Biodegradation of Emerging Pharmaceuticals from Domestic Wastewater by Membrane Bioreactor: The Effect of Solid Retention Time
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of the MBR System and Its Operation
2.2. Samples Preparation
2.3. Solid-Phase Extraction Method
2.4. Analytical Methods
2.5. Metagenomic Analysis
2.6. DNA Isolation
2.7. Next Generation Sequencing (NGS)
3. Results
3.1. Determination of the SPE Method
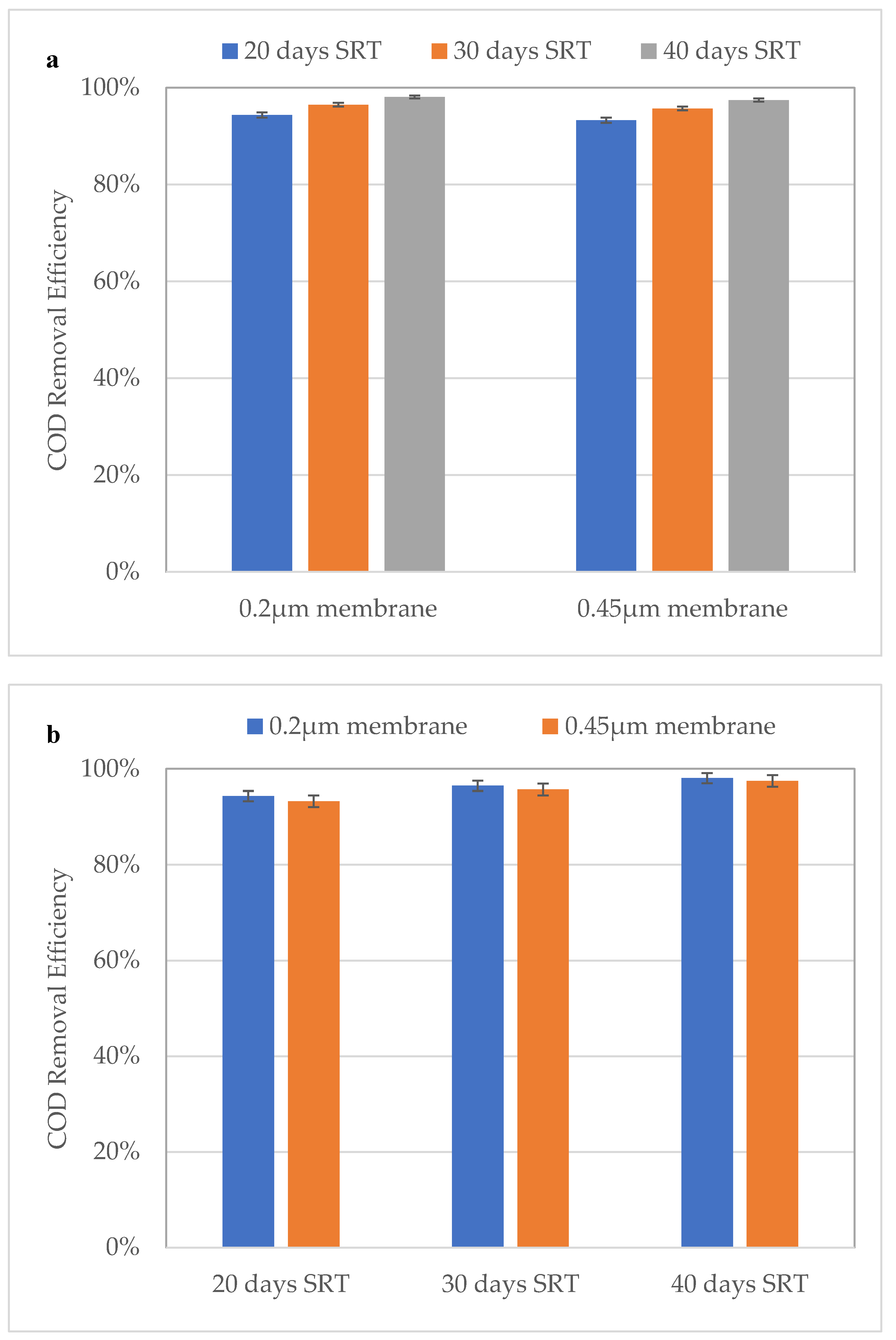
3.2. Effect of SRT on COD Removal and MLSS Concentration
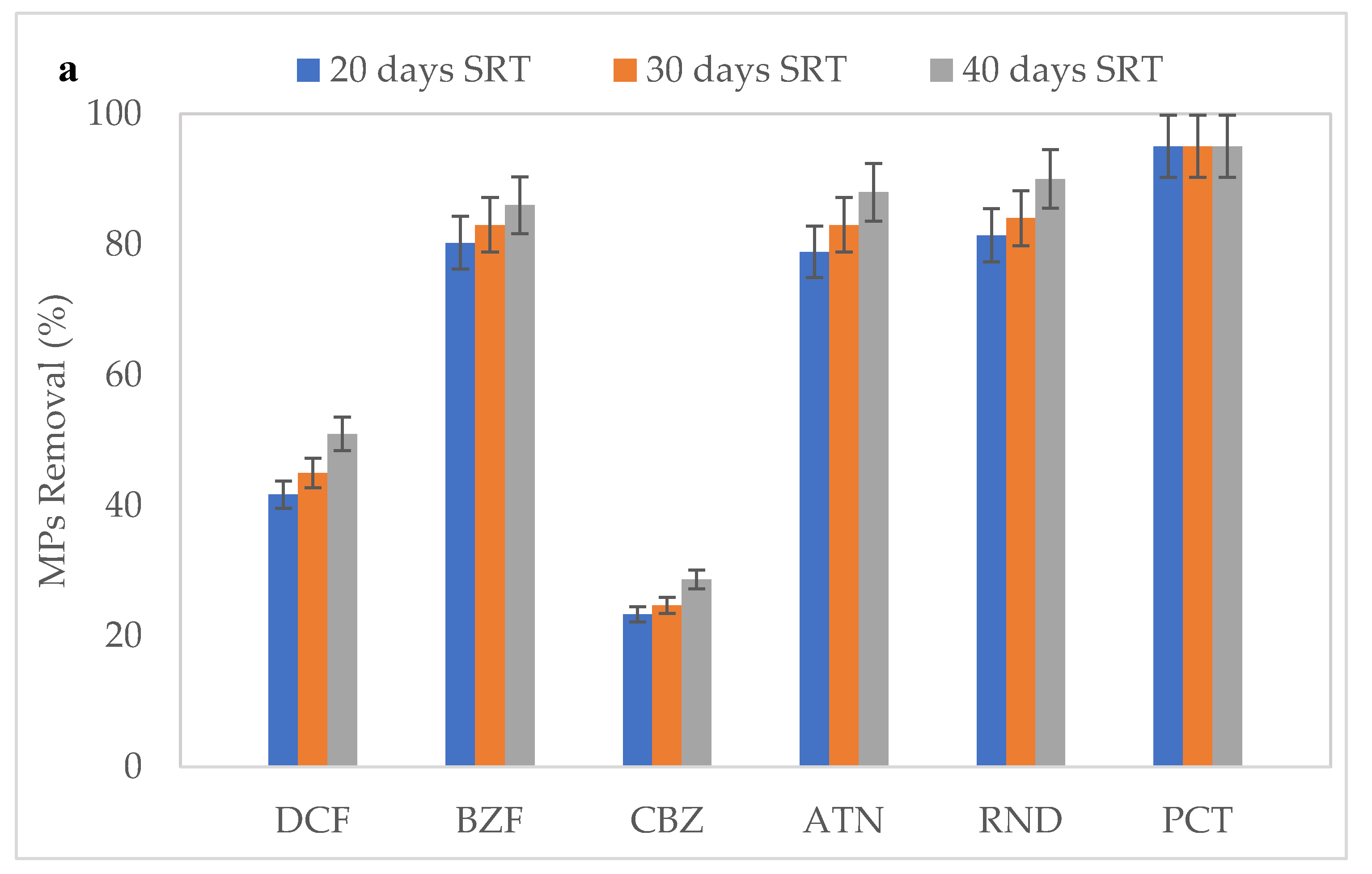
3.3. Effect of SRT on the Removal of MPs
3.4. Removal Mechanism and Possible Pathways
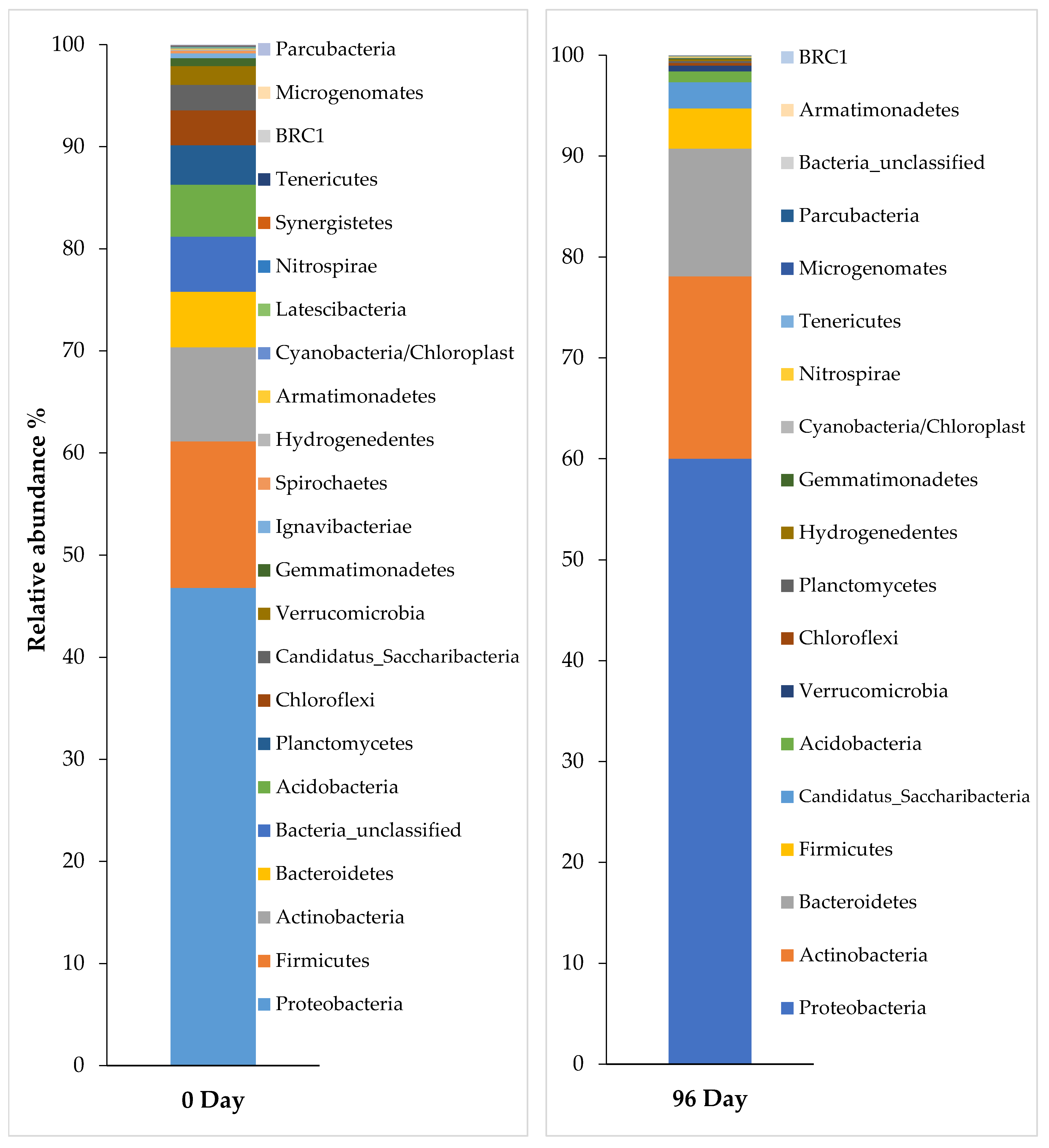
3.5. Bacterial Community Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kondor, A.C.; Jakab, G.; Vancsik, A.; Filep, T.; Szeberényi, J.; Szabó, L.; Maász, G.; Ferincz, Á.; Dobosy, P.; Szalai, Z. Occurrence of pharmaceuticals in the Danube and drinking water wells: Efficiency of riverbank filtration. Environ. Pollut. 2020, 265, 114893. [Google Scholar] [CrossRef]
- Van Driezum, I.H.; Derx, J.; Oudega, T.J.; Zessner, M.; Naus, F.L.; Saracevic, E.; Kirschner, A.K.; Sommer, R.; Farnleitner, A.H.; Blaschke, A.P. Spatiotemporal resolved sampling for the interpretation of micropollutant removal during riverbank filtration. Sci. Total Environ. 2019, 649, 212–223. [Google Scholar] [CrossRef]
- Kruglova, A.; Kråkström, M.; Riska, M.; Mikola, A.; Rantanen, P.; Vahala, R.; Kronberg, L. Comparative study of emerging micropollutants removal by aerobic activated sludge of large laboratory-scale membrane bioreactors and sequencing batch reactors under low-temperature conditions. Bioresour. Technol. 2016, 214, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Marszałek, A.; Bohdziewicz, J.; Puszczało, E. Co-Treatment of Municipal Landfill Leachate with Dairy Wastewater in Membrane Bioreactor. Ecol. Chem. Eng. S 2020, 27, 139–149. [Google Scholar] [CrossRef]
- Taheran, M.; Brar, S.K.; Verma, M.; Surampalli, R.; Zhang, T.; Valero, J. Membrane processes for removal of pharmaceutically active compounds (PhACs) from water and wastewaters. Sci. Total Environ. 2016, 547, 60–77. [Google Scholar] [CrossRef]
- Luo, Y.; Jiang, Q.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Price, W.E.; Wang, J.; Guo, W. Evaluation of micropollutant removal and fouling reduction in a hybrid moving bed biofilm reactor-membrane bioreactor system. Bioresour. Technol. 2015, 191, 355–359. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Hai, F.I.; Kang, J.; Nghiem, L.D.; Price, W.E.; Guo, W.; Ngo, H.H.; Tung, K.-L. Comparison between sequential and simultaneous application of activated carbon with membrane bioreactor for trace organic contaminant removal. Bioresour. Technol. 2013, 130, 412–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goswami, L.; Kumar, R.V.; Borah, S.N.; Manikandan, N.A.; Pakshirajan, K.; Pugazhenthi, G. Membrane bioreactor and integrated membrane bioreactor systems for micropollutant removal from wastewater: A review. J. Water Process. Eng. 2018, 26, 314–328. [Google Scholar] [CrossRef]
- Kamaz, M.; Wickramasinghe, S.R.; Eswaranandam, S.; Zhang, W.; Jones, S.M.; Watts, M.J.; Qian, X. Investigation into Micropollutant Removal from Wastewaters by a Membrane Bioreactor. Int. J. Environ. Res. Public Health 2019, 16, 1363. [Google Scholar] [CrossRef] [Green Version]
- Kamaz, M.; Jones, S.M.; Qian, X.; Watts, M.J.; Zhang, W.; Wickramasinghe, S.R. Atrazine Removal from Municipal Wastewater Using a Membrane Bioreactor. Int. J. Environ. Res. Public Health 2020, 17, 2567. [Google Scholar] [CrossRef]
- Cirja, M.; Ivashechkin, P.; Schäffer, A.; Corvini, P.F.X. Factors affecting the removal of organic micropollutants from wastewater in conventional treatment plants (CTP) and membrane bioreactors (MBR). Rev. Environ. Sci. Biotechnol. 2007, 7, 61–78. [Google Scholar] [CrossRef]
- Dogan, A.; Płotka-Wasylka, J.; Kempińska-Kupczyk, D.; Namieśnik, J.; Kot-Wasik, A. Detection, identification and determination of chiral pharmaceutical residues in wastewater: Problems and challenges. TRAC Trends Anal. Chem. 2020, 122, 115710. [Google Scholar] [CrossRef]
- Butkovskyi, A.; Leal, L.H.; Zeeman, G.; Rijnaarts, H. Micropollutants in source separated wastewater streams and recovered resources of source separated sanitation. Environ. Res. 2017, 156, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Beijer, K.; Björlenius, B.; Shaik, S.; Lindberg, R.H.; Brunström, B.; Brandt, I. Removal of pharmaceuticals and unspecified contaminants in sewage treatment effluents by activated carbon filtration and ozonation: Evaluation using biomarker responses and chemical analysis. Chemosphere 2017, 176, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Altamirano, M.; Maza-Márquez, P.; Peña-Herrera, J.; Rodelas, B.; Osorio, F.; Pozo, C. Removal of anti-inflammatory/analgesic pharmaceuticals from urban wastewater in a pilot-scale A2O system: Linking performance and microbial population dynamics to operating variables. Sci. Total Environ. 2018, 643, 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Botero-Coy, A.; Martínez-Pachón, D.; Boix, C.; Rincón, R.; Castillo, N.; Arias-Marín, L.; Manrique-Losada, L.; Torres-Palma, R.; Moncayo-Lasso, A.; Hernández, F. ‘An investigation into the occurrence and removal of pharmaceuticals in Colombian wastewater’. Sci. Total Environ. 2018, 642, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Gros, M.; Petrović, M.; Barceló, D. Development of a multi-residue analytical methodology based on liquid chromatography–tandem mass spectrometry (LC–MS/MS) for screening and trace level determination of pharmaceuticals in surface and wastewaters. Talanta 2006, 70, 678–690. [Google Scholar] [CrossRef]
- Kumar, R.; Sarmah, A.K.; Padhye, L.P. Fate of pharmaceuticals and personal care products in a wastewater treatment plant with parallel secondary wastewater treatment train. J. Environ. Manag. 2019, 233, 649–659. [Google Scholar] [CrossRef]
- Lahti, M. The Fate Aspects of Pharmaceuticals in the Environment: Biotransformation, Sedimentation and Exposure of Fish; University of Jyväskylä: Jyväskylä, Finland, 2012. [Google Scholar]
- APHA; AWWA. Standard Methods for the Examination of Water and Wastewater, 22th ed.; APHA: Washington, DC, USA, 2012. [Google Scholar]
- Bozcal, E.; Dagdeviren, M. Bacterial metagenome analysis of Mytilus galloprovincialis collected from Istanbul and Izmir coastal stations of Turkey. Environ. Monit. Assess. 2020, 192, 1–18. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broeck, R.V.D.; Van Dierdonck, J.; Nijskens, P.; Dotremont, C.; Krzeminski, P.; Van Der Graaf, J.; Van Lier, J.; Van Impe, J.; Smets, I. The influence of solids retention time on activated sludge bioflocculation and membrane fouling in a membrane bioreactor (MBR). J. Membr. Sci. 2012, 401–402, 48–55. [Google Scholar] [CrossRef]
- Delgado, S.; Villarroel, R.; Gonzalez, E.; Morales, M. Aerobic Membrane Bioreactor for Wastewater Treatment—Performance Under Substrate-Limited Conditions. Biomass Detect. Prod. Usage 2011. [Google Scholar] [CrossRef] [Green Version]
- Chtourou, M.; Mallek, M.; Dalmau, M.; Mamo, J.; Santos-Clotas, E.; Ben Salah, A.; Walha, K.; Salvadó, V.; Monclús, H. Triclosan, carbamazepine and caffeine removal by activated sludge system focusing on membrane bioreactor. Process. Saf. Environ. Prot. 2018, 118, 1–9. [Google Scholar] [CrossRef]
- Mousel, D.; Bastian, D.; Firk, J.; Palmowski, L.; Pinnekamp, J. Removal of pharmaceuticals from wastewater of health care facilities. Sci. Total Environ. 2021, 751, 141310. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Guerra, P.; Shah, A.; Parsa, M.; Alaee, M.; Smyth, S.A. Removal of pharmaceuticals and personal care products in a membrane bioreactor wastewater treatment plant. Water Sci. Technol. 2014, 69, 2221–2229. [Google Scholar] [CrossRef]
- Trinh, T.; Akker, B.V.D.; Coleman, H.M.; Stuetz, R.M.; Drewes, J.E.; Le-Clech, P.; Khan, S.J. Seasonal variations in fate and removal of trace organic chemical contaminants while operating a full-scale membrane bioreactor. Sci. Total Environ. 2016, 550, 176–183. [Google Scholar] [CrossRef]
- Tiwari, B.; Sellamuthu, B.; Piché-Choquette, S.; Drogui, P.; Tyagi, R.D.; Vaudreuil, M.A.; Sauvé, S.; Buelna, G.; Dubé, R. Acclimatization of microbial community of submerged membrane bioreactor treating hospital wastewater. Bioresour. Technol. 2021, 319, 124223. [Google Scholar] [CrossRef]
- Gurung, K.; Ncibi, M.C.; Sillanpää, M. Removal and fate of emerging organic micropollutants (EOMs) in municipal wastewater by a pilot-scale membrane bioreactor (MBR) treatment under varying solid retention times. Sci. Total Environ. 2019, 667, 671–680. [Google Scholar] [CrossRef]
- Tambosi, J.L.; De Sena, R.F.; Favier, M.; Gebhardt, W.; José, H.J.; Schröder, H.F.; Moreira, R.D.F.P.M. Removal of pharmaceutical compounds in membrane bioreactors (MBR) applying submerged membranes. Desalination 2010, 261, 148–156. [Google Scholar] [CrossRef]
- Quintana, J.B.; Weiss, S.; Reemtsma, T. Pathways and metabolites of microbial degradation of selected acidic pharmaceutical and their occurrence in municipal wastewater treated by a membrane bioreactor. Water Res. 2005, 39, 2654–2664. [Google Scholar] [CrossRef]
- Vieno, N.; Sillanpää, M. Fate of diclofenac in municipal wastewater treatment plant—A review. Environ. Int. 2014, 69, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Vo, H.N.P.; Ngo, H.H.; Guo, W.; Nguyen, T.M.H.; Li, J.; Liang, H.; Deng, L.; Chen, Z. Poly-and perfluoroalkyl substances in water and wastewater: A comprehensive review from sources to remediation. J. Water Process. Eng. 2020, 36, 101393. [Google Scholar] [CrossRef]
- Tahir, K.; Miran, W.; Nawaz, M.; Jang, J.; Shahzad, A.; Moztahida, M.; Kim, B.; Azam, M.; Jeong, S.E.; Jeon, C.O.; et al. Investigating the role of anodic potential in the biodegradation of carbamazepine in bioelectrochemical systems. Sci. Total Environ. 2019, 688, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Geng, J.; Shi, Y.; Wang, L.; Xu, K.; Ren, H. Comparison of diclofenac transformation in enriched nitrifying sludge and heterotrophic sludge: Transformation rate, pathway, and role exploration. Water Res. 2020, 184, 116158. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Duan, L.; Wei, J.; Qian, F.; Hermanowicz, S.W. Carbamazepine removal from wastewater and the degradation mechanism in a submerged forward osmotic membrane bioreactor. Bioresour. Technol. 2020, 314, 123732. [Google Scholar] [CrossRef]
- Mhemid, R.K.S.; Akmirza, I.; Shihab, M.S.; Turker, M.; Alp, K. Ethanethiol gas removal in an anoxic bio-scrubber. J. Environ. Manag. 2019, 233, 612–625. [Google Scholar] [CrossRef]
- Demir, M.N. Determination of microbial community in a pilot scale two-stage step-feed biological nutrient removal process. Glob. NEST J. 2019, 21, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Żur, J.; Piński, A.; Marchlewicz, A.; Hupert-Kocurek, K.; Wojcieszyńska, D.; Guzik, U. Organic micropollutants paracetamol and ibuprofen—Toxicity, biodegradation, and genetic background of their utilization by bacteria. Environ. Sci. Pollut. Res. 2018, 25, 21498–21524. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, S. Removal of pharmaceuticals and personal care products (PPCPs) from wastewater: A review. J. Environ. Manag. 2016, 182, 620–640. [Google Scholar] [CrossRef]
- Santos, I.J.S.; Grossman, M.J.; Sartoratto, A.; Ponezi, A.N.; Durrant, L.R. Degradation of the recalcitrant pharmaceuticals car-bamazepine and 17α-ethinylestradiol by ligninolytic fungi. Chem. Eng. 2012, 27, 169–174. [Google Scholar] [CrossRef]
- Popa, C.; Favier, L.; Dinica, R.; Semrany, S.; Djelal, H.; Amrane, A.; Bahrim, G. Potential of newly isolated wild Streptomycesstrains as agents for the biodegradation of a recalcitrant pharmaceutical, carbamazepine. Environ. Technol. 2014, 35, 3082–3091. [Google Scholar] [CrossRef] [PubMed]



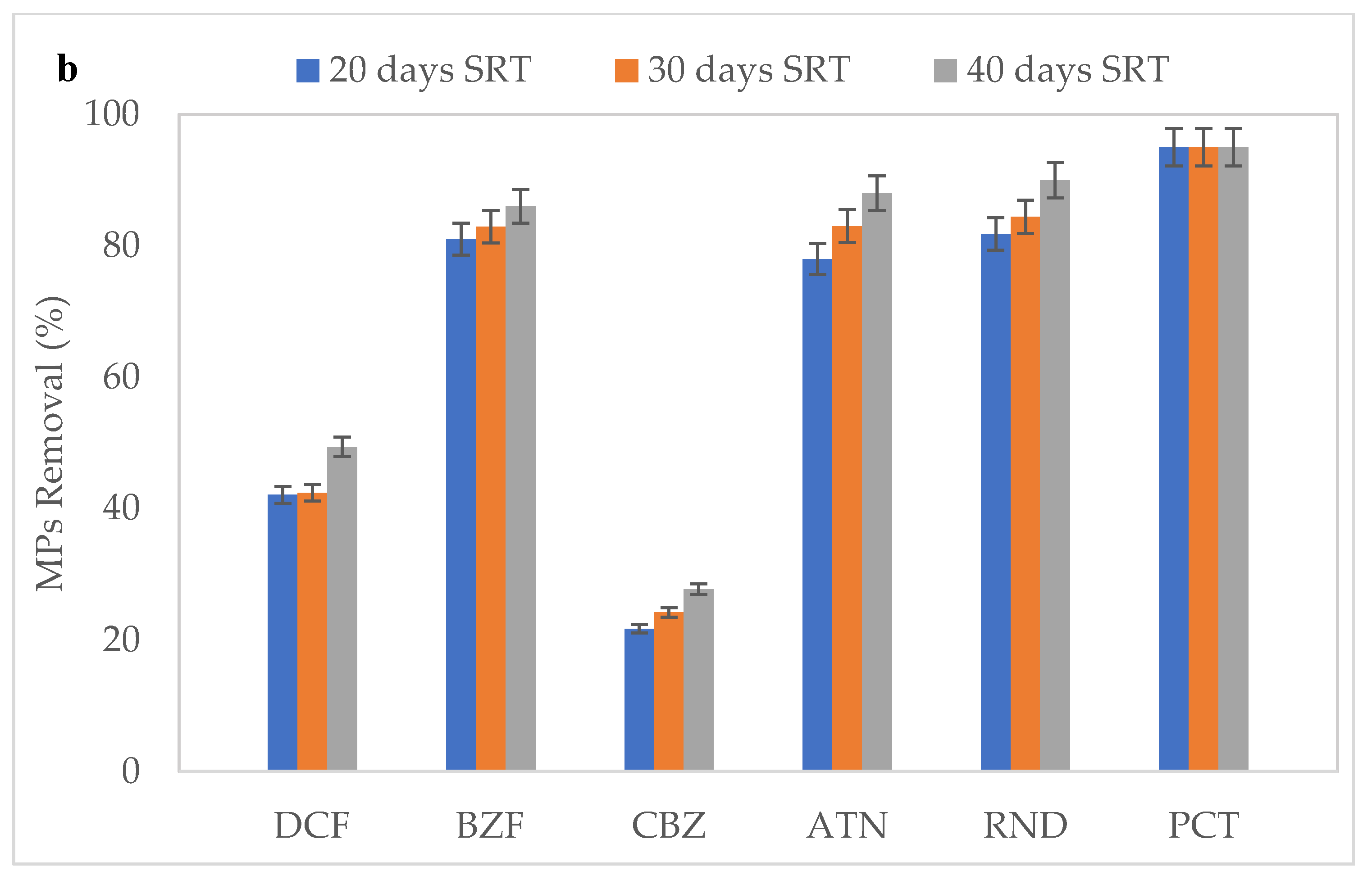
| Carbamazepine | Acetaminophen (Paracetamol) | Diclofenac (Sodium Salt) | Bezafibrate | Ranitidine Hydrochloride | Atenolol | |
|---|---|---|---|---|---|---|
| Applications | Antiepileptic | Therapeutic | Anti-inflammatory | Lipid regulator | Anti-histamine | β-Blockers |
| CAS Number | 298-46-4 | 103-90-2 | 15307-86-5 | 41859-67-0 | 66357-59-3 | 29122-68-7 |
| Formula | C15H12N2O | C8H9NO2 | C14H10Cl2NNaO2 | C19H20ClNO4 | C13H22N4O3S·HCl | C14H22N2O3 |
| Log Kow | 2.45 | 0.46 | 4.51 | 4.25 | 0.27 | 0.16 |
| pKa | 13.9 | 9.4 | 4.15 | 3.61 | 8 | 9.6 |
| Log D | 1.89 | 0.47 | 1.77 | −0.93 | −0.63 | −2.09 |
| Henry Constant (atm-m3/mole) | 1.1 × 10−12 | 6.42 × 10−13 | 4.7×10−12 | 2.12 × 10−15 | 3.42 × 10−15 | 1.37 × 10−18 |
| M.W. | 236.27 | 151.17 | 318.14 | 361.82 | 350.86 | 266.34 |
| Molecular Structure |  |  |  |  | 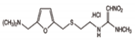 |  |
| Applications | SPE Method 1 | SPE Method 2 | SPE Method 3 | SPE Method 4 |
|---|---|---|---|---|
| Types of cartridge | Oasis HLB cartridge 200 mg, 6 mL | Oasis HLB cartridge 200 mg, 6 mL | C18 cartridge, 500 mg, 6 mL | C18 cartridge, 500 mg, 6 mL |
| Conditioning | 5 mL MeOH, 5 mL water | 5 mL acetone, 5 mL MeOH, 5 mL water | 5 mL MeOH, 5 mL water | 5 mL acetone, 5 mL MeOH, 5 mL water |
| Percolation | 100 mL | 100 mL | 100 mL | 100 mL |
| Washing | 5 mL water | 5 mL water | 5 mL water | 5 mL water |
| Drying | 15 min. | 15 min. | 15 min. | 15 min. |
| Elution | 8 mL MeOH | 8 mL MeOH | 8 mL MeOH | 8 mL MeOH |
| 40-Day SRT | 30-Day SRT | 20-Day SRT | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comp. | R2 | LOD | LOQ | Inf. | 0.2 µm mem. MBR Eff. | RSD% (n = 3) | R.E. % | 0.45 µm mem. MBR Eff. | RSD % (n = 3) | R.E.% | Inf. | 0.2 µm mem. MBR Eff. | RSD % (n = 3) | R.E.% | 0.45 µm mem. MBR Eff. | RSD % (n = 3) | R.E% | Inf. | 0.2 µm mem. MBR Eff. | RSD % (n = 3) | R.E% | 0.45 µm mem. MBR Eff. | RSD % (n = 3) | R.E% |
| DCF | 0.997 | 1.3 | 4.39 | 20.76 ± 0.11 | 10.173 ± 0.19 | 1.875 | 51 | 10.49 ± 0.38 | 3.713 | 49.4 | 20.48 ± 0.34 | 11.23 ± 0.17 | 1.496 | 45 | 11.8 ± 0.16 | 1.4 | 42.4 | 20.82 ± 0.23 | 12.13 ± 0.16 | 1.281 | 41.7 | 12.05 ± 0.134 | 1.112 | 42.1 |
| BZF | 0.997 | 0.134 | 0.445 | 12.48 ± 0.21 | 1.72 ± 0.01 | 0.723 | 86 | 1.75 ± 0.02 | 1.4 | 86 | 12.44 ± 0.18 | 2.093 ± 0.01 | 0.225 | 83 | 2.126 ± 0.038 | 1.814 | 82.9 | 12.31 ± 0.15 | 2.43 ± 0.01 | 0.455 | 80.26 | 2.34 ± 0.025 | 0.752 | 81 |
| CBZ | 0.991 | 1.17 | 3.9 | 12.583 ± 0.15 | 8.96 ± 0.1 | 1.112 | 28.7 | 9.09 ± 0.08 | 0.933 | 27.7 | 12.693 ± 0.15 | 9.55 ± 0.15 | 1.539 | 24.7 | 9.62 ± 0.237 | 2.466 | 24.2 | 12.428 ± 0.17 | 9.529 ± 0.09 | 0.891 | 23.32 | 9.731 ± 0.067 | 0.688 | 21.7 |
| ATN | 0.992 | 0.64 | 2.13 | 20.81 ± 0.12 | 2.41 ± 0.04 | 1.79 | 88 | 2.51 ± 0.06 | 2.28 | 88 | 20.87 ± 0.04 | 3.52 ± 0.03 | 0.877 | 83 | 3.5 ± 0.054 | 1.55 | 83 | 20.81 ± 0.26 | 4.385 ± 0.02 | 0.522 | 78.9 | 4.58 ± 0.037 | 0.814 | 78 |
| RND | 0.997 | 0.665 | 2.22 | 20.48 ± 0.07 | 2.02 ± 0.03 | 1.4 | 90 | 2.028 ± 0.05 | 2.43 | 90 | 20.6 ± 0.27 | 3.28 ± 0.03 | 0.995 | 84 | 3.21 ± 0.088 | 2.77 | 84.4 | 20.52 ± 0.18 | 3.81 ± 0.01 | 0.228 | 81.4 | 3.73 ± 0.012 | 0.342 | 81.8 |
| PCT | 0.999 | 5.3 | 17.7 | 103.96 ± 0.81 | n.d | ≥95 | n.d | ≥95 | 104.033 ± 1.95 | n.d | ≥95 | n.d | ≥95 | 103.247 ± 1.26 | n.d | ≥95 | n.d | ≥95 | ||||||
| PPCPs | Transformation Products | m/z | Structure | Formula |
|---|---|---|---|---|
| Diclofenac | DCF-TP1 DCF-lactam or 1-(2,6-dichlorophenyl)-1,3-dihydro-2H-indol-2-one | 280.095 | 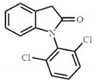 | C14H10NOCl2 |
| DCF-TP2 DCF-benzoic acid | 283.036 | 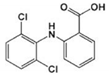 | C13H9Cl2NO2 | |
| Bezafibrate | 4-Chlorobenzoic acid | 113.05 | 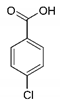 | C7H5ClO2 |
| Carbamazepine | CBZ-TP1 | 253.097 |  | C15H12N2O2 |
| CBZ-TP2 | 210.015 | 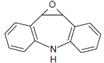 | C14H11NO | |
| CBZ-TP3 | 224.125 | 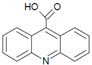 | C14H9NO2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
ALOBAIDI, R.A.K.; ULUCAN-ALTUNTAS, K.; MHEMID, R.K.S.; MANAV-DEMIR, N.; CINAR, O. Biodegradation of Emerging Pharmaceuticals from Domestic Wastewater by Membrane Bioreactor: The Effect of Solid Retention Time. Int. J. Environ. Res. Public Health 2021, 18, 3395. https://doi.org/10.3390/ijerph18073395
ALOBAIDI RAK, ULUCAN-ALTUNTAS K, MHEMID RKS, MANAV-DEMIR N, CINAR O. Biodegradation of Emerging Pharmaceuticals from Domestic Wastewater by Membrane Bioreactor: The Effect of Solid Retention Time. International Journal of Environmental Research and Public Health. 2021; 18(7):3395. https://doi.org/10.3390/ijerph18073395
Chicago/Turabian StyleALOBAIDI, Raghad Asad Kadhim, Kubra ULUCAN-ALTUNTAS, Rasha Khalid Sabri MHEMID, Neslihan MANAV-DEMIR, and Ozer CINAR. 2021. "Biodegradation of Emerging Pharmaceuticals from Domestic Wastewater by Membrane Bioreactor: The Effect of Solid Retention Time" International Journal of Environmental Research and Public Health 18, no. 7: 3395. https://doi.org/10.3390/ijerph18073395






