Tetracycline-Resistant Bacteria Selected from Water and Zebrafish after Antibiotic Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Culture and Exposure
2.2. Culture-Dependent Analyses
2.2.1. Bacterial Counts and Strains Isolation and Identification
2.2.2. Detection of Tetracycline Resistance and Integrase Encoding Genes
2.2.3. Antibiotic Susceptibility Testing
2.2.4. Zebrafish Pathogenicity Test
2.3. Culture-Independent Analyses
2.3.1. Environmental DNA Extraction
2.3.2. Quantitative Polymerase Chain Reaction (qPCR)
2.4. Statistical Analysis
3. Results and Discussion
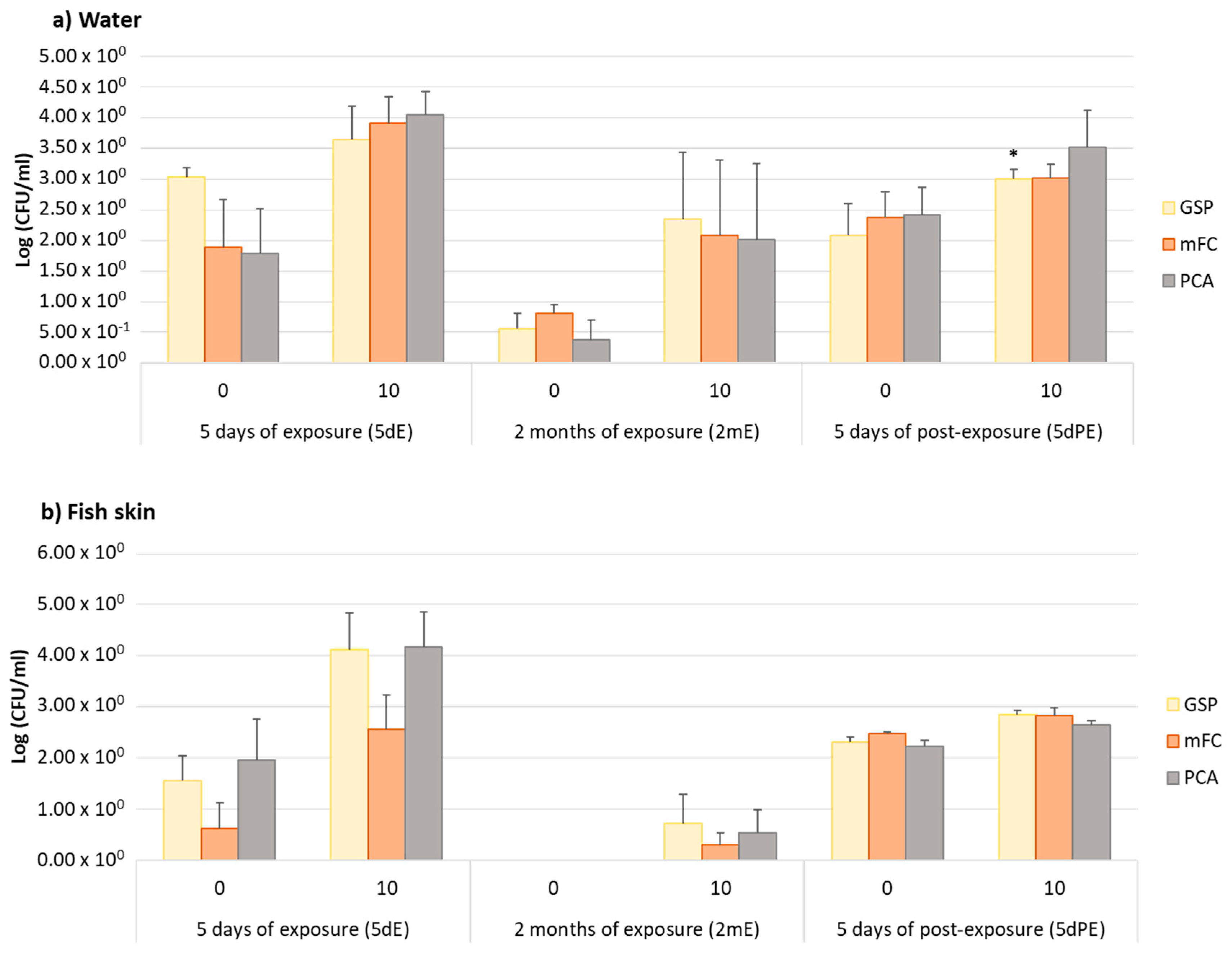
3.1. Effects of OTC in the Abundance of Tetracycline-Resistant Bacteria
3.2. Taxonomic Affiliation of Isolates
3.3. Antibiotic Resistance Profile and Resistance Genes
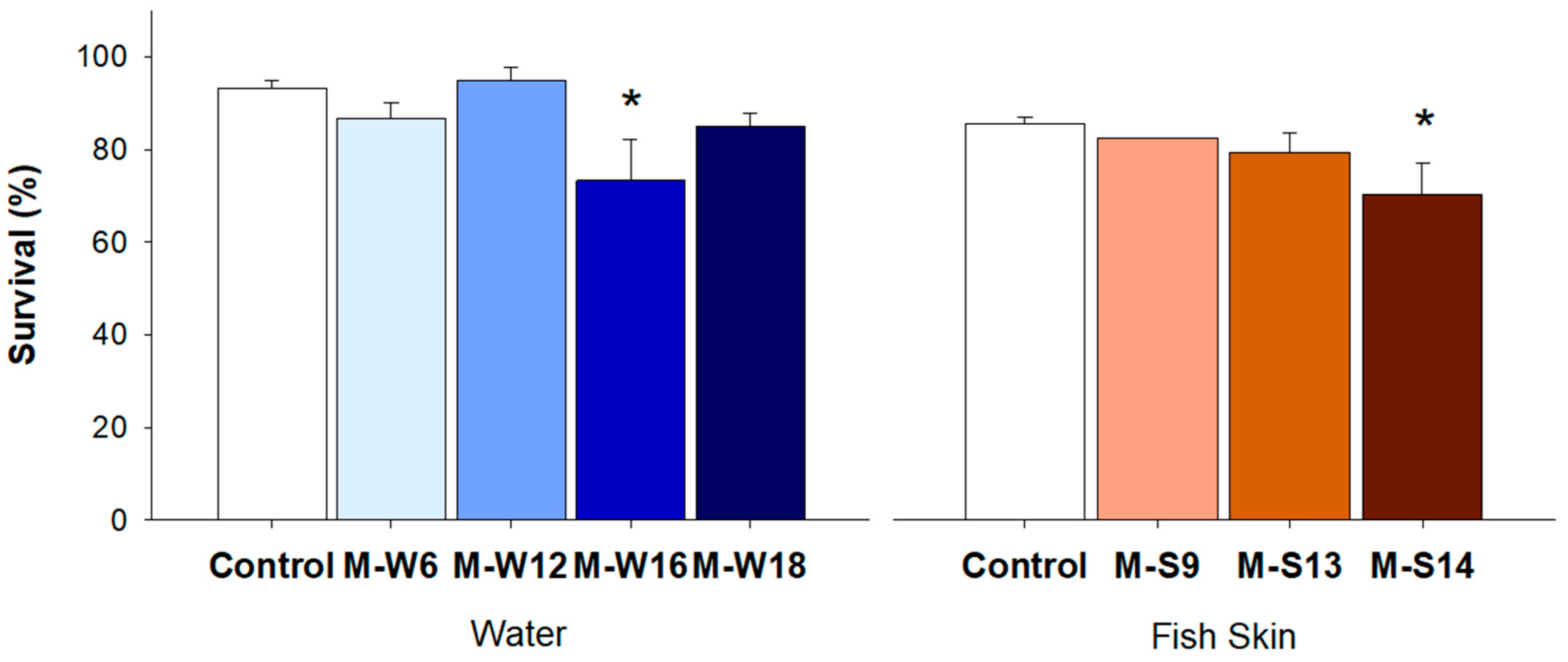
3.4. Pathogenicity Test
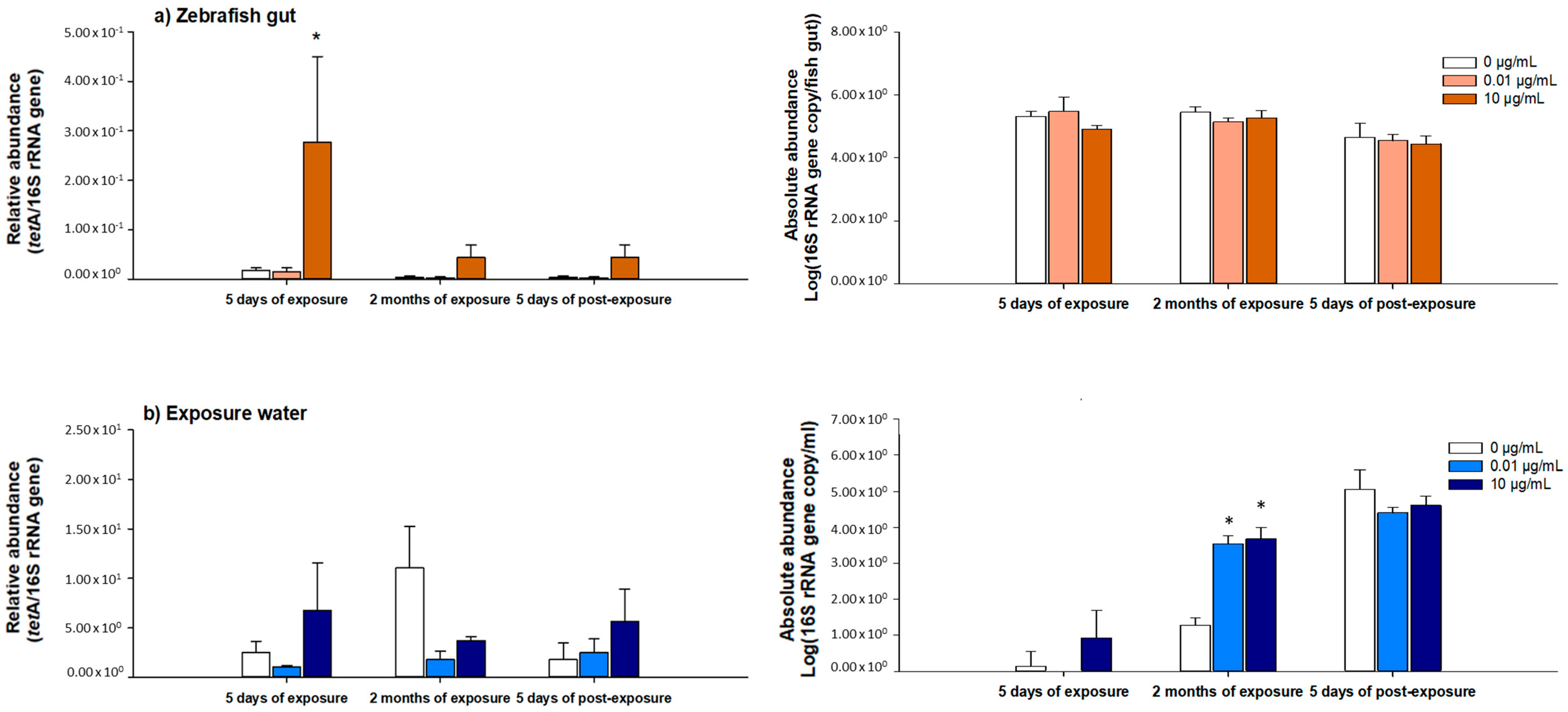
3.5. Abundance of tetA gene in Water and Fish Gut
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Føre, M.; Frank, K.; Norton, T.; Svendsen, E.; Arve, J.; Dempster, T.; Eguiraun, H.; Watson, W.; Stahl, A.; Magne, L.; et al. Precision fish farming: A new framework to improve production in aquaculture. Biosyst. Eng. 2018, 173, 176–193. [Google Scholar] [CrossRef]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2017. Trends from 2010 to 2017. (Ema/294674/2019). 2019. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2017_en.pdf (accessed on 5 February 2020).
- Cravedi, J.-P.; Choubert, G.; Delous, G. Digestibility of chloramphenicol, oxolinic acid and oxytetracycline in rainbow trout and influence of these antibiotics on lipid digestibility. Aquaculture 1987, 60, 133–141. [Google Scholar] [CrossRef]
- Chen, H.; Liu, S.; Xu, X.R.; Liu, S.S.; Zhou, G.J.; Sun, K.F.; Zhao, J.L.; Ying, G.G. Antibiotics in typical marine aquaculture farms surrounding Hailing Island, South China: Occurrence, bioaccumulation and human dietary exposure. Mar. Pollut. Bull. 2015, 90, 181–187. [Google Scholar] [CrossRef]
- Monteiro, S.H.; Francisco, J.G.; Andrade, G.C.R.M.; Botelho, R.G.; Figueiredo, L.A.; Tornisielo, V.L. Study of spatial and temporal distribution of antimicrobial in water and sediments from caging fish farms by on-line SPE-LC-MS/MS. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2016, 51, 634–643. [Google Scholar] [CrossRef]
- Harnisz, M.; Korzeniewska, E.; Gołaś, I. The impact of a freshwater fish farm on the community of tetracycline-resistant bacteria and the structure of tetracycline resistance genes in river water. Chemosphere 2015, 128, 134–141. [Google Scholar] [CrossRef]
- Zou, S.; Xu, W.; Zhang, R.; Tang, J.; Chen, Y.; Zhang, G. Occurrence and distribution of antibiotics in coastal water of the Bohai Bay, China: Impacts of river discharge and aquaculture activities. Environ. Pollut. 2011, 159, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.; Graham, D.W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2009, 44, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Zhao, Y.; Li, B.; Huang, C.L.; Zhang, S.Y.; Yu, S.; Chen, Y.S.; Zhang, T.; Gillings, M.R.; Su, J.Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Guo, C.; Luo, Y.; Lv, J.; Zhang, Y.; Lin, H.; Wang, L.; Xu, J. Occurrence and distribution of antibiotics, antibiotic resistance genes in the urban rivers in Beijing, China. Environ. Pollut. 2016, 213, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, Y.B.; Xu, J.X.; Ling, J.Y.; Chen, J.L.; Zhou, J.L.; Zheng, L.; Du, Q.P. Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere 2017, 168, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef]
- Tacão, M.; Correia, A.; Henriques, I. Resistance to broad-spectrum antibiotics in aquatic systems: Anthropogenic activities modulate the dissemination of blaCTX-M-like genes. Appl. Environ. Microbiol. 2012, 78, 4134–4140. [Google Scholar] [CrossRef]
- Seyfried, E.E.; Newton, R.J.; Rubert, K.F., IV; Pedersen, J.A.; McMahon, K.D. Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 2010, 59, 799–807. [Google Scholar] [CrossRef]
- Maruzani, R.; Canali, A.; Sera, V.; Pantoja, L.; Shah, A.J.; Perito, B.; Marvasi, M. Effect of anthropogenic pollution on the fitness of tetracycline sensitive Shigella flexneri in Thames river water. J. Environ. Chem. Eng. 2018, 6, 19–27. [Google Scholar] [CrossRef]
- Maruzani, R.; Pathak, A.; Ward, M.; Serafim, V. Antibiotic selective pressure in microcosms: Pollution influences the persistence of multidrug resistant Shigella flexneri 2a YSH6000 strain in polluted river water samples. Environ. Technol. Innov. 2020, 19, 100821. [Google Scholar] [CrossRef]
- Jia, J.; Cheng, M.; Xue, X.; Guan, Y.; Wang, Z. Characterization of tetracycline effects on microbial community, antibiotic resistance genes and antibiotic resistance of Aeromonas spp. in gut of goldfish Carassius auratus Linnaeus. Ecotoxicol. Environ. Saf. 2020, 191, 110182. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, X.; Yang, Q.; Chen, Y.; Du, B. Evolution of microbial community and drug resistance during enrichment of tetracycline-degrading bacteria. Ecotoxicol. Environ. Saf. 2019, 171, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Mo, W.Y.; Chen, Z.; Leung, H.M.; Leung, A.O.W. Application of veterinary antibiotics in China’s aquaculture industry and their potential human health risks. Environ. Sci. Pollut. Res. 2017, 24, 8978–8989. [Google Scholar] [CrossRef]
- Osman, K.M.; Luiz, O.; Saad, A.; Hamed, M. Nile tilapia (Oreochromis niloticus) as an aquatic vector for Pseudomonas species of medical importance: Antibiotic resistance association with biofilm formation, quorum sensing and virulence. Aquaculture 2021, 532. [Google Scholar] [CrossRef]
- Serrano, P.H. Responsible Use of Antibiotics in Aquaculture; FAO Fisheries Technical Paper; Cambridge University Press: Rome, Italy, 2005. [Google Scholar] [CrossRef]
- Almeida, A.R.; Tacão, M.; Machado, A.L.; Golovko, O.; Zlabek, V.; Domingues, I.; Henriques, I. Long-term effects of oxytetracycline exposure in zebrafish: A multi-level perspective. Chemosphere 2019, 222, 333–344. [Google Scholar] [CrossRef]
- Almeida, A.R.; Alves, M.; Domingues, I.; Henriques, I. The impact of antibiotic exposure in water and zebrafish gut microbiomes: A 16S rRNA gene-based metagenomic analysis. Ecotoxicol. Environ. Saf. 2019, 186, 109771. [Google Scholar] [CrossRef]
- Huerta, B.; Marti, E.; Gros, M.; López, P.; Pompêo, M.; Armengol, J.; Barceló, D.; Balcázar, J.L.; Rodríguez-Mozaz, S.; Marcé, R. Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci. Total Environ. 2013, 456–457, 161–170. [Google Scholar] [CrossRef]
- Silva, I.; Tacão, M.; Tavares, R.D.S.; Miranda, R.; Araújo, S.; Manaia, C.M.; Henriques, I. Fate of cefotaxime-resistant Enterobacteriaceae and ESBL-producers over a full-scale wastewater treatment process with UV disinfection. Sci. Total Environ. 2018, 639, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Domingues, I.; Oliveira, R.; Soares, A.M.V.M.; Amorim, M.J.B. Effects of ivermectin on Danio rerio: A multiple endpoint approach: Behaviour, weight and subcellular markers. Ecotoxicology 2016, 25, 491–499. [Google Scholar] [CrossRef] [PubMed]
- OECD. Test No. 230: 21-day Fish Assay, OECD Guidelines for the Testing of Chemicals, OECD Guidelines for the Testing of Chemicals, Section 2. OECD Publishing. 2009. Available online: http://www.oecd-ilibrary.org/environment/test-no-230-21-day-fish-assay_9789264076228-en (accessed on 5 October 2017).
- Matuschek, E.; Brown, D.F.J.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 25 March 2020).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Milligan-Myhre, K.; Charette, J.R.; Phennicie, R.T.; Stephens, W.Z.; Rawls, J.F.; Guillemin, K.; Kim, C.H. Study of Host–Microbe Interactions in Zebrafish. In Methods in Cell Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 87–116. [Google Scholar] [CrossRef]
- Almeida, A.R.; Domingues, I.; Henriques, I. Zebrafish and water microbiome recovery after oxytetracycline exposure. Environ. Pollut. 2021, 272, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Henriques, I.S.; Almeida, A.; Cunha, A.; Correia, A. Molecular sequence analysis of prokaryotic diversity in the middle and outer sections of the Portuguese estuary Ria de Aveiro. FEMS Microbiol. Ecol. 2004, 49, 269–279. [Google Scholar] [CrossRef]
- Silva, I.; Tacao, M.; Henriques, I. Selection of antibiotic resistance by metals in a riverine bacterial community. Chemosphere 2021, 263, 127936. [Google Scholar] [CrossRef]
- Tavares, R.D.S.; Tacão, M.; Figueiredo, A.S.; Duarte, A.S.; Esposito, F.; Lincopan, N.; Manaia, C.M.; Henriques, I. Genotypic and phenotypic traits of blaCTX-M-carrying Escherichia coli strains from an UV-C-treated wastewater effluent. Water Res. 2020, 184. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Gao, P.; Mao, D.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef]
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.C.; Kolvenbach, B.A.; Nunes, O.C.; Corvini, P.F.X. Biodegradation of antibiotics: The new resistance determinants—Part I. New Biotechnol. 2020, 54, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Rosado, D.; Xavier, R.; Severino, R.; Tavares, F.; Cable, J.; Pérez-Losada, M. Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Geng, Y.; Wang, K.; Chen, D.; Huang, X.; He, M.; Yin, Z. Stenotrophomonas maltophilia, an emerging opportunist pathogen for cultured channel catfish, Ictalurus punctatus, in China. Aquaculture 2010. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Stenström, T.A.; Okoh, A.I. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: Looking beyond contemporary antibiotic therapy. Front. Microbiol. 2017, 8, 2276. [Google Scholar] [CrossRef]
- Benhamed, S.; Guardiola, F.A.; Mars, M.; Esteban, M.Á. Pathogen bacteria adhesion to skin mucus of fishes. Vet. Microbiol. 2014, 171, 1–12. [Google Scholar] [CrossRef]
- Boutin, S.; Bernatchez, L.; Audet, C.; Derôme, N. Network analysis highlights complex interactions between pathogen, host and commensal microbiota. PLoS ONE 2013, 8, e84772. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, T.; Marcelino, J.; Ricardo, F.; Soares, A.M.V.M.; Calado, R. Bacterial communities 16S rDNA fingerprinting as a potential tracing tool for cultured seabass Dicentrarchus labrax. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ozaktas, T.; Taskin, B.; Gozen, A.G. High level multiple antibiotic resistance among fish surface associated bacterial populations in non-aquaculture freshwater environment. Water Res. 2012, 46, 6382–6390. [Google Scholar] [CrossRef]
- Chang, Y.T.; Lin, C.Y.; Chen, Y.H.; Hsueh, P.R. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 2015, 6, 893. [Google Scholar] [CrossRef] [PubMed]
- Rizek, C.F.; Jonas, D.; Garcia Paez, J.I.; Rosa, J.F.; Perdigão Neto, L.V.; Martins, R.R.; Moreno, L.Z.; Rossi Junior, A.; Levin, A.S.; Costa, S.F. Multidrug-resistant Stenotrophomonas maltophilia: Description of new MLST profiles and resistance and virulence genes using whole-genome sequencing. J. Glob. Antimicrob. Resist. 2018, 15, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef]
- Nicodemo, A.C.; Paez, J.I.G. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 229–237. [Google Scholar] [CrossRef]
- Bassetti, M.; Castaldo, N.; Cattelan, A.; Mussini, C.; Righi, E.; Tascini, C.; Menichetti, F.; Maria, C.; Tumbarello, M.; Grossi, P.; et al. Ceftolozane/tazobactam for the treatment of serious Pseudomonas aeruginosa infections: A multicentre nationwide clinical experience. Int. J. Antimicrob. Agents 2019, 53, 408–415. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, 1–52. [Google Scholar] [CrossRef]
- Agersø, Y.; Bruun, M.S.; Dalsgaard, I.; Larsen, J.L. The tetracycline resistance gene tet(E) is frequently occurring and present on large horizontally transferable plasmids in Aeromonas spp. from fish farms. Aquaculture 2007, 266, 47–52. [Google Scholar] [CrossRef]
- Henriques, I.S.; Fonseca, F.; Alves, A.; Saavedra, M.J.; Correia, A. Tetracycline-resistance genes in Gram-negative isolates from estuarine waters. Lett. Appl. Microbiol. 2008, 47, 526–533. [Google Scholar] [CrossRef]
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef] [PubMed]
- Lundström, S.V.; Östman, M.; Bengtsson-Palme, J.; Rutgersson, C.; Thoudal, M.; Sircar, T.; Blanck, H.; Eriksson, K.M.; Tysklind, M.; Flach, C.F.; et al. Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Sci. Total Environ. 2016, 553, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.C.; Mostowy, S. The case for modeling human infection in zebrafish. Trends Microbiol. 2020, 28, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Torraca, V.; Mostowy, S. Zebrafish infection: From pathogenesis to cell biology. Trends Cell Biol. 2018, 28, 143–156. [Google Scholar] [CrossRef]
- Rowe, H.M.; Withey, J.H.; Neely, M.N. Zebrafish as a model for zoonotic aquatic pathogens. Dev. Comp. Immunol. 2014, 46, 96–107. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef]
- Zhang, M.Q.; Yuan, L.; Li, Z.H.; Zhang, H.C.; Sheng, G.P. Tetracycline exposure shifted microbial communities and enriched antibiotic resistance genes in the aerobic granular sludge. Environ. Int. 2019, 130, 104902. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, A.R.; Tacão, M.; Soares, J.; Domingues, I.; Henriques, I. Tetracycline-Resistant Bacteria Selected from Water and Zebrafish after Antibiotic Exposure. Int. J. Environ. Res. Public Health 2021, 18, 3218. https://doi.org/10.3390/ijerph18063218
Almeida AR, Tacão M, Soares J, Domingues I, Henriques I. Tetracycline-Resistant Bacteria Selected from Water and Zebrafish after Antibiotic Exposure. International Journal of Environmental Research and Public Health. 2021; 18(6):3218. https://doi.org/10.3390/ijerph18063218
Chicago/Turabian StyleAlmeida, Ana Rita, Marta Tacão, Joana Soares, Inês Domingues, and Isabel Henriques. 2021. "Tetracycline-Resistant Bacteria Selected from Water and Zebrafish after Antibiotic Exposure" International Journal of Environmental Research and Public Health 18, no. 6: 3218. https://doi.org/10.3390/ijerph18063218
APA StyleAlmeida, A. R., Tacão, M., Soares, J., Domingues, I., & Henriques, I. (2021). Tetracycline-Resistant Bacteria Selected from Water and Zebrafish after Antibiotic Exposure. International Journal of Environmental Research and Public Health, 18(6), 3218. https://doi.org/10.3390/ijerph18063218






