Bio-Mercury Remediation Suitability Index: A Novel Proposal That Compiles the PGPR Features of Bacterial Strains and Its Potential Use in Phytoremediation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Obtaining the Mercury-Tolerant Strains
2.2. Identification of the Bacterial Isolates
2.3. Determination of PGPR Capacity
2.4. Maximum Bactericidal Concentration of Hg (MBC)
2.5. Bio-Mercury Remediation Suitability Index (BMRSI)
2.6. Processing the Information
3. Results
3.1. Selection of Mercury-Tolerant Strains and Assessment of PGPR Activity
3.2. Maximum Bactericidal Concentration to Hg (MBC)
3.3. Bio-Mercury Remediation Suitability Index (BMRSI)
- Typology I: Strains with values equal to or greater than 6.5 in the BMRSI.
- Typology II: IAA producing strains above 5.5 µg/mL.
- Typology III: Strains producing three or more PGPR activities simultaneously.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Das, S.; Dash, H.; Chakraborty, J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbial. Biotechnol. 2016, 100, 2967–2984. [Google Scholar] [CrossRef]
- Gil-Hernández, F.; Gómez-Fernández, A.R.; la Torre-Aguilar, M.J.; Pérez-Navero, J.L.; Flores-Rojas, K.; Martín-Borreguero, P.; Gil-Campos, M. Neurotoxicity by mercury is not associated with autism spectrum disorders in Spanish children. Ital. J. Pediatr. 2020, 46, 19. [Google Scholar] [CrossRef]
- Wohlgemuth, L.; Osterwalder, S.; Joseph, C.; Kahmen, A.; Hoch, G.; Alewell, C.; Jiskra, M. A bottom-up quantification of foliar mercury uptake fluxes across Europe. Biogeosciences 2020, 17, 6441–6456. [Google Scholar] [CrossRef]
- Hao, X.; Xie, P.; Johnstone, L.; Miller, S.J.; Rensing, C.; Wei, G. Genomesequence and mutationalanalysis of plant-growth-promotingbacterium Agrobacterium tumefaciens CCNWGS0286 isolatedfrom a zinc-lead mine tailing. Appl. Environ. Microbiol. 2012, 78, 5384–5394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Jiménez, E.; Vázquez, S.; Carpena-Ruiz, R.O.; Esteban, E.; Peñalosa, J.M. Using Mediterranean shrubs for the phytoremediation of a soil impacted by pyritic wastes in Southern Spain. A field experiment. Environ. Manag. J. 2011, 92, 1584–1590. [Google Scholar] [CrossRef] [Green Version]
- Souza de, M.P.; Huang, C.P.A.; Chee, N.; Terry, N. Rhizosphere bacteria enhance the accumulation of selenium and mercury in wetland plants. Planta 1999, 209, 259–263. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Schroth, M.N. Plant growth-promoting rhizobacteria in radish. In Proceedings of the 4th International Conference of Plant Pathogenic Bacteria, Tours, France, 27 August–2 September 1978; pp. 879–882. [Google Scholar]
- Le Cloirec, P.; Andrès, Y.; Glass, D. Bioremediation of Heavy Metals Using Microorganisms. Bioremediat. Aquat. Terr. Ecosyst. 2005, 97–140. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Das, S.; Raj, R.; Mangwani, N.; Dash, H.R.; Chakraborty, J. Heavy metals and hydrocarbons: Adverse effects and mechanism of toxicity. Microb. Biodegrad. Bioremediat. 2014, 23–54. [Google Scholar]
- Osborn, A.M.; Bruce, K.D.; Strike, P.; Ritchie, D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. 1997, 19, 239–262. [Google Scholar] [CrossRef]
- Neville, F. Replication of Staphylococcal multiresistance plasmids. J. Bacteriol. 2000, 182, 2170–2178. [Google Scholar]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef] [Green Version]
- Verma, T.; Srinath, T.; Gadpayle, R.U.; Ramteke, P.W.; Hans, R.K.; Garg, S.K. Chromate tolerant bacteria isolated from tannery effluent. Bioresoul Technol. 2001, 78, 31–35. [Google Scholar] [CrossRef]
- Woegerbauer, M.; Zeinzinger, J.; Gottsberger, R.A.; Pascher, K.; Hufnagl, P.; Indra, A.; Fuchs, R.; Hofrichter, J.; Kopacka, I.; Korschineck, I.; et al. Antibiotic resistance marker genes as environmental pollutants in GMO-pristine agricultural soils in Austria. Environ. Pollut. 2015, 206, 342–351. [Google Scholar] [CrossRef]
- Gerhardt, K.E.; Huang, X.D.; Glick, B.R.; Greenberg, B.M. Phytoremediation and rhizoremediation of organic soil contaminants: Potential and challenges. Plant Sci. 2009, 176, 20–30. [Google Scholar] [CrossRef]
- Millán, R.; Carpena, R.O.; Sierra, M.J.; Moreno, E.; Peñalosa, J.; Gamarra, R.; Esteban, E. Rehabilitación de suelos contaminados con mercurio: Estrategias aplicables en el área de Almadén. Ecosistemas 2007, 16, 56–66. [Google Scholar]
- Garcia-Villarco, A.; Probanza, A.; Gutiérrez, J.R.; Ramos, B.; Lucas, J.A. Functional diversity of rhizosphere microorganisms from different genotypes of Arabidopsis thaliana. Community Ecol. 2009, 10, 111–119. [Google Scholar] [CrossRef]
- Mathema, V.B.; Thakuri, B.K.C.; Sillanpää, M.; Shrestha, R.A. Study of mercury (II) tolerant bacterial isolates from Baghmati River with estimation of plasmid size and growth variation for the high mercury (II) resistant Enterobacter spp. J. Biotech. Res. 2011, 3, 72–77. [Google Scholar]
- Suneja, P.; Dudeja, S.S.; Dahiya, P. Deciphering the phylogenetic relationships among rhizobia nodulating chickpea: A Review. J. Appl. Biol. 2016, 4, 61–70. [Google Scholar]
- Weisberg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Nicklenn, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Ehmann, A. The van URK-Salkowski reagent—A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. A 1977, 132, 267–276. [Google Scholar] [CrossRef]
- Glick, B.R. The enhancement of plant growth by freeliving bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils. 1991, 12, 39–45. [Google Scholar] [CrossRef]
- De Freitas, J.; Banerjee, M.; Germida, J. Phosphate-solubilizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.). Biol. Fertil. Soils 1997, 24, 358–364. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Zak, J.C.; Willig, M.R.; Moorhead, D.L.; Wildman, H.G. Functional diversity of microbial communities: A quantitative approach. Soil Biol. Biochem. 1994, 26, 1101–1108. [Google Scholar] [CrossRef]
- Hill, T.C.J.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L. Plant growth-promoting bacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. 2012, 35, 1044–1051. [Google Scholar]
- Manoj, S.R.; Karthik, C.; Kadirvelu, K. Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: A review. J. Environ. Manag. 2020, 254, 109779. [Google Scholar] [CrossRef]
- Chari, K.D.; Reddy, R.S.; Triveni, S.; Trimurtulu, N.; Rani, C.V.D.; Sreedhar, M. Isolation and Characterization of Abiotic Stress Tolerant Plant Growth Promoting Bacillus Spp. from Different Rhizospheric Soils of Telangana. Biosci. Biotech. Res. Asia 2018, 15, 485–494. [Google Scholar] [CrossRef]
- Hindersah, R.; Nurhabibah, G.; Asmiran, P.; Pratiwi, E. Antibiotic Resistance of Azotobacter Isolated from Mercury-Contaminated Area. Anim. Sci. J. 2019, 7, 70–81. [Google Scholar] [CrossRef]
- Kang, S.-M.; Asaf, S.; Khan, A.L.; Lubna; Khan, A.; Mun, B.-G.; Khan, M.A.; Gul, H.; Lee, I.-J. Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions. Microorganisms 2020, 8, 382. [Google Scholar] [CrossRef] [Green Version]
- Naguib, M.M.; Khairalla, A.S.; El-Gendy, A.O.; Elkhatib, W.F. Isolation and characterization of mercury-resistant bacteria from wastewater sources in Egypt. Can. J. Microbiol. 2019, 65, 308–321. [Google Scholar] [CrossRef]
- Kour, D. Rhizospheric Microbiomes: Biodiversity, Mechanisms of Plant Growth Promotion, and Biotechno-logical Applications for Sustainable Agriculture. In Plant Growth Promoting Rhizobac-teria for Agricultural Sustainability; Kumar, A., Meena, V., Eds.; Springer: Singapore, 2019; Chapter 2; pp. 19–56. [Google Scholar] [CrossRef]
- Abbas, S.; Yee, C.; Hossain, K.; Ahmad, A.; Rafatullah, M. Isolation and characterization of mercury-resistant bacteria from industrial wastewater. Desalin. Water Treat 2019, 138, 128–133. [Google Scholar] [CrossRef]
- Mariano, C.; Mello, I.S.; Barros, B.M.; Da Silva, G.F.; Terezo, A.J.; Soares, M.A. Mercury alters the rhizobacterial community in Brazilian wetlands and it can be bioremediated by the plant-bacteria association. Environ. Sci. Pollut. Res. 2020, 27, 13550–13564. [Google Scholar] [CrossRef] [PubMed]
- Nonnoi, F.; Chinnaswamy, A.; de la Torre, V.S.G.; de la Peña, T.C.; Lucas, M.M.; Pueyo, J.J. Metal tolerance of rhizobial strains isolated from nodules of herbaceous legumes (Medicago spp. and Trifolium spp.) growing in mercury-contaminated soils. Appl. Soil Ecol. 2012, 61, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Bekuzarova, S.A.; Bekmurzov, A.D.; Datieva, I.A.; Lushchenko, G.V.; Salbieva, M.G. Clover nodule bacteria as bioindicators of soils contaminated with heavy metals. IOP Conf. Ser. Earth Environ. Sci. 2020, 421, 062043. [Google Scholar] [CrossRef]
- Boyd, E.S.; Ebarkay, T. The Mercury Resistance Operon: From an Origin in a Geothermal Environment to an Efficient Detoxification Machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhutani, N.; Maheshwari, R.; Negi, M.; Suneja, P. Optimization of IAA production by endophytic Bacillus spp. from Vigna radiata for their potential use as plant growth promoters. Isr. J. Plant Sci. 2018, 65, 83–96. [Google Scholar] [CrossRef]
- Shokri, D.; Emtiazi, G. Indole-3-Acetic Acid (IAA) Production in Symbiotic and Non-Symbiotic Nitrogen-Fixing Bacteria and its Optimization by Taguchi Design. Curr. Microbiol. 2010, 61, 217–225. [Google Scholar] [CrossRef]
- Lurthy, T.; Cantat, C.; Jeudy, C.; Declerck, P.; Gallardo, K.; Barraud, C.; Leroy, F.; Ourry, A.; Lemanceau, P.; Salon, C.; et al. Impact of Bacterial Siderophores on Iron Status and Ionome in Pea. Front. Plant Sci. 2020, 11, 12. [Google Scholar] [CrossRef]
- Jin, C.W.; Li, G.X.; Yu, X.H.; Zheng, S.J. Plant Fe status affects the composition of siderophore–secreting microbes in the rhizosphere. Ann. Bot. 2010, 105, 835–841. [Google Scholar] [CrossRef]
- Baldi, F.; Gallo, M.; Battistel, D.; Barbaro, E.; Gambaro, A.; Daniele, S. A broad mercury resistant strain of Pseudomonas putida secretes pyoverdine under limited iron conditions and high mercury concentrations. BioMetals 2016, 29, 1097–1106. [Google Scholar] [CrossRef]
- Lewis, R.W.; Islam, A.; Opdahl, L.; Davenport, J.R.; Sullivan, T.S. Comparative Genomics, Siderophore Production, and Iron Scavenging Potential of Root Zone Soil Bacteria Isolated from ‘Concord’ Grape Vineyards. Microb. Ecol. 2019, 78, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Jiménez, E.; Gamarra, R.; Carpena-Ruiz, R.O.; Millan, R.; Peñalosa, J.M.; Esteban, E. Mercury bioaccumulation by phytotoxicity in two wild palnt species in Almadén area. Chemosphere 2006, 63, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Penrose, D.M.; Li, J. A model for the lowering of plant ethylene concentrations by plant-growthpromoting bacteria. J. Theor. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef]
- Glick, B.R.; Todorovic, B.; Czarny, J.; Cheng, Z.; Duan, J.; McConkey, B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Liang, Z.; Soranno, P.A.; Wagner, T. The role of phosphorus and nitrogen on chlorophyll a: Evidence from hundreds of lakes. Water Res. 2020, 185, 116236. [Google Scholar] [CrossRef]
- Rodriguez, H.; Fraga, R.; Gonzalez, T.; Bashan, Y. Genetics of phosphate solubilization and its potential applications for improving plant growth- promoting bacteria. Plant Soil 2006, 287, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Ortíz-Castro, R.; Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; López-Bucio, J. The role of microbial signals in plant growth and development. Plant Signal. Behav. 2009, 4, 701–712. [Google Scholar]
- Emami, T.; Mirzaeiheydari, M.; Maleki, A.; Bazgir, M. Effect of native growth promoting bacteria and commercial biofertilizers on growth and yield of wheat (Triticum aestivum) and barley (Hordeum vulgare) under salinity stress conditions. Cell Mol. Biol. 2019, 65, 22–27. [Google Scholar] [CrossRef]
- Jiménez, G.; Urdiain, M.; Cifuentes, A.; López-López, A.; Blanch, A.R.; Tamames, J.; Kämpfer, P.; Kolstø, A.B.; Ramón, D.; Martínez, J.F.; et al. Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst. Appl. Microbiol. 2013, 36, 383–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerrouk, I.Z.; Rahmoune, B.; Auer, S.; Rößler, S.; Lin, T.; Baluska, F.; Dobrev, P.I.; Motyka, V.; Ludwig-Müller, J. Growth and aluminum tolerance of maize roots mediated by auxin- and cytokinin-producing Bacillus toyonensis requires polar auxin transport. Environ. Exp. Bot. 2020, 176, 104064. [Google Scholar] [CrossRef]
- Baldi, F.; Marchetto, D.; Gallo, M.; Fani, R.; Maida, I.; Covelli, S.; Fajon, V.; Žižek, S.; Hines, M.; Horvat, M. Chlor-alkali plant contamination of Aussa River sediments induced a large Hg-resistant bacterial community. Estuar. Coast. Shelf Sci. 2012, 113, 96–104. [Google Scholar] [CrossRef]
- Khezrinejad, N.; Khodakaramian, G.; Shahryari, F. Characterization of potential plant growth-promoting rhizobacteria isolated from sunflower (Helianthus annuus L.) in Iran. Biol. Futur. 2019, 70, 268–277. [Google Scholar] [CrossRef]
- Mahmood, S.; Daur, I.; Al-Solaimani, S.G.; Ahmad, S.; Madkour, M.H.; Yasir, M.; Hirt, H.; Ali, S.; Ali, Z. Plant Growth Promoting Rhizobacteria and Silicon Synergistically Enhance Salinity Tolerance of Mung Bean. Front. Plant Sci. 2016, 7, 876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arikan, S.; Pirlak, L. Effects of Plant Growth Promoting Rhizobacteria (PGPR) on Physiological Parameters against Salinity in Apple Cultivar “Fuji”. Sak. Univ. J. Sci. 2020, 24, 281–286. [Google Scholar] [CrossRef]
- Mahler, I.; Levinson, H.S.; Wang, Y.; Halvorson, H.O. Cadmium-and mercury-resistant Bacillus strains from a salt marsh and from Boston Harbor. Appl. Environ. Microbiol. 1986, 52, 1293–1298. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Ka, J.O.; Song, H.G. Growth promotion of Xanthium italicum by application of rhizobacterial isolates of Bacillus aryabhattai in microcosm soil. J. Microbiol. 2012, 50, 45–49. [Google Scholar] [CrossRef]
- Park, Y.G.; Mun, B.G.; Yun, B.W. Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS ONE 2017, 12, e0173203. [Google Scholar] [CrossRef] [Green Version]
- Arnold, D.L.; Lovell, H.C.; Jackson, R.W.; Mansfield, J.W. Pseudomonas syringae pv. phaseolicola: From ‘has bean’ to supermodel. Mol. Plant Pathol. 2011, 12, 617–627. [Google Scholar]
- Zachow, C.; Müller, H.; Tilcher, R.; Donat, C.; Berg, G. Catch the Best: Novel Screening Strategy to Select Stress Protecting Agents for Crop Plants. Agronomy 2013, 3, 794–815. [Google Scholar] [CrossRef]
- Scarlett, C.M.; Fletcher, J.T.; Roberts, P.; Lelliott, R.A. Tomato pith necrosis caused by Pseudomonas corrugata n. sp. Ann. Appl. Biol. 1978, 88, 105–114. [Google Scholar] [CrossRef]
- Sang, M.; Kim, K.D. Biocontrol activity and root colonization by Pseudomonas corrugata strains CCR04 and CCR80 against Phytophthora blight of pepper. BioControl 2014, 59, 437. [Google Scholar] [CrossRef]
- Guo, Y.; Zheng, H.; Yang, Y.; Wang, H. Characterization of Pseudomonas corrugata strain P94 isolated from soil in Beijing as a potential biocontrol agent. Curr. Microbiol. 2007, 55, 247–253. [Google Scholar] [CrossRef]
- Gamez, R.; Cardinale, M.; Montes, M.; Ramirez, S.; Schnell, S.; Rodriguez, F. Screening, plant growth promotion and root colonization pattern of two rhizobacteria (Pseudomonas fluorescens Ps006 and Bacillus amyloliquefaciens Bs006) on banana cv. Williams (Musa acuminata Colla). Microbiol. Res. 2019, 220, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Heng, S.; Munis, M.F.H.; Fahad, S.; Yang, X. Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria: A review. Environ. Exp. Bot. 2015, 117, 28–40. [Google Scholar] [CrossRef]
- Sivasakthi, S.; Usharani, G.; Saranraj, P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)—Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]
- MacLean, A.; Bley, A.M.; Appanna, V.P.; Appanna, V.D. Metabolic manipulation by Pseudomonas fluorescens: A powerful stratagem against oxidative and metal stress. J. Med. Microbiol. 2020, 69, 339–346. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, D.; Pan, X.; Lee, D.-J.; Al-Misned, F.A.; Mortuza, M.G.; Gadd, G.M. Aerobic and anaerobic biosynthesis of nano-selenium for remediation of mercury contaminated soil. Chemosphere 2017, 170, 266–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
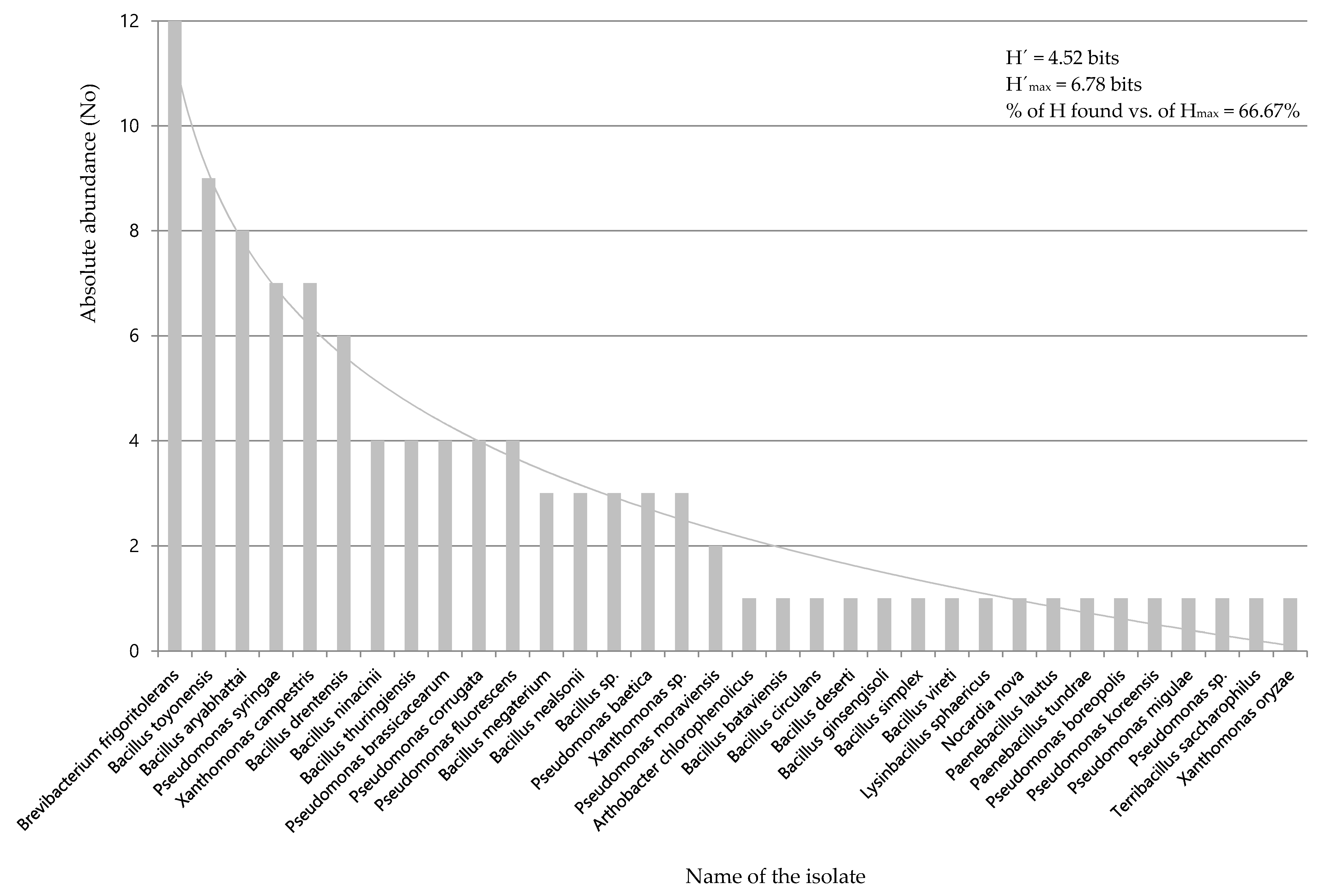
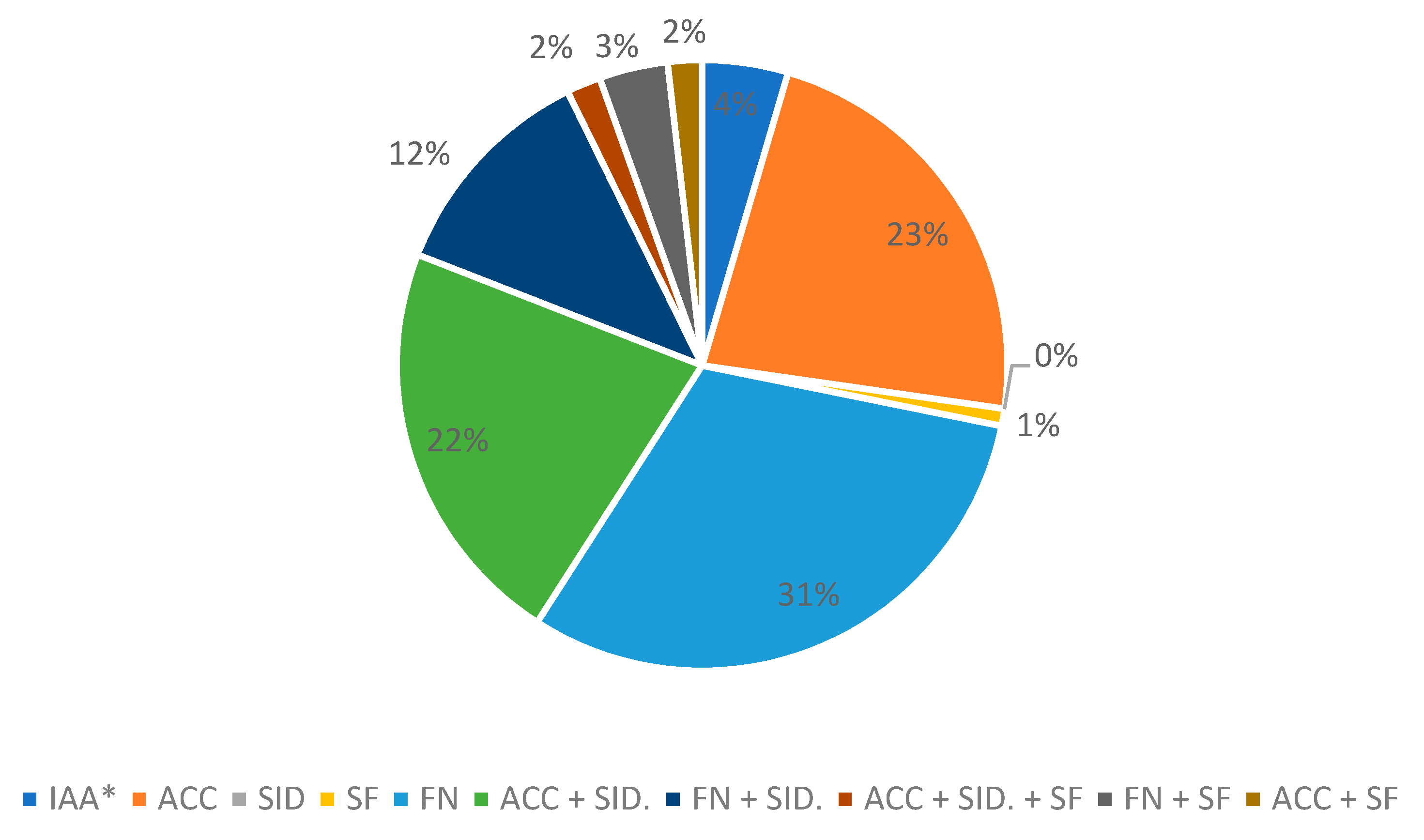
| Strain | 69-II | 80 | 74 | 130 | 146 | 25 | 18 | 69-I | 211 | 212 | 11 | 43 | 95 | 20 | 79 |
| BMRSI | 8.51 | 8.42 | 8.07 | 8.01 | 7.99 | 7.89 | 7.87 | 7.85 | 7.74 | 7.73 | 7.69 | 7.68 | 7.57 | 7.55 | 7.55 |
| Strain | 10 | 31 | 57 | 55 | 21 | 50 | 175 | 37 | 98 | 76 | 23 | 204 | 1 | 48 | 173 |
| BMRSI | 7.42 | 7.4 | 7.26 | 7.23 | 7.21 | 7.08 | 7.08 | 7.07 | 7.05 | 7.04 | 6.97 | 6.8 | 6.68 | 6.62 | 6.6 |
| Strain | 122 | 9 | 58 | 56 | 159 | 70 | 214 | 114 | 160 | 75 | 149 | 186 | 35 | 168 | 166 |
| BMRSI | 6.59 | 6.56 | 6.46 | 6.43 | 6.38 | 6.35 | 6.34 | 6.32 | 6.32 | 6.3 | 6.26 | 6.23 | 6.21 | 6.09 | 6.03 |
| Strain | 178 | 167 | 217 | 104 | 26 | 133 | 213 | 19 | 22 | 118 | 121 | 151 | 155 | 112 | 161 |
| BMRSI | 6.00 | 5.93 | 5.93 | 5.86 | 5.84 | 5.83 | 5.82 | 5.81 | 5.75 | 5.71 | 5.69 | 5.63 | 5.61 | 5.61 | 5.6 |
| Strain | 47 | 14 | 16 | 154 | 200 | 88 | 223 | 203 | 174 | 190 | 199 | 206 | 195 | 126 | 68 |
| BMRSI | 5.58 | 5.51 | 5.47 | 5.46 | 5.46 | 5.41 | 5.35 | 5.34 | 5.33 | 5.33 | 5.32 | 5.31 | 5.3 | 5.29 | 5.25 |
| Strain | 224 | 30 | 189 | 128 | 162 | 137 | 117 | 216 | 5 | 197 | 191 | 196 | 109 | 180 | 192 |
| BMRSI | 5.23 | 5.23 | 5.2 | 5.2 | 5.2 | 5.17 | 5.16 | 5.15 | 5.11 | 5.05 | 5.00 | 4.94 | 4.91 | 4.9 | 4.86 |
| Strain | 201 | 124 | 134 | 45 | 106 | 135 | 96 | 108 | 142 | 145 | 82 | 153 | 91 | 143 | 210 |
| BMRSI | 4.82 | 4.79 | 4.79 | 4.77 | 4.76 | 4.75 | 4.73 | 4.71 | 4.69 | 4.55 | 4.53 | 4.52 | 4.47 | 4.44 | 4.39 |
| Strain | 125 | 132 | 139 | 188 | 4 | ||||||||||
| BMRSI | 4.34 | 4.34 | 4.32 | 4.3 | 4.26 |
| No. | RF/SL | MBC (µg/mL) | BMRSI | IAA (µg/mL) | ACCd (p/a) | SID. (cm) | SOL. PO43− | IDENTIFICATION |
|---|---|---|---|---|---|---|---|---|
| 1 | SL | 50 | 6.68 | 4.63 | - | 1 | - | Bacillus toyonensis |
| 9 | SL | 75 | 6.56 | 5.59 | + | - | - | Bacillus toyonensis |
| 10 | SL | 200 | 7.42 | 6.12 | - | 1.1 | - | ND |
| 11 | SL | 87.5 | 7.69 | 5.61 | - | 1 | - | Bacillus toyonensis |
| 18 | SL | 100 | 7.87 | 6.28 | + | 0.5 | - | Bacillus toyonensis |
| 20 | SL | 100 | 7.55 | 5.96 | + | 0.5 | - | Bacillus toyonensis |
| 21 | SL | 100 | 7.21 | 5.31 | + | 0.8 | - | Bacillus toyonensis |
| 22 | SL | 87.5 | 5.75 | 4.57 | + | 0.1 | - | Bacillus toyonensis |
| 23 | SL | 175 | 6.97 | 4.89 | + | 0.9 | - | Pseudomonas moraviensis |
| 25 | SL | 150 | 7.89 | 5.85 | + | 0.9 | - | Bacillus toyonensis |
| 31 | A | 100 | 7.4 | 5.6 | + | 0.7 | - | Pseudomonas brassicacearum subsp. brassicacearum |
| 37 | A | 87.5 | 7.07 | 5.58 | - | 0.5 | - | Bacillus aryabhattai |
| 43 | A | 87.5 | 7.68 | 5.7 | + | 0.9 | - | Bacillus toyonensis |
| 48 | A | 100 | 6.62 | 4.92 | + | 0.6 | - | ND |
| 50 | A | 100 | 7.08 | 5.29 | + | 0.7 | - | Bacillus toyonensis |
| 55 | A | 87.5 | 7.23 | 5.56 | - | 0.8 | - | Pseudomonas brassicacearum sbups. neoaurantiaca |
| 56 | B | 200 | 6.43 | 4.43 | + | 0.8 | - | Pseudomonas brassicacearum subsp. brassicacearum |
| 57 | B | 175 | 7.26 | 6.38 | + | 0.6 | - | Pseudomonas syringae pv. phaseolicola |
| 58 | B | 100 | 6.46 | 5.56 | + | 0.7 | - | Pseudomonas brassicacearum subsp. brassicacearum |
| 69-I | B | 75 | 7.85 | 6.08 | - | 0.7 | - | Pseudomonas corrugata |
| 69-II | B | 350 | 8.51 | 5.71 | + | 0.7 | + | Pseudomonas corrugata |
| 74 | B | 100 | 8.07 | 6.27 | + | 0.7 | - | Pseudomonas syringae pv. phaseolicola |
| 76 | B | 350 | 7.04 | 4.99 | + | 0.7 | - | Pseudomonas syringae pv. phaseolicola |
| 79 | B | 87.5 | 7.55 | 5.27 | + | 0.4 | - | Pseudomonas syringae pv. phaseolicola |
| 80 | B | 80 | 8.42 | 6.47 | + | 0.8 | - | Pseudomonas syringae pv. phaseolicola |
| 95 | C | 80 | 7.57 | 4.69 | - | 2.8 | - | Brevibacterium frigoritolerans |
| 98 | C | 160 | 7.05 | 5.29 | + | 0.6 | - | Pseudomonas baetica |
| 112 | C | 150 | 5.61 | 4.36 | + | 0.1 | - | Pseudomonas corrugata |
| 122 | D | 87.5 | 6.59 | 4.51 | + | - | + | Brevibacterium frigoritolerans |
| 130 | D | 160 | 8.01 | 5.85 | + | 1 | - | Pseudomonas corrugata |
| 146 | E | 80 | 7.99 | 6.09 | + | 0.8 | - | Pseudomonas fluorescens |
| 151 | E | 87.5 | 5.63 | 4.38 | + | 0.2 | - | Bacillus aryabhattai |
| 168 | A | 87.5 | 6.09 | 4.00 | + | - | + | Bacillus aryabhattai |
| 173 | A | 175 | 6.6 | 5.53 | + | - | - | Bacillus toyonensis |
| 175 | A | 80 | 7.08 | 6.00 | + | - | - | ND |
| 204 | D | 80 | 6.8 | 5.72 | - | - | + | ND |
| 211 | D | 80 | 7.74 | 6.16 | + | 0.5 | - | Bacillus drentensis |
| 212 | D | 80 | 7.73 | 6.16 | + | 0.4 | - | Bacillus drentensis |
| 217 | E | 100 | 5.93 | 4.88 | + | 2 | + | Bacillus nealsonii |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robas, M.; Jiménez, P.A.; González, D.; Probanza, A. Bio-Mercury Remediation Suitability Index: A Novel Proposal That Compiles the PGPR Features of Bacterial Strains and Its Potential Use in Phytoremediation. Int. J. Environ. Res. Public Health 2021, 18, 4213. https://doi.org/10.3390/ijerph18084213
Robas M, Jiménez PA, González D, Probanza A. Bio-Mercury Remediation Suitability Index: A Novel Proposal That Compiles the PGPR Features of Bacterial Strains and Its Potential Use in Phytoremediation. International Journal of Environmental Research and Public Health. 2021; 18(8):4213. https://doi.org/10.3390/ijerph18084213
Chicago/Turabian StyleRobas, Marina, Pedro A. Jiménez, Daniel González, and Agustín Probanza. 2021. "Bio-Mercury Remediation Suitability Index: A Novel Proposal That Compiles the PGPR Features of Bacterial Strains and Its Potential Use in Phytoremediation" International Journal of Environmental Research and Public Health 18, no. 8: 4213. https://doi.org/10.3390/ijerph18084213
APA StyleRobas, M., Jiménez, P. A., González, D., & Probanza, A. (2021). Bio-Mercury Remediation Suitability Index: A Novel Proposal That Compiles the PGPR Features of Bacterial Strains and Its Potential Use in Phytoremediation. International Journal of Environmental Research and Public Health, 18(8), 4213. https://doi.org/10.3390/ijerph18084213









