] [CrossRef]
- Majumder, M.S.; Rivers, C.; Lofgren, E.; Fisman, D. Estimation of MERS-Coronavirus Reproductive Number and Case Fatality Rate for the Spring 2014 Saudi Arabia Outbreak: Insights from Publicly Available Data. PLoS Curr. 2014, 6. [Google Scholar] [CrossRef]
- Chien, T.W. The First Case in UK on 31 January 2020. Available online: http://www.healthup.org.tw/kpiall/wuhen2abc.asp?mid=UK&mtype=1&mlog=1 (accessed on 28 March 2020).
- Schiffmann, A. nCoV2019.live. Available online: https://ncov2019.live/data (accessed on 29 March 2020).
- Linacre, J.M. The Efficacy of Warm’s Weighted Mean Likelihood Estimate (WLE) Correction to Maximum Likelihood Estimate (MLE) Bias. Rasch Meas. Trans. 2009, 23, 1188–1189. [Google Scholar]
- Linacre, J.M. Estimating Rasch measures with known polytomous (or rating scale) item difficulties: Anchored Maximum Likelihood Estimation (AMLE). Rasch Meas. Trans. 1998, 12, 638. [Google Scholar]
- Warm, T.A. Weighted likelihood estimation of ability in item response theory. Psychometrik 1989, 54, 427–450. [Google Scholar] [CrossRef]
- Mercatelli, D.; Giorgi, F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef] [PubMed]
- Makarenkov, V.; Mazoure, B.; Rabusseau, G.; Legendre, P. Horizontal gene transfer and recombination analysis of SARS-CoV-2 genes helps discover its close relatives and shed light on its origin. BMC Ecol. Evol. 2021, 21, 5. [Google Scholar] [CrossRef]


| Area | n | Mean | SD | Different (p < 0.05) | Stage |
|---|---|---|---|---|---|
| From Area i | |||||
| (3) CHINA | 31 | −2.5382 | 3.1201 | (1)(2)(4)(5)(7)(8) | I |
| (6) OCEANIA | 15 | −1.6913 | 2.4752 | (2)(4)(5)(7)(8) | |
| (1) AFRICA | 53 | 1.0080 | 2.3828 | (3)(4) | II |
| (2) ASIA | 44 | 1.7731 | 3.1458 | (3)(4)(6) | |
| (5) N. AMERICA | 36 | 1.9385 | 3.4053 | (3)(4)(6) | |
| (8) US | 61 | 2.6096 | 2.5807 | (3)(6) | |
| (7) S. AMERICA | 12 | 2.6469 | 1.3944 | (3)(6) | |
| (4) EUROPE | 49 | 4.2287 | 1.9927 | (1)(2)(3)(5)(6) | III |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yie, K.-Y.; Chien, T.-W.; Yeh, Y.-T.; Chou, W.; Su, S.-B. Using Social Network Analysis to Identify Spatiotemporal Spread Patterns of COVID-19 around the World: Online Dashboard Development. Int. J. Environ. Res. Public Health 2021, 18, 2461. https://doi.org/10.3390/ijerph18052461
Yie K-Y, Chien T-W, Yeh Y-T, Chou W, Su S-B. Using Social Network Analysis to Identify Spatiotemporal Spread Patterns of COVID-19 around the World: Online Dashboard Development. International Journal of Environmental Research and Public Health. 2021; 18(5):2461. https://doi.org/10.3390/ijerph18052461
Chicago/Turabian StyleYie, Kyent-Yon, Tsair-Wei Chien, Yu-Tsen Yeh, Willy Chou, and Shih-Bin Su. 2021. "Using Social Network Analysis to Identify Spatiotemporal Spread Patterns of COVID-19 around the World: Online Dashboard Development" International Journal of Environmental Research and Public Health 18, no. 5: 2461. https://doi.org/10.3390/ijerph18052461
APA StyleYie, K.-Y., Chien, T.-W., Yeh, Y.-T., Chou, W., & Su, S.-B. (2021). Using Social Network Analysis to Identify Spatiotemporal Spread Patterns of COVID-19 around the World: Online Dashboard Development. International Journal of Environmental Research and Public Health, 18(5), 2461. https://doi.org/10.3390/ijerph18052461






