Anaerobic Ammonium Oxidation Bacteria in a Freshwater Recirculating Pond Aquaculture System
Abstract
1. Introduction
2. Materials and Methods
2.1. RPAS Description
2.2. Sample Collection and Physicochemical Analyses
2.3. DNA Extraction and Nested PCR
2.4. Quantitative PCR Assay
2.5. Cloning, Sequencing and Phylogenetic Analysis
2.6. Nucleotide Sequence Accession Numbers
2.7. Statistical Analysis
3. Results
3.1. Physicochemical Characteristics of the RPAS
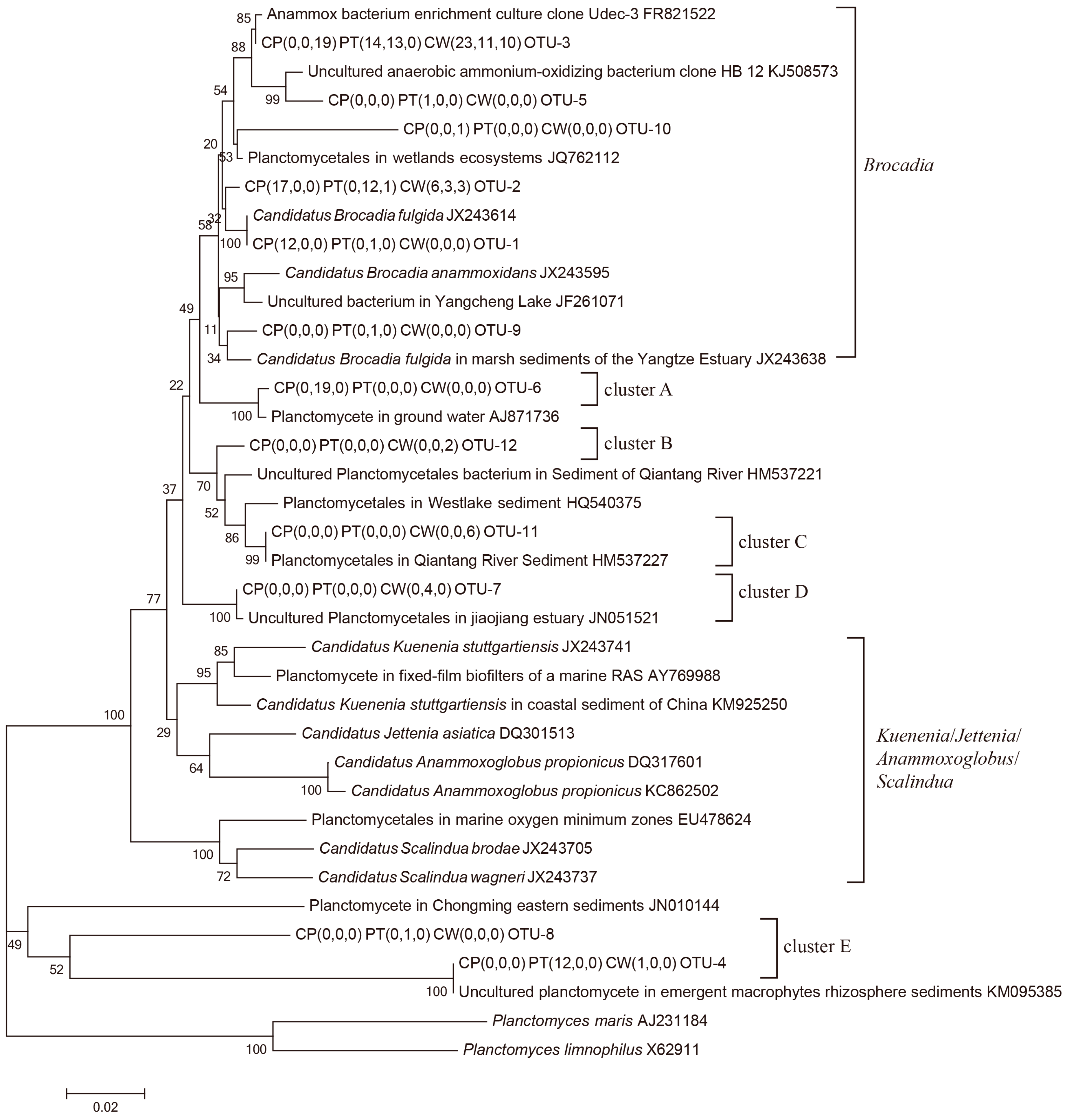
3.2. Diversity and Distribution of Anammox Bacteria in RPAS
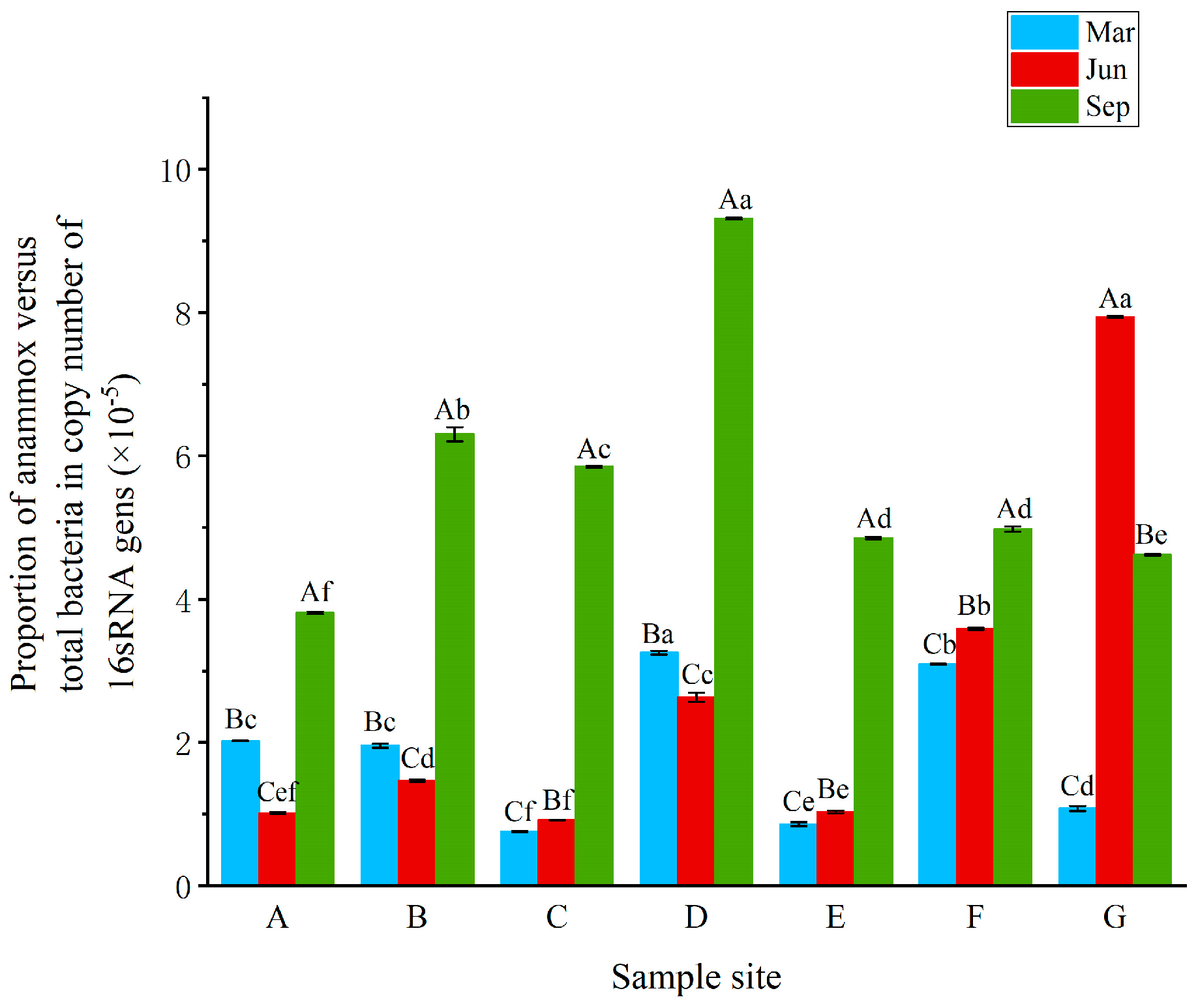
3.3. Abundance of Anammox Bacteria in RPAS
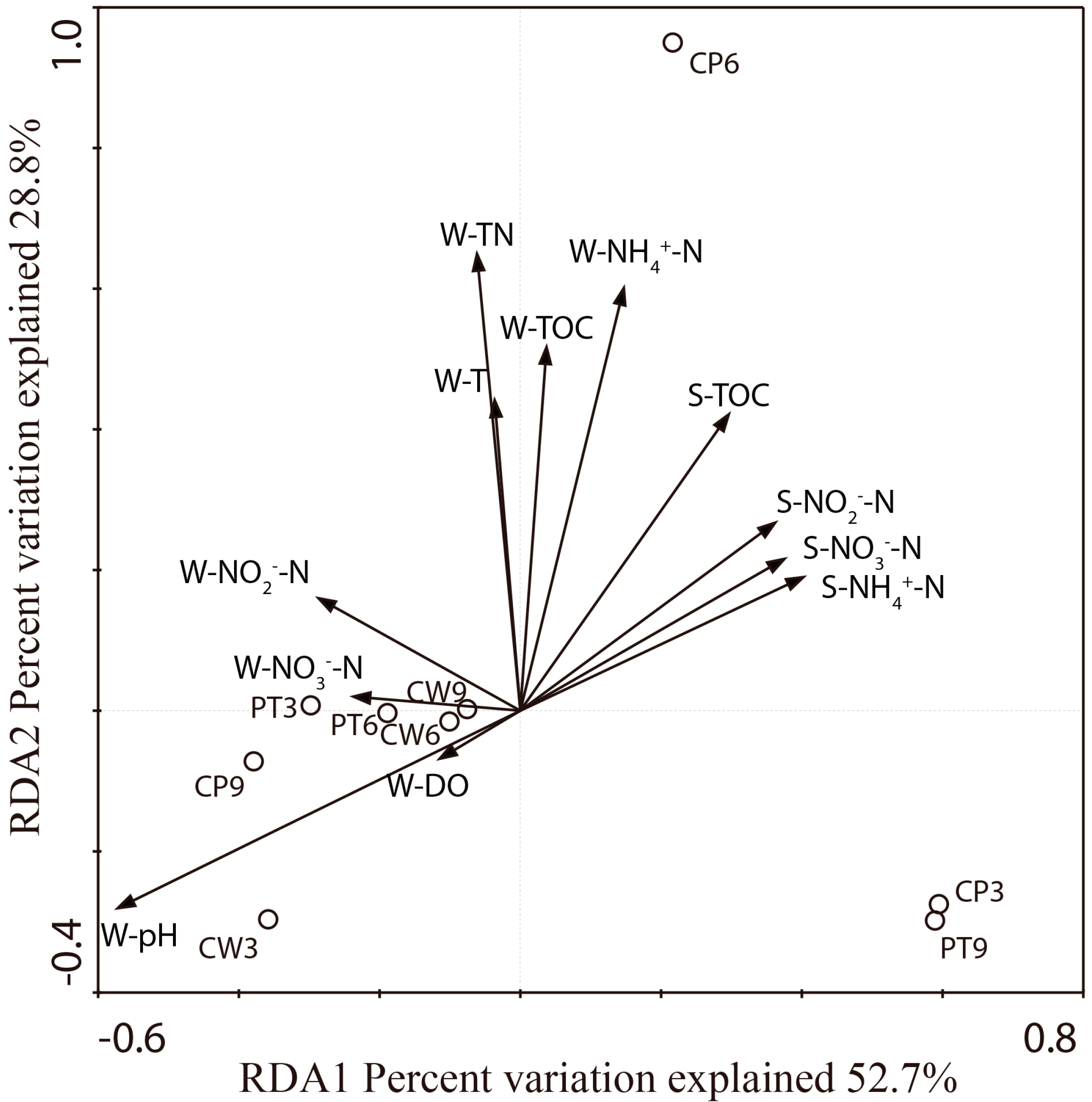
3.4. Correlations of Anammox Bacteria with Environmental Factors
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Mulder, A.; van de Graaf, A.A.; Robertson, L.A.; Kuenen, J.G. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 1995, 16, 177–183. [Google Scholar] [CrossRef]
- Trimmer, M.; Nicholls, J.C.; Deflandre, B. Anaerobic ammonium oxidation measured in sediments along the Thames estuary, United Kingdom. Appl. Environ. Microbiol. 2003, 69, 6447–6454. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, G.; Peng, Y.; Jetten, M.S.; Yin, C. Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River estuary. Environ. Sci. Technol. 2012, 46, 8834–8842. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Wang, Y.; Zhang, G.; Gu, J. Distribution patterns of ammonia-oxidizing bacteria and anammox bacteria in the freshwater marsh of Honghe wetland in Northeast China. Ecotoxicology 2014, 23, 1930–1942. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, Y.; Ma, X.; Gu, J. Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl. Microbiol. Biot. 2013, 97, 7919–7934. [Google Scholar] [CrossRef]
- Sun, W.; Xia, C.; Xu, M.; Guo, J.; Wang, A.; Sun, G. Diversity and distribution of planktonic anaerobic ammonium-oxidizing bacteria in the Dongjiang River, China. Microbiol. Res. 2014, 169, 897–906. [Google Scholar] [CrossRef]
- Humbert, S.; Tarnawski, S.; Fromin, N.; Mallet, M.P.; Aragno, M.; Zopfi, J. Molecular detection of anammox bacteria in terrestrial ecosystems: Distribution and diversity. ISME J. 2010, 4, 450–454. [Google Scholar] [CrossRef]
- Sato, Y.; Ohta, H.; Yamagishi, T.; Guo, Y.; Nishizawa, T.; Rahman, M.H.; Kuroda, H.; Kato, T.; Saito, M.; Yoshinaga, I.; et al. Detection of anammox activity and 16S rRNA genes in ravine paddy field soil. Microbes Environ. 2012, 27, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.D.; Liu, S.; Lou, L.P.; Liu, W.P.; Xu, X.Y.; Zheng, P.; Hu, B.L. Broad distribution of diverse anaerobic Ammonium-Oxidizing bacteria in Chinese agricultural soils. Appl. Environ. Microbiol. 2013, 79, 6167–6172. [Google Scholar] [CrossRef]
- Tal, Y.; Watts, J.; Schreier, H.J. Anaerobic ammonium-oxidizing (anammox) bacteria and associated activity in fixed-film biofilters of a marine recirculating aquaculture system. Appl. Environ. Microbiol. 2006, 72, 2896–2904. [Google Scholar] [CrossRef]
- Van Kessel, M.A.H.J.; Harhangi, H.R.; Flik, G.; Jetten, M.S.M.; Klaren, P.H.M.; Op Den Camp, H.J.M. Anammox bacteria in different compartments of recirculating aquaculture systems. Biochem. Soc. Trans. 2011, 39, 1817–1821. [Google Scholar] [CrossRef]
- Amano, T.; Yoshinaga, I.; Yamagishi, T.; Thuoc, C.V.; Thu, P.T.; Ueda, S.; Kato, K.; Sako, Y.; Suwa, Y. Contribution of anammox bacteria to benthic nitrogen cycling in a mangrove forest and shrimp ponds, haiphong, vietnam. Microbes Environ. 2011, 26, 1–6. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization of the United Nations, Fisheries and Aquaculture Department). The State of World Fisheries and Aquaculture 2018: Meeting the Sustainable Development Goals; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018. [Google Scholar]
- Liu, X.G.; Che, X.; Wang, X.D.; Zhu, H.; Cheng, G.F.; Gu, Z.J.; Liu, C. The technology and method for green and clean pond aquaculture. Chin. Fish. Qual. Stand. 2018, 8, 1–8. [Google Scholar]
- Liu, X. Study on Pond Aquaculture Pollution and Ecological Engineering Regulation Techniques; Nanjing Agricultural University: Nanjing, China, 2011. [Google Scholar]
- Bao, S.D. Chemical Analysis for Agricultural Soil; China Agriculture Press: Beijing, China, 2000. (In Chinese) [Google Scholar]
- Zhu, G.; Wang, S.; Wang, Y.; Wang, C.; Risgaard-Petersen, N.; Jetten, M.S.M.; Yin, C. Anaerobic mmonia oxidation in a fertilized paddy soil. ISME J. 2011, 5, 1905–1912. [Google Scholar] [CrossRef] [PubMed]
- Juretschko, S.; Timmermann, G.; Schmid, M.; Schleifer, K.H.; Pommerening-Roser, A.; Koops, H.P. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 1998, 64, 3042–3051. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Walsh, K.; Webb, R.; Rijpstra, W.; van de Pas-Schoonen, K.; Verbruggen, M.J.; Hill, T.; Moffett, B.; Fuerst, J.; Schouten, S.; et al. Candidatus “Scalindua brodae”, sp nov., Candidatus “Scalindua wagneri”, sp nov., Two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 2003, 26, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Hamersley, M.R.; Lavik, G.; Woebken, D.; Rattray, J.E.; Lam, P.; Hopmans, E.C. Anaerobic ammonium oxidation in the Peruvian oxygen minimum zone. Limnol. Oceanogr. 2007, 52, 923–933. [Google Scholar] [CrossRef]
- Dionisi, H.M.; Harms, G.; Layton, A.C.; Gregory, I.R.; Parker, J.; Hawkins, S.A.; Robinson, K.G.; Sayler, G.S. Power analysis for real-time PCR quantification of genes in activated sludge and analysis of the variability introduced by DNA extraction. Appl. Environ. Microbiol. 2003, 69, 6597–6604. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Ter Braak, C.; Smilauer, P. CANOCO Reference Manual and Canodraw for Windows User’s Guide: Software for Canonical Community Ordination (Version 4.5); Microcomputer Power: Ithaca, NY, USA, 2002. [Google Scholar]
- Ministry of Agriculture of the People’s Republic of China. Requirements for Water Discharge from Freshwater Aquaculture Pond; Fisheries industry standard of the people’s Republic of China, SC/T 9101-2007; China Agriculture Press: Beijing, China, 2007. [Google Scholar]
- Hirsch, M.D.; Long, Z.T.; Song, B. Anammox bacterial diversity in various aquatic ecosystems based on the detection of hydrazine oxidase genes (hzoA/hzoB). Microb. Ecol. 2011, 61, 264–276. [Google Scholar] [CrossRef]
- Humbert, S.; Zopfi, J.; Tarnawski, S. Abundance of anammox bacteria in different wetland soils. Environ. Microbiol. Rep. 2012, 4, 484–490. [Google Scholar] [CrossRef]
- Wenk, C.B.; Blees, J.; Zopfi, J.; Veronesi, M.; Bourbonnais, A.; Schubert, C.J.; Niemann, H.; Lehmann, M.F. Anaerobic ammonium oxidation (anammox) bacteria and sulfide-dependent denitrifiers coexist in the water column of a meromictic south-alpine lake. Limnol. Oceanogr. 2013, 58, 1–12. [Google Scholar] [CrossRef]
- Gori, F.; Tringe, S.G.; Kartal, B.; Marchiori, E.; Jetten, M.S.M. The metagenomic basis of anammox metabolism in Candidatus ‘Brocadia fulgida’ (vol 39, pg 1799, 2011). Biochem. Soc. Trans. 2012, 40, 295. [Google Scholar] [CrossRef][Green Version]
- Bryant, J.A.; Stewart, F.J.; Eppley, J.M.; DeLong, E.F. Microbial community phylogenetic and trait diversity declines with depth in a marine oxygen minimum zone. Ecology 2012, 93, 1659–1673. [Google Scholar] [CrossRef] [PubMed]
- Sonthiphand, P.; Neufeld, J.D. Evaluating primers for profiling anaerobic ammonia oxidizing bacteria within freshwater environments. PLoS ONE 2013, 8, e57242. [Google Scholar] [CrossRef]

| Time | Units | Overlying Water/Interstitial Water a | Sediments | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T (℃) | DO (mg/L) | pH | TOC (mg/L) | TN (mg/L) | NH4+-N (mg/L) | NO2−-N (mg/L) | NO3−-N (mg/L) | TOC (g/kg) | NH4+-N (mg/kg) | NO2−-N (mg/kg) | NO3−-N (mg/kg) | ||
| March | CP | 11.42 ± 0.02 | 2.47 ± 0.43 | 7.02 ± 0.08 | nd | nd | nd | nd | nd | 2.87 ± 0.31 | 117.47 ± 23.50 | 0.19 ± 0.01 | 4.21 ± 0.42 |
| PT | 13.43 ± 0.35 | 8.94 ± 0.68 | 7.77 ± 0.16 | nd | nd | nd | nd | nd | 3.53 ± 0.72 | 120.75 ± 31.20 | 0.13 ± 0.01 | 3.93 ± 0.99 | |
| CW | 11.15 ± 0.21 | 1.95 ± 0.78 | 7.52 ± 0.11 | nd | nd | nd | nd | nd | – | – | – | – | |
| June | CP | 24.87 ± 0.23 | 2.54 ± 0.61 | 7.20 ± 0.17 | 11.17 ± 0.29 | 16.44 ± 1.93 | 0.79 ± 0.02 | 0.03 ± 0.00 | 0.11 ± 0.03 | 3.75 ± 0.37 | 136.77 ± 17.19 | 0.18 ± 0.02 | 3.63 ± 0.42 |
| PT | 27.10 ± 0.21 | 6.06 ± 1.35 | 7.60 ± 0.05 | 10.60 ± 0.30 | 14.89 ± 2.19 | 0.50 ± 0.02 | 0.02 ± 0.00 | 0.04 ± 0.01 | 2.93 ± 0.19 | 143.54 ± 22.70 | 0.19 ± 0.04 | 2.30 ± 0.28 | |
| CW | 24.44 ± 0.69 | 1.74 ± 0.83 | 7.65 ± 0.04 | 8.92 ± 0.19 | 13.46 ± 1.39 | 0.33 ± 0.03 | 0.02 ± 0.01 | 0.19 ± 0.10 | – | – | – | – | |
| September | CP | 23.35 ± 0.02 | 2.93 ± 0.04 | 7.54 ± 0.05 | 8.62 ± 0.33 | 9.24 ± 0.69 | 0.58 ± 0.03 | 0.10 ± 0.00 | 0.56 ± 0.02 | 2.16 ± 0.06 | 128.50 ± 40.53 | 0.16 ± 0.01 | 3.57 ± 0.49 |
| PT | 22.58 ± 0.18 | 5.03 ± 1.88 | 7.63 ± 0.11 | 8.16 ± 0.19 | 5.71 ± 1.54 | 0.55 ± 0.02 | 0.02 ± 0.00 | 0.17 ± 0.02 | 2.20 ± 0.08 | 164.26 ± 75.35 | 0.13 ± 0.00 | 2.93 ± 0.24 | |
| CW | 23.20 ± 0.85 | 1.07 ± 0.49 | 7.50 ± 0.08 | 6.97 ± 0.14 | 6.45 ± 0.57 | 0.21 ± 0.05 | 0.02 ± 0.01 | 0.17 ± 0.03 | – | – | – | – | |
| Time | Units | Clones | Unique Sequences | OTUs | Coverage (%) | Shannon | Simpson | SACE | Chaol |
|---|---|---|---|---|---|---|---|---|---|
| March | CP | 29 | 9 | 2 | 100.0 | 0.68 | 0.50 | 0.00 | 2.00 |
| PT | 27 | 14 | 3 | 96.3 | 0.82 | 0.45 | 0.00 | 3.00 | |
| CW | 30 | 23 | 3 | 96.7 | 0.64 | 0.62 | 4.76 | 3.00 | |
| June | CP | 19 | 9 | 1 | 100.0 | 0.00 | 1.00 | 0.00 | 1.00 |
| PT | 17 | 10 | 5 | 76.5 | 0.87 | 0.57 | 0.00 | 11.00 | |
| CW | 18 | 15 | 3 | 100.0 | 0.93 | 0.42 | 3.00 | 3.00 | |
| September | CP | 20 | 13 | 2 | 95.0 | 0.20 | 0.90 | 0.00 | 2.00 |
| PT | 21 | 7 | 1 | 100.0 | 0.00 | 1.00 | 0.00 | 1.00 | |
| CW | 21 | 16 | 4 | 100.0 | 1.21 | 0.30 | 4.00 | 4.00 |
| Total Numbers | Relative Abundance (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| March | June | September | |||||||||
| OTUs | Clones | CP | PT | CW | CP | PT | CW | CP | PT | CW | |
| Brocadia | 6 | 157 | 100 | 100 | 100 | 0 | 100 | 77.78 | 100 | 100 | 61.9 |
| Cluster A | 1 | 19 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 |
| Cluster B | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9.52 |
| Cluster C | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28.57 |
| Cluster D | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 22.22 | 0 | 0 | 0 |
| March | ||||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | ||
| Abundance (copies/ng DNA) | Anammox Bacteria (×101) | 10.02 ± 0.66ac | 9.45 ± 4.13ac | 4.04 ± 1.82a | 18.83 ± 4.18b | 3.89 ± 0.43a | 11.31 ± 2.96c | 4.18 ± 2.22a |
| Total Bacteria (×106) | 4.81 ± 0.10a | 4.81 ± 0.47a | 6.11 ± 0.44b | 5.89 ± 0.75bc | 4.86 ± 0.53b | 3.61 ± 0.45c | 3.69 ± 0.39c | |
| Abundance (copies/g DW) | Anammox Bacteria (×105) | 17.76 ± 1.16a | 15.29 ± 6.68a | 7.57 ± 3.42b | 22.11 ± 4.91a | 4.93 ± 0.54b | – | – |
| Total Bacteria (×1010) | 8.52 ± 0.18a | 7.78 ± 0.76ad | 11.45 ± 0.82c | 6.91 ± 0.88bd | 6.15 ± 0.67b | – | – | |
| June | ||||||||
| A | B | C | D | E | F | G | ||
| Abundance (copies/ng DNA) | Anammox Bacteria (×101) | 8.31 ± 0.92ab | 7.96 ± 1.21ab | 3.33 ± 0.05a | 11.16 ± 2.92ab | 3.66 ± 2.33a | 16.83 ± 4.09b | 41.84 ± 9.46c |
| Total Bacteria (×106) | 7.81 ± 0.12a | 5.41 ± 0.34b | 3.33 ± 0.09c | 4.35 ± 0.15d | 3.58 ± 0.22c | 4.61 ± 0.39d | 5.19 ± 0.57b | |
| Abundance (copies/g DW) | Anammox Bacteria (×105) | 23.88 ± 2.63a | 13.15 ± 2.00b | 5.53 ± 0.08b | 34.94 ± 9.15c | 9.20 ± 5.85b | – | – |
| Total Bacteria (×1010) | 22.44 ± 0.33a | 8.94 ± 0.56b | 5.53 ± 0.15c | 13.61 ± 0.46d | 9.00 ± 0.56b | – | – | |
| September | ||||||||
| A | B | C | D | E | F | G | ||
| Abundance (copies/ng DNA) | Anammox Bacteria (×101) | 4.32 ± 0.61a | 19.54 ± 4.55b | 14.08 ± 4.00bc | 19.53 ± 0.20b | 10.08 ± 2.83ac | 15.82 ± 3.77ab | 13.11 ± 4.65ab |
| Total Bacteria (×106) | 1.13 ± 0.00a | 3.12 ± 0.20b | 2.41 ± 0.29c | 2.07 ± 0.28c | 2.06 ± 0.11c | 3.10 ± 0.26b | 2.82 ± 0.17b | |
| Abundance (copies/g DW) | Anammox Bacteria (×105) | 49.86 ± 7.06a | 83.24 ± 19.38a | 85.86 ± 24.42a | 121.80 ± 1.22b | 48.32 ± 13.58a | – | – |
| Total Bacteria (×1010) | 13.07 ± 0.06a | 13.28 ± 0.86a | 14.70 ± 1.75a | 12.93 ± 1.73a | 9.89 ± 0.51b | – | – | |
| Physicochemical Parameters | OTUs | Shannon | Simpson | SACE | Chaol | Abundance |
|---|---|---|---|---|---|---|
| W-Temperature | 0.124 | −0.136 | 0.274 | −0.178 | 0.325 | 0.314 |
| W-DO | 0.132 | −0.048 | 0.043 | −0.585 | 0.292 | −0.223 |
| W-pH | 0.384 | 0.226 | −0.152 | 0.168 | 0.280 | 0.446 |
| W-TOC | −0.075 | −0.350 | 0.394 | −0.619 | 0.278 | −0.455 |
| W-TN | 0.129 | −0.024 | 0.046 | −0.266 | 0.326 | −0.175 |
| W-NH4+-N | −0.642 | −0.876 * | 0.900 * | −0.872 * | −0.267 | −0.597 |
| W-NO2−-N | −0.267 | −0.377 | 0.387 | −0.357 | −0.264 | −0.152 |
| W-NO3−-N | −0.308 | −0.278 | 0.256 | −0.118 | −0.425 | 0.140 |
| S-TOC | 0.113 | 0.267 | −0.307 | 0.000 | 0.055 | −0.680 |
| S-NH4+-N | −0.155 | −0.513 | 0.628 | 0.000 | 0.090 | 0.396 |
| S-NO2−-N | 0.300 | 0.233 | −0.170 | 0.000 | 0.416 | −0.855 * |
| S-NO3−-N | −0.491 | −0.007 | −0.217 | 0.000 | −0.699 | −0.069 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.-G.; Wang, J.; Wu, Z.-F.; Cheng, G.-F.; Gu, Z.-J. Anaerobic Ammonium Oxidation Bacteria in a Freshwater Recirculating Pond Aquaculture System. Int. J. Environ. Res. Public Health 2021, 18, 4941. https://doi.org/10.3390/ijerph18094941
Liu X-G, Wang J, Wu Z-F, Cheng G-F, Gu Z-J. Anaerobic Ammonium Oxidation Bacteria in a Freshwater Recirculating Pond Aquaculture System. International Journal of Environmental Research and Public Health. 2021; 18(9):4941. https://doi.org/10.3390/ijerph18094941
Chicago/Turabian StyleLiu, Xing-Guo, Jie Wang, Zong-Fan Wu, Guo-Feng Cheng, and Zhao-Jun Gu. 2021. "Anaerobic Ammonium Oxidation Bacteria in a Freshwater Recirculating Pond Aquaculture System" International Journal of Environmental Research and Public Health 18, no. 9: 4941. https://doi.org/10.3390/ijerph18094941
APA StyleLiu, X.-G., Wang, J., Wu, Z.-F., Cheng, G.-F., & Gu, Z.-J. (2021). Anaerobic Ammonium Oxidation Bacteria in a Freshwater Recirculating Pond Aquaculture System. International Journal of Environmental Research and Public Health, 18(9), 4941. https://doi.org/10.3390/ijerph18094941







