Fourier-Transform Infrared Spectroscopy as a Discriminatory Tool for Myotonic Dystrophy Type 1 Metabolism: A Pilot Study
Abstract
:1. Introduction
2. Material and Methods
2.1. DM1-Derived Fibroblast and Controls
2.2. Cell Culture
2.3. FTIR Measurements
2.4. FTIR Data Analysis
2.5. Intensity Ratio Evaluation
3. Results
3.1. FTIR Spectral Analysis of Fibroblasts from Coriell Institute
3.2. Multivariate Analysis of the Spectroscopic Data of Fibroblasts from Coriell Institute
3.3. Intensity Ratios Obtained from Spectroscopic Data of Fibroblasts from Coriell Institute
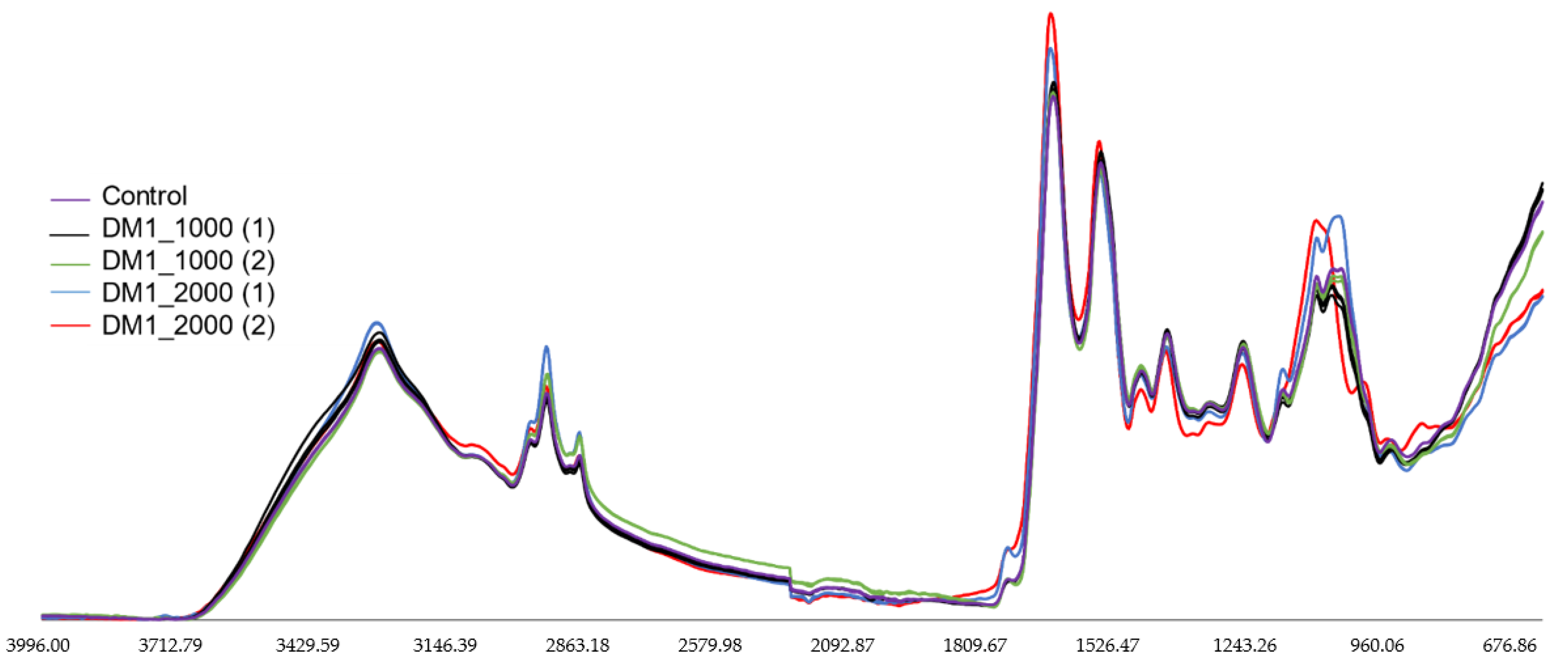
3.4. FTIR Spectral Analysis of DM1-Derived Fibroblast from Neurolab UA
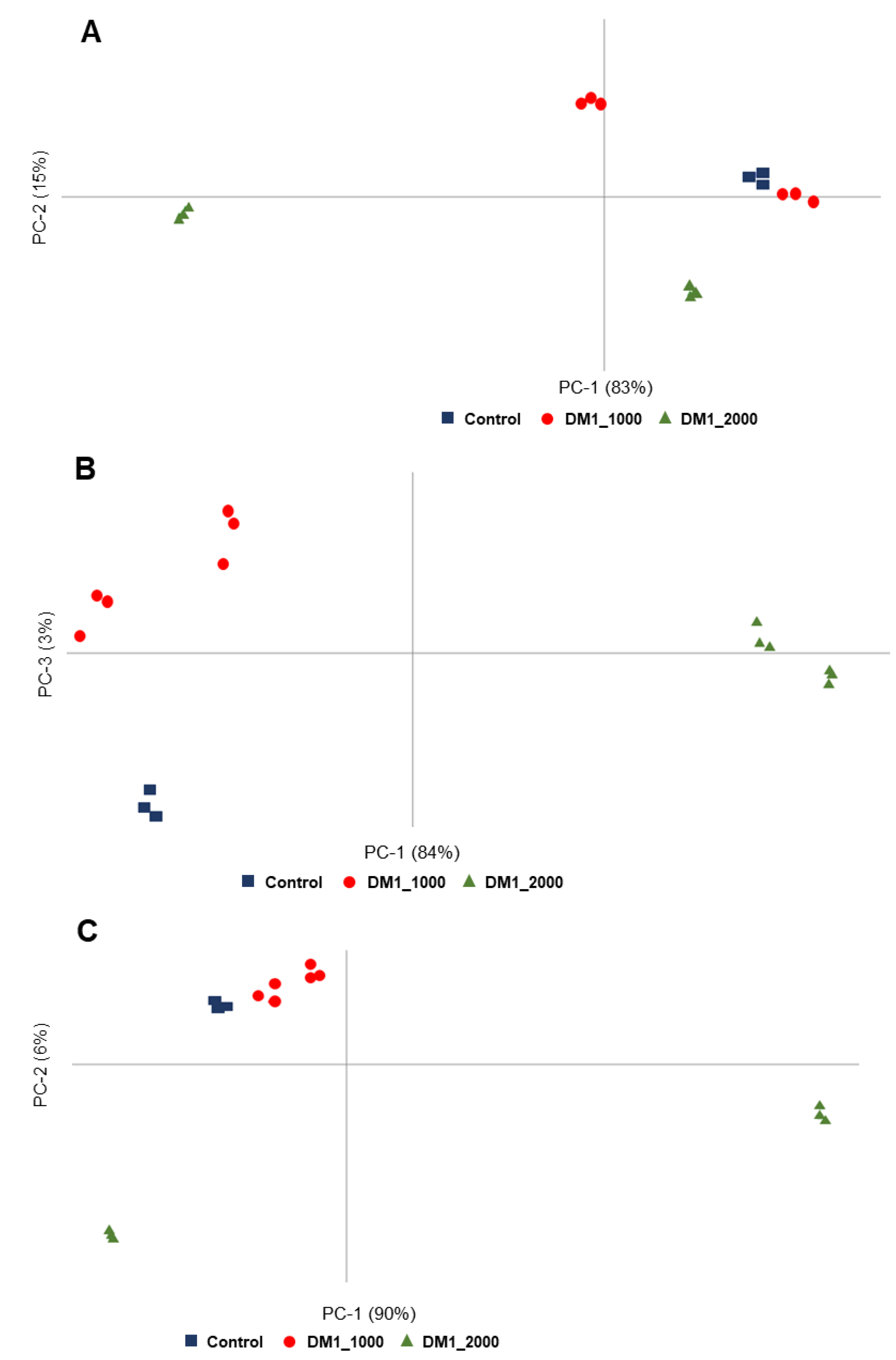
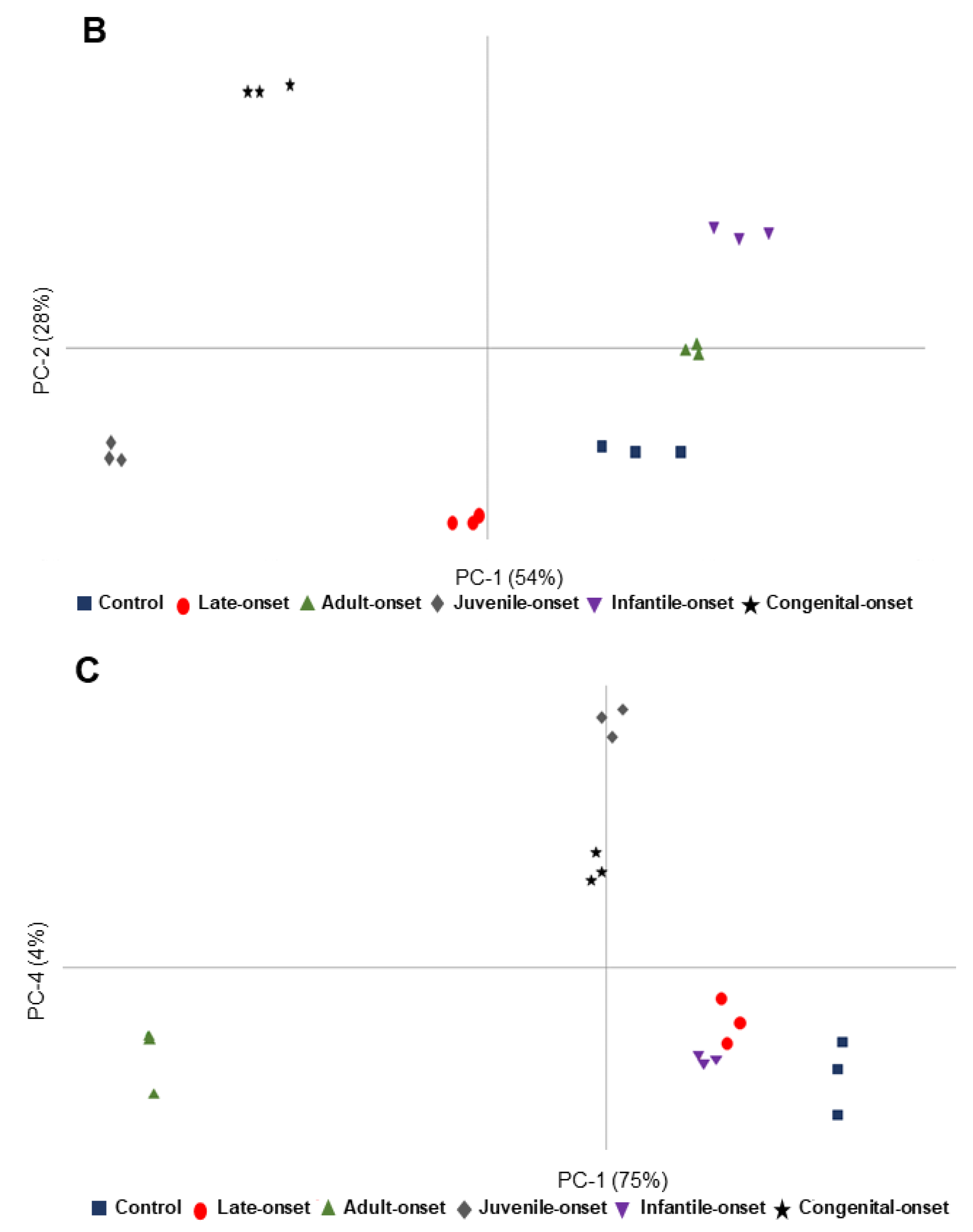
3.5. Multivariate Analysis of the Spectroscopic Data of Fibroblasts Cultured at Neurolab UA
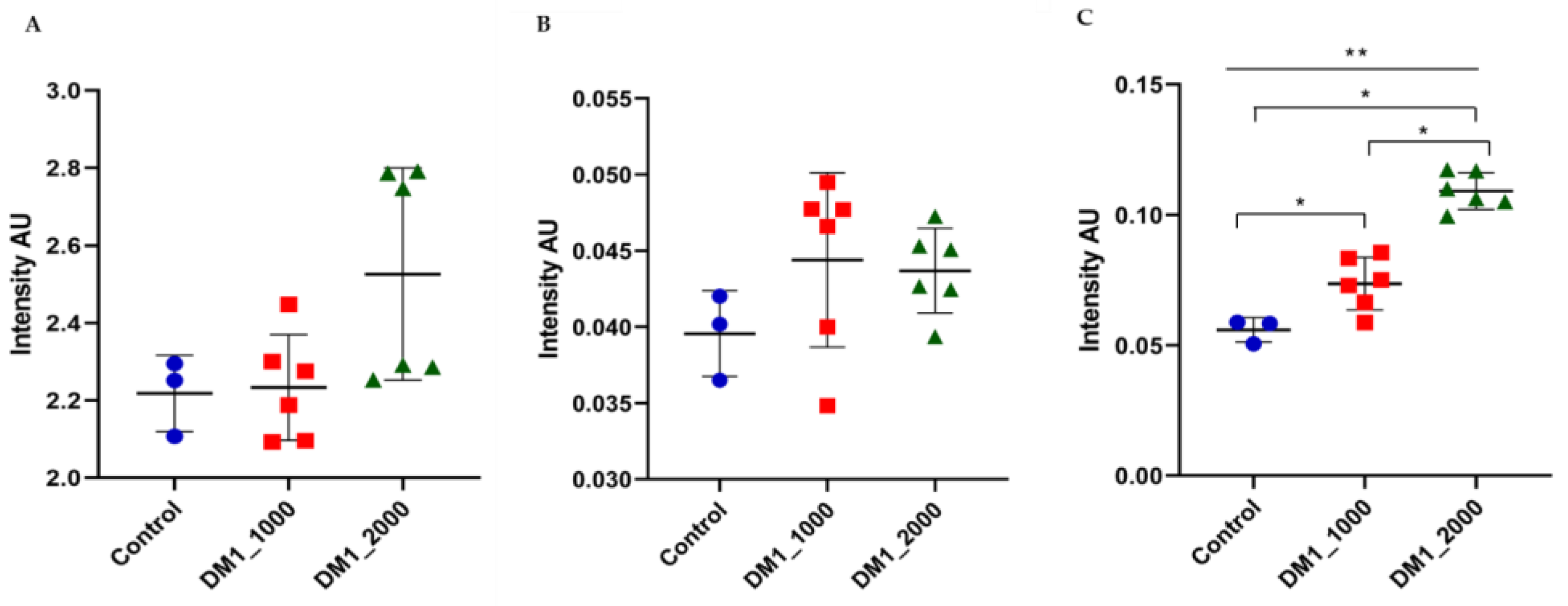
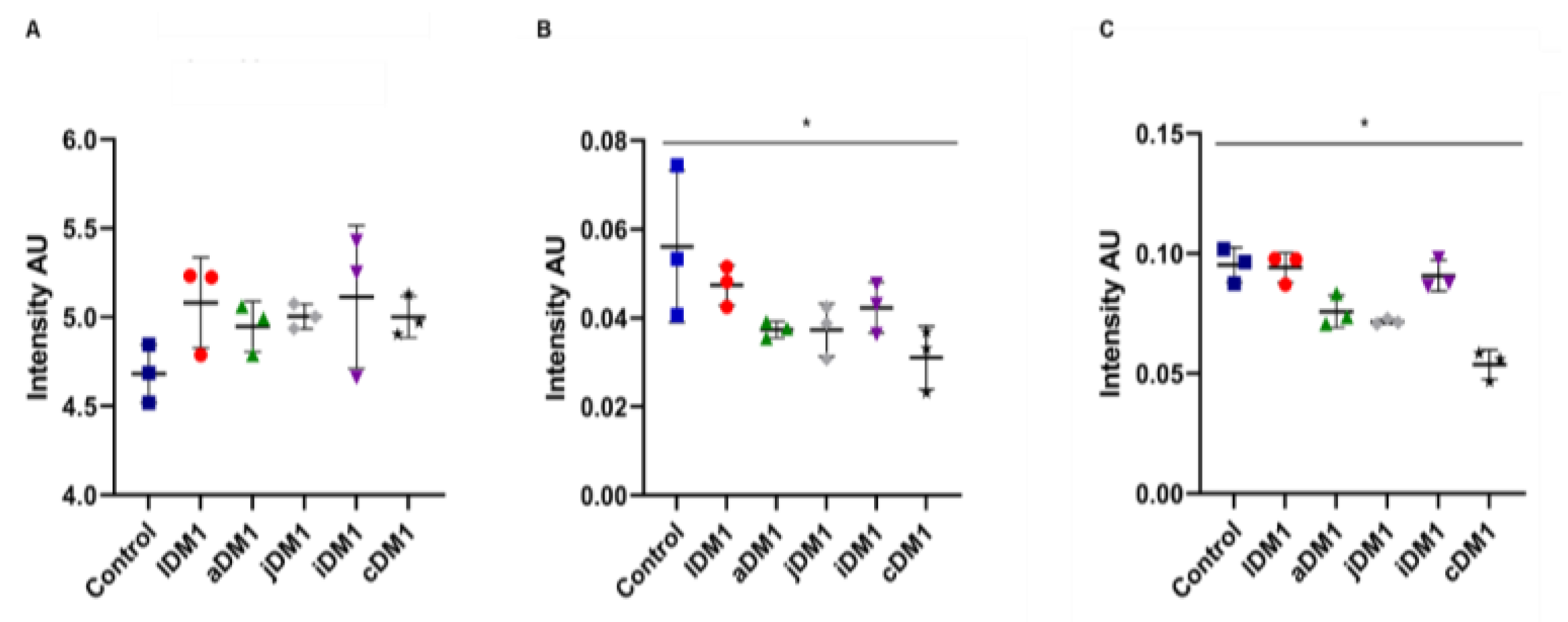
3.6. Intensity Ratios Obtained from Spectroscopic Data of DM1-Derived Fibroblasts Cultured at Neurolab UA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, N.E. Myotonic Muscular Dystrophies. Contin. Lifelong Learn. Neurol. 2019, 25, 1682–1695. [Google Scholar] [CrossRef] [PubMed]
- Bozovic, I.; Peric, S.; Pesovic, J.; Bjelica, B.; Brkusanin, M.; Basta, I.; Bozic, M.; Sencanic, I.; Marjanovic, A.; Brankovic, M.; et al. Myotonic Dystrophy Type 2 – Data from the Serbian Registry. J. Neuromuscul. Dis. 2018, 5, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, N.; Rastelli, E.; Antonini, G.; Bianchi, M.L.E.; Botta, A.; Bucci, E.; Casali, C.; Costanzi-Porrini, S.; Giacanelli, M.; Gibellini, M.; et al. An Age-Standardized Prevalence Estimate and a Sex and Age Distribution of Myotonic Dystrophy Types 1 and 2 in the Rome Province, Italy. Neuroepidemiology 2016, 46, 191–197. [Google Scholar] [CrossRef]
- Rodríguez, R.; Hernández-Hernández, O.; Magaña, J.J.; González-Ramírez, R.; García-López, E.S.; Cisneros, B. Altered nuclear structure in myotonic dystrophy type 1-derived fibroblasts. Mol. Biol. Rep. 2015, 42, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Cè, E.; Rampichini, S.; Monti, E.; Limonta, E.; Fossati, B.; Meola, G. Electromechanical delays during a fatiguing exercise and recovery in patients with myotonic dystrophy type 1. Eur. J. Appl. Physiol. 2017, 117, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Magaña, J.J.; Suárez-Sánchez, R.; Leyva-García, N.; Cisneros, B.; Hernández-Hernández, O. Myotonic Dystrophy Protein Kinase: Structure, Function and Its Possible Role in the Pathogenesis of Myotonic Dystrophy Type 1. In Advances in Protein Kinases; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- Cho, D.H.; Tapscott, S.J. Myotonic dystrophy: Emerging mechanisms for DM1 and DM2. Biochim. Biophys. Acta Mol. Basis Dis. 2007, 1772, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Thornton, C.A. Myotonic Dystrophy. Neurol. Clin. 2014, 32, 705–719. [Google Scholar] [CrossRef] [Green Version]
- De Antonio, M.; Dogan, C.; Hamroun, D.; Mati, M.; Zerrouki, S.; Eymard, B.; Katsahian, S.; Bassez, G. Unravelling the myotonic dystrophy type 1 clinical spectrum: A systematic registry-based study with implications for disease classification. Rev. Neurol. (Paris). 2016, 172, 572–580. [Google Scholar] [CrossRef]
- Vujnic, M.; Peric, S.; Popovic, S.; Raseta, N.; Ralic, V.; Dobricic, V.; Novakovic, I.; Rakocevic-Stojanovic, V. Metabolic syndrome in patients with myotonic dystrophy type 1. Muscle Nerve 2015, 52, 273–277. [Google Scholar] [CrossRef]
- Mateus, T.; Martins, F.; Nunes, A.; Herdeiro, M.T. Metabolic Alterations in Myotonic Dystrophy Type 1 and Their Correlation with Lipin. Int. J. Environ. Res. Public Health 2021, 18, 1794. [Google Scholar] [CrossRef]
- Ben Hamou, A.; Espiard, S.; Do Cao, C.; Ladsous, M.; Loyer, C.; Moerman, A.; Boury, S.; Kyheng, M.; Dhaenens, C.-M.; Tiffreau, V.; et al. Systematic thyroid screening in myotonic dystrophy: Link between thyroid volume and insulin resistance. Orphanet J. Rare Dis. 2019, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Renna, L.V.; Bosè, F.; Brigonzi, E.; Fossati, B.; Meola, G.; Cardani, R. Aberrant insulin receptor expression is associated with insulin resistance and skeletal muscle atrophy in myotonic dystrophies. PLoS ONE 2019, 14, e0214254. [Google Scholar] [CrossRef]
- Renna, L.V.; Bosè, F.; Iachettini, S.; Fossati, B.; Saraceno, L.; Milani, V.; Colombo, R.; Meola, G.; Cardani, R. Receptor and post-receptor abnormalities contribute to insulin resistance in myotonic dystrophy type 1 and type 2 skeletal muscle. PLoS ONE 2017, 12, e0184987. [Google Scholar] [CrossRef] [Green Version]
- Daniele, A.; De Rosa, A.; De Cristofaro, M.; Monaco, M.L.; Masullo, M.; Porcile, C.; Capasso, M.; Tedeschi, G.; Oriani, G.; Di Costanzo, A. Decreased concentration of adiponectin together with a selective reduction of its high molecular weight oligomers is involved in metabolic complications of myotonic dystrophy type 1. Eur. J. Endocrinol. 2011, 165, 969–975. [Google Scholar] [CrossRef] [Green Version]
- Shieh, K.; Gilchrist, J.M.; Promrat, K. Frequency and predictors of nonalcoholic fatty liver disease in myotonic dystrophy. Muscle Nerve 2010, 41, 197–201. [Google Scholar] [CrossRef]
- Johansson, A.; Olsson, T.; Cederquist, K.; Forsberg, H.; Holst, J.; Ahren, B. Abnormal release of incretins and cortisol after oral glucose in subjects with insulin-resistant myotonic dystrophy. Eur. J. Endocrinol. 2002, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Johansson, A.; Boman, K.; Cederquist, K.; Forsberg, H.; Olsson, T. Increased levels of tPA antigen and tPA/PAI-1 complex in myotonic dystrophy. J. Intern. Med. 2001, 249, 503–510. [Google Scholar] [CrossRef] [Green Version]
- Cacciatore, S.; Loda, M. Innovation in metabolomics to improve personalized healthcare. Ann. N. Y. Acad. Sci. 2015, 1346, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Ellis, D.I.; Dunn, W.B.; Griffin, J.L.; Allwood, J.W.; Goodacre, R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 2007, 8, 1243–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derenne, A.; Vandersleyen, O.; Goormaghtigh, E. Lipid quantification method using FTIR spectroscopy applied on cancer cell extracts. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- Stuart, B.H. Infrared Spectroscopy of Biological Applications: An Overview. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd: Chichester, UK, 2012. [Google Scholar]
- Bujok, J.; Gąsior-Głogowska, M.; Marszałek, M.; Trochanowska-Pauk, N.; Zigo, F.; Pavľak, A.; Komorowska, M.; Walski, T. Applicability of FTIR-ATR Method to Measure Carbonyls in Blood Plasma after Physical and Mental Stress. Biomed Res. Int. 2019, 2019, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, B.H. Infrared Spectroscopy: Fundamentals and Applications; John. Wiley & Sons, Ltd: LtdHoboken, NJ, USA, 2005; Volume 8, ISBN 9780470011140. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Lopes, J.; Correia, M.; Martins, I.; Henriques, A.G.; Delgadillo, I.; Da Cruz E Silva, O.; Nunes, A. FTIR and Raman Spectroscopy Applied to Dementia Diagnosis Through Analysis of Biological Fluids. J. Alzheimer’s Dis. 2016, 52, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Oleszko, A.; Hartwich, J.; Wójtowicz, A.; Gąsior-Głogowska, M.; Huras, H.; Komorowska, M. Comparison of FTIR-ATR and Raman spectroscopy in determination of VLDL triglycerides in blood serum with PLS regression. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 183, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Mizaikoff, B. Application of multivariate data-analysis techniques to biomedical diagnostics based on mid-infrared spectroscopy. Anal. Bioanal. Chem. 2008, 391, 1641–1654. [Google Scholar] [CrossRef]
- Santos, F.; Magalhães, S.; Henriques, M.C.; Silva, B.; Valença, I.; Ribeiro, D.; Fardilha, M.; Nunes, A. Understanding Prostate Cancer Cells Metabolome: A Spectroscopic Approach. Curr. Metabolomics 2019, 6, 218–224. [Google Scholar] [CrossRef]
- Igci, N.; Sharafi, P.; Ozel Demiralp, D.; Demiralp, C.; Yuce, A.; Dokmeci (Emre), S. Application of Fourier transform infrared spectroscopy to biomolecular profiling of cultured fibroblast cells from Gaucher disease patients: A preliminary investigation. Adv. Clin. Exp. Med. 2017, 26, 1053–1061. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.D.; Martins, F.; Santos, M.; Müeller, T.; da Cruz e Silva, O.A.B.; Rebelo, S. Nuclear Accumulation of LAP1:TRF2 Complex during DNA Damage Response Uncovers a Novel Role for LAP1. Cells 2020, 9, 1804. [Google Scholar] [CrossRef]
- Martins, F.; Serrano, J.B.; Müller, T.; da Cruz e Silva, O.A.B.; Rebelo, S. BRI2 Processing and Its Neuritogenic Role Are Modulated by Protein Phosphatase 1 Complexing. J. Cell. Biochem. 2017, 118, 2752–2763. [Google Scholar] [CrossRef]
- Staniszewska-Slezak, E.; Wiercigroch, E.; Fedorowicz, A.; Buczek, E.; Mateuszuk, L.; Baranska, M.; Chlopicki, S.; Malek, K. A possible Fourier transform infrared-based plasma fingerprint of angiotensin-converting enzyme inhibitor-induced reversal of endothelial dysfunction in diabetic mice. J. Biophotonics 2018, 11, e201700044. [Google Scholar] [CrossRef]
- Felgueiras, J.; Silva, J.V.; Nunes, A.; Fernandes, I.; Patrício, A.; Maia, N.; Pelech, S.; Fardilha, M. Investigation of spectroscopic and proteomic alterations underlying prostate carcinogenesis. J. Proteomics 2020, 226, 103888. [Google Scholar] [CrossRef] [PubMed]
- Yonar, D.; Ocek, L.; Tiftikcioglu, B.I.; Zorlu, Y.; Severcan, F. Relapsing-Remitting Multiple Sclerosis diagnosis from cerebrospinal fluids via Fourier transform infrared spectroscopy coupled with multivariate analysis. Sci. Rep. 2018, 8, 1025. [Google Scholar] [CrossRef]
- Oleszko, A.; Olsztyńska-Janus, S.; Walski, T.; Grzeszczuk-Kuć, K.; Bujok, J.; Gałecka, K.; Czerski, A.; Witkiewicz, W.; Komorowska, M. Application of FTIR-ATR Spectroscopy to Determine the Extent of Lipid Peroxidation in Plasma during Haemodialysis. Biomed Res. Int. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Cline, M.S.; Osborne, R.J.; Tuttle, D.L.; Clark, T.A.; Donohue, J.P.; Hall, M.P.; Shiue, L.; Swanson, M.S.; Thornton, C.A.; et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010, 17, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.T.; Treacy, D.; Eichinger, K.; Struck, A.; Estabrook, J.; Olafson, H.; Wang, T.T.; Bhatt, K.; Westbrook, T.; Sedehizadeh, S.; et al. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum. Mol. Genet. 2019, 28, 1312–1321. [Google Scholar] [CrossRef]
- Chen, Y.; Rui, B.-B.; Tang, L.-Y.; Hu, C.-M. Lipin Family Proteins - Key Regulators in Lipid Metabolism. Ann. Nutr. Metab. 2015, 66, 10–18. [Google Scholar] [CrossRef]
- Okuno, H.; Okuzono, H.; Hayase, A.; Kumagai, F.; Tanii, S.; Hino, N.; Okada, Y.; Tachibana, K.; Doi, T.; Ishimoto, K. Lipin-1 is a novel substrate of protein phosphatase PGAM5. Biochem. Biophys. Res. Commun. 2019, 509, 886–891. [Google Scholar] [CrossRef]
- Péterfy, M.; Phan, J.; Reue, K. Alternatively Spliced Lipin Isoforms Exhibit Distinct Expression Pattern, Subcellular Localization, and Role in Adipogenesis. J. Biol. Chem. 2005, 280, 32883–32889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finck, B.N.; Gropler, M.C.; Chen, Z.; Leone, T.C.; Croce, M.A.; Harris, T.E.; Lawrence, J.C.; Kelly, D.P. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 2006, 4, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Srinivasan, A.; Nikolajeff, F. Role of Infrared Spectroscopy and Imaging in Cancer Diagnosis. Curr. Med. Chem. 2017, 25, 1055–1072. [Google Scholar] [CrossRef] [PubMed]
- Perczel, A.; Gáspári, Z.; Csizmadia, I.G. Structure and stability of β-pleated sheets. J. Comput. Chem. 2005, 26, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.M.; Krym, M.C.; Thornton, C.A. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007, 17, 242–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravel-Chapuis, A.; Bélanger, G.; Yadava, R.S.; Mahadevan, M.S.; DesGroseillers, L.; Côté, J.; Jasmin, B.J. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012, 196, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Ramana, K.V.; Srivastava, S.; Singhal, S.S. Lipid Peroxidation Products in Human Health and Disease. Oxid. Med. Cell. Longev. 2013, 2013, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Wills, E.D. Mechanisms of lipid peroxide formation in animal tissues. Biochem. J. 1966, 99, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014. [Google Scholar] [CrossRef]


| 3000–2800 cm−1 Region | |||
| Discrimination across PC-2 | λ (cm−1) | Vibrational Mode | Assignments |
| PC-2 – DM1_2000 (1 and 2) | 2953 | CH3 asymmetric stretching | Lipid (long chain fatty acids, phospholipids) |
| 2916 | CH2 and CH3 stretching of phospholipids | ||
| 2849 | CH2 symmetric stretching | ||
| PC-2 + Control and DM1_1000 (1 and 2) | 2925 | CH2 asymmetric stretching | |
| 2874 | CH3 symmetric stretching | ||
| 1800–1500 cm−1 Region | |||
| Discrimination across PC-1 | λ (cm−1) | Vibrational Mode | Assignments |
| PC-1 – Control and DM1_1000 (1 and 2) | 1747 | C=O stretching | Triacylglycerol, cholesterol esters, glycerophospholipids |
| 1736 | |||
| 1696 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: anti-parallel β-sheets (peptide, protein) | |
| 1682 | Amide-I: anti-parallel β- sheets (peptide, protein) | ||
| 1651 | Amide-I: α- helices | ||
| 1554 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1523 | |||
| PC-1 + DM1_2000 (1 and 2) | 1628 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: parallel β- sheets (peptide, protein) |
| 1537 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1509 | CH2 bending | Lipid, protein | |
| 1800–1500 cm−1 Region (Q2 and Q3) | |||
| Quadrant-2 | λ (cm−1) | Vibrational Mode | Assignments |
| DM1_1000 (1 and 2) | 1693 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: anti-parallel β-sheets (peptide, protein) |
| 1639 | Amide-I: parallel β- sheets (peptide, protein) | ||
| Quadrant-3 | λ (cm−1) | Vibrational mode | Assignments |
| Control samples | 1747-1743 | C=O stretching | Triacylglycerol, cholesterol esters, glycerophospholipids |
| 1682 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: anti-parallel β- sheets (peptide, protein) | |
| 1651 | Amide-I: α- helices | ||
| 1554 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1543 | |||
| 1200–900 cm−1 Region | |||
| Discrimination across PC-2 | λ (cm−1) | Vibrational Mode | Assignments |
| PC-2 – DM1_2000 (1 and 2) | 1172 | C–O stretching | Carbohydrates/glycogen, nucleic acids |
| 1013 | C–O stretching and C–OH bending | DNA and RNA, oligosaccharides, polysaccharides (e.g., glucose) | |
| 991 | C–O stretching | DNA and RNA ribose | |
| 914 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA | |
| PC-2 + Control and DM1_1000 (1 and 2) | 1152 | C–O stretching, C–O–H bending | Carbohydrates |
| 1104 | PO2− symmetrical stretching | DNA, RNA, phospholipid, phosphorylated protein | |
| 1079 | |||
| 1053 | C–O stretching and C–OH bending | DNA and RNA, oligosaccharides, polysaccharides (e.g., glucose) | |
| 968 | PO32− stretching | DNA and RNA ribose | |
| 928 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA | |
| 3000–2800 cm−1 Region | |||
| Discrimination across PC-1 | λ (cm−1) | Vibrational Mode | Assignments |
| PC-1 − Control, lDM1, aDM1 | 2871 | CH3 symmetric stretching | Lipid (long chain fatty acids, phospholipids) |
| PC-1 + jDM1, iDM1 and cDM1 | 2959 | CH3 asymmetric stretching | |
| 2919 | CH2 asymmetric stretching | ||
| 2851 | CH2 symmetric stretching | ||
| 3000–2800 cm−1 region (Q1, Q3 and Q4) | |||
| Quadrant-1 | λ (cm−1) | Vibrational Mode | Assignments |
| jDM1 | No peaks | NA | Lipid (long chain fatty acids, phospholipids) |
| Quadrant-3 | λ (cm−1) | Vibrational mode | Assignments |
| aDM1 | No peaks | NA | Lipid (long chain fatty acids, phospholipids) |
| Quadrant-4 | λ (cm−1) | Vibrational mode | Assignments |
| iDM1 and cDM1 | 2956 | CH3 asymmetric stretching | Lipid (long chain fatty acids, phospholipids) |
| 2922 | CH2 asymmetric stretching | ||
| 2854 | CH2 symmetric stretching | ||
| Mid-IR bands at 1800–1500 cm−1 Region | |||
| Discrimination across PC-2 | λ (cm−1) | Vibrational Mode | Assignments |
| PC-2 − Control, lDM1, aDM1 and jDM1 | 1648 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: α- helices |
| 1628 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: parallel β- sheets (peptide, protein) | |
| 1551 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1537 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1512 | CH2 bending | Lipid, protein | |
| PC-2 + iDM1 and cDM1 | 1747 | C=O stretching | Triacylglycerol, cholesterol esters, glycerophospholipids |
| 1696 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: anti-parallel β-sheets (peptide, protein) | |
| 1682 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: anti-parallel β-sheets (peptide, protein) | |
| 1662 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: α- helices | |
| 1639 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: parallel β- sheets (peptide, protein) | |
| 1523 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1800–1500 cm−1 (Q1, Q2, Q3 and Q4) | |||
| Quadrant-1 | λ (cm−1) | Vibrational Mode | Assignments |
| iDM1 | No peak | NA | NA |
| Quadrant-2 | λ (cm−1) | Vibrational mode | Assignments |
| cDM1 | 1747 | C=O stretching | Triacylglycerol, cholesterol esters, glycerophospholipids |
| 1736 | |||
| 1696 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: anti-parallel β-sheets (peptide, protein) | |
| 1682 | |||
| 1639 | Amide-I: parallel β- sheets (peptide, protein) | ||
| Quadrant-3 | λ (cm−1) | Vibrational mode | Assignments |
| lDM1 and jDM1 | No peak | NA | NA |
| Quadrant-4 | λ (cm−1) | Vibrational mode | Assignments |
| Control and aDM1 | 1631 | 80% C=O stretching, 10% N–H bending, 10% C–N stretching | Amide-I: parallel β- sheets (peptide, protein) |
| 1534 | 60% N–H bending, 40% C–N stretching | Amide II (proteins) | |
| 1515 | CH2 bending | Lipid, protein | |
| 1200–900 cm−1 Region | |||
| Discrimination across PC-4 | λ (cm−1) | Vibrational Mode | Assignments |
| PC-4 − Control, lDM1, aDM1, iDM1 | 1155 | C–O stretching, C–O–H bending | Carbohydrates |
| 1076 | PO2− symmetrical stretching | DNA, RNA, phospholipid, phosphorylated protein | |
| 1025 | C–O stretching and C–OH bending | DNA and RNA, oligosaccharides, polysaccharides (e.g., glucose) | |
| 923 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA | |
| PC-4 + jDM1 and cDM1 | 1121 | Phosphodiester groups of PO2− | RNA |
| 1084 | PO2− symmetrical stretching | DNA, RNA, phospholipid, phosphorylated protein | |
| 1047 | C–O stretching and C–OH bending | DNA and RNA, oligosaccharides, polysaccharides (e.g., glucose) | |
| 994 | C–O stretching | DNA and RNA ribose | |
| 968 | PO32− stretching | DNA and RNA ribose | |
| 931 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA | |
| 914 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA | |
| 1200–900 cm−1 Region (Q1, Q2, Q3 and Q4) | |||
| Quadrant-1 | λ (cm−1) | Vibrational Mode | Assignments |
| jDM1 | 1124 | Phosphodiester groups of PO2− | RNA |
| 996 | C–O stretching | DNA and RNA ribose | |
| Quadrant-2 | λ (cm−1) | Vibrational mode | Assignments |
| cDM1 | 914 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA |
| Quandrant-3 | λ (cm−1) | Vibrational mode | Assignments |
| aDM1 | 1112 | Phosphodiester groups of PO2− | RNA |
| Quadrant-4 | λ (cm−1) | Vibrational mode | Assignments |
| Control, lDM1 and iDM1 | 1155 | C–O stretching, C–O–H bending | Carbohydrates |
| 1079 | PO2− symmetrical stretching | DNA, RNA, phospholipid, phosphorylated protein | |
| 1042 | C–O stretching and C–OH bending | DNA and RNA, oligosaccharides, polysaccharides (e.g., glucose) | |
| 1022 | |||
| 926 | C–N+–C stretching | DNA and RNA ribose-phosphate chain vibration of RNA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mateus, T.; Almeida, I.; Costa, A.; Viegas, D.; Magalhães, S.; Martins, F.; Herdeiro, M.T.; da Cruz e Silva, O.A.B.; Fraga, C.; Alves, I.; et al. Fourier-Transform Infrared Spectroscopy as a Discriminatory Tool for Myotonic Dystrophy Type 1 Metabolism: A Pilot Study. Int. J. Environ. Res. Public Health 2021, 18, 3800. https://doi.org/10.3390/ijerph18073800
Mateus T, Almeida I, Costa A, Viegas D, Magalhães S, Martins F, Herdeiro MT, da Cruz e Silva OAB, Fraga C, Alves I, et al. Fourier-Transform Infrared Spectroscopy as a Discriminatory Tool for Myotonic Dystrophy Type 1 Metabolism: A Pilot Study. International Journal of Environmental Research and Public Health. 2021; 18(7):3800. https://doi.org/10.3390/ijerph18073800
Chicago/Turabian StyleMateus, Tiago, Idália Almeida, Adriana Costa, Diana Viegas, Sandra Magalhães, Filipa Martins, Maria Teresa Herdeiro, Odete A. B. da Cruz e Silva, Carla Fraga, Ivânia Alves, and et al. 2021. "Fourier-Transform Infrared Spectroscopy as a Discriminatory Tool for Myotonic Dystrophy Type 1 Metabolism: A Pilot Study" International Journal of Environmental Research and Public Health 18, no. 7: 3800. https://doi.org/10.3390/ijerph18073800








