Mitigating the Adverse Effects of Polychlorinated Biphenyl Derivatives on Estrogenic Activity via Molecular Modification Techniques
Abstract
1. Introduction
2. Materials and Methods
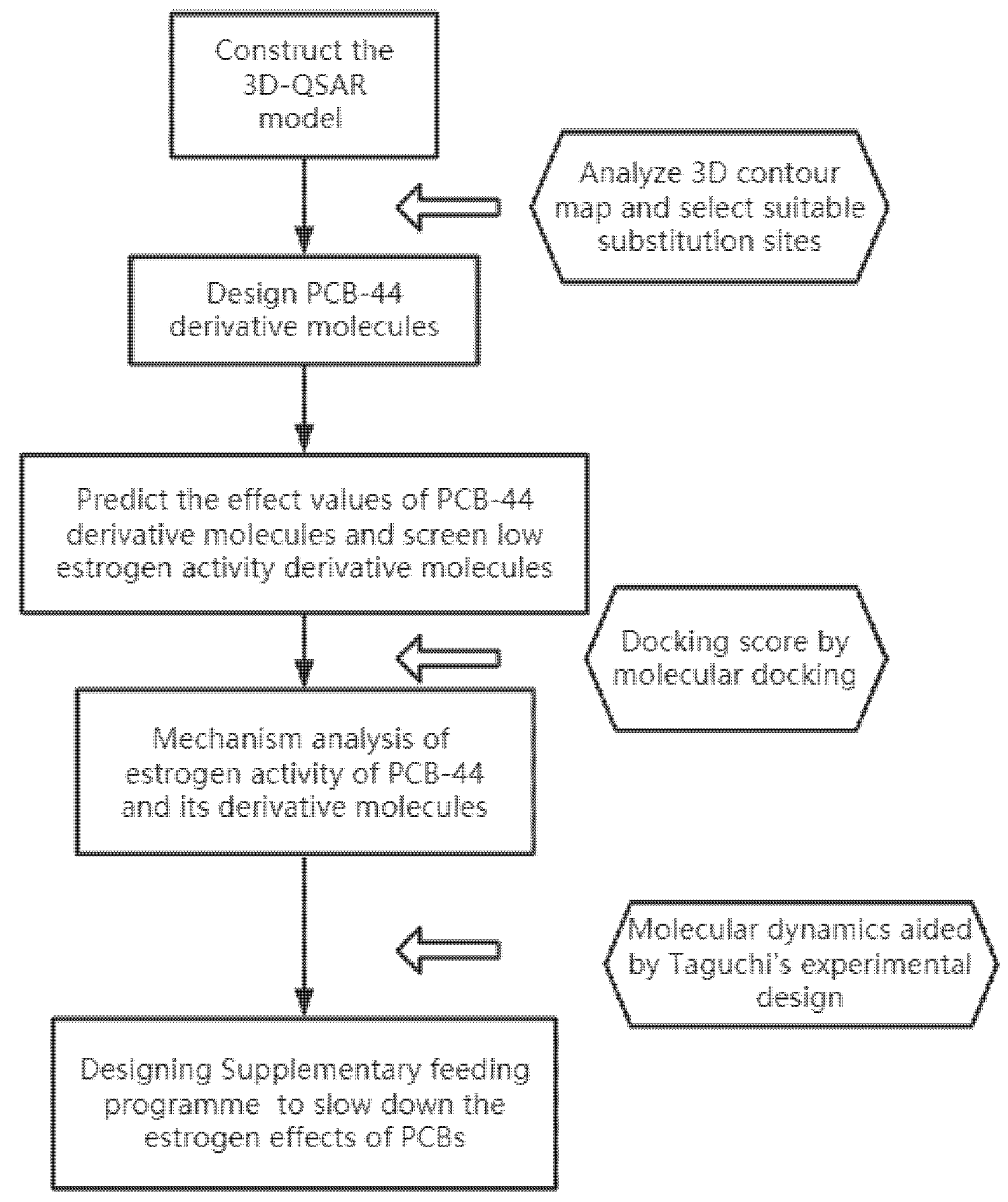
2.1. Construction of a 3D-QSAR Model and Molecular Modification of PCBs with Low Estrogenic Activity
2.2. Mechanism Analysis Method for Determining the Anti-Oestrogenic Activity of PCBs before and after Modification Based on Molecular Docking
3. Results and Discussion
3.1. Molecular Substitution Design of Low Anti-Oestrogenic Activity of PCB Derivatives Based on the CoMSIA Model 3D Contour Map
3.1.1. Construction and Verification of the CoMSIA Model of Anti-Oestrogenic Activity of PCBs
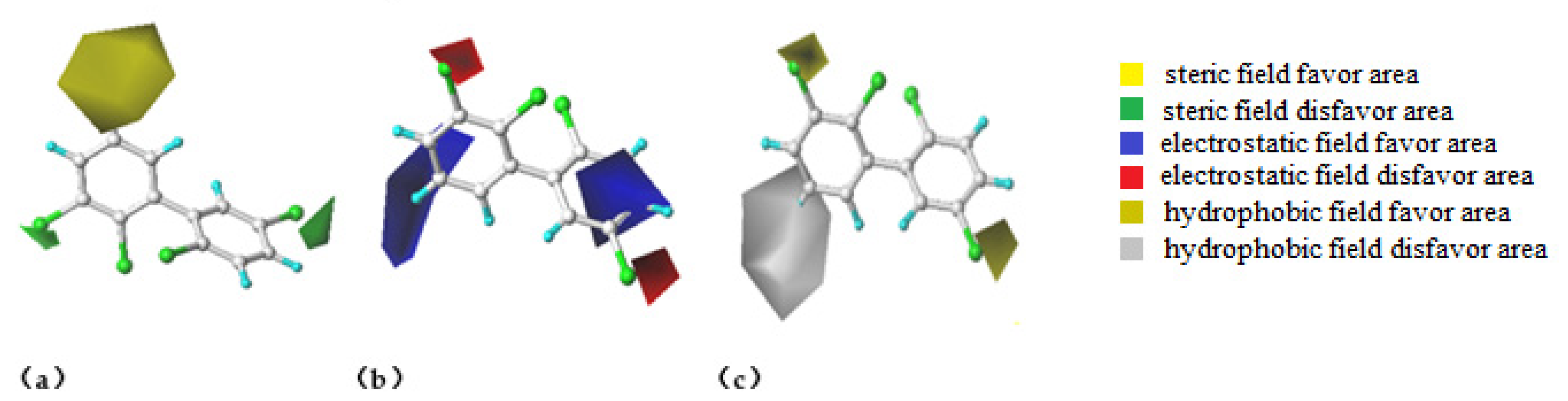
3.1.2. Analysis of Factors Affecting the Anti-Oestrogenic Activity of PCB Molecules Based on the 3D Contour Map of the CoMSIA Model and the Modification Design of PCB Derivatives with a Low Stimulating Hormone
3.1.3. Prediction of the Anti-Oestrogenic Activity of PCB Derivatives and Evaluation of POPs
3.2. Analysis of the Underlying Mechanism That Caused Changes in the Anti-Oestrogenic Activity of PCB Derivatives Based on Molecular Docking Techniques
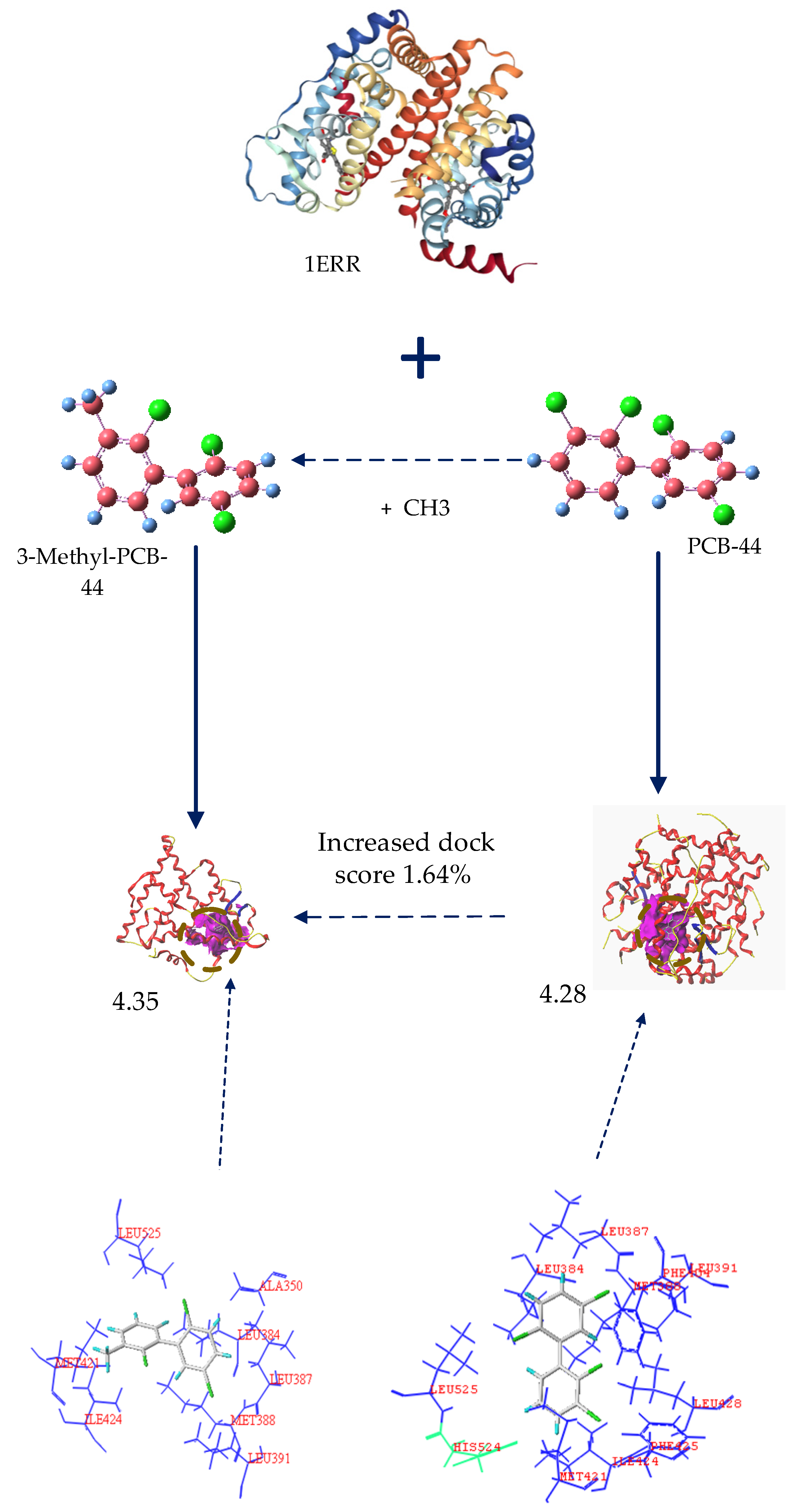
3.2.1. Analysis of the Molecular Docking Effect of PCB-44 Bound to hERα before and after Modification
3.2.2. Analysis of Amino Acid Residues Docked to hERα before and after Molecular Modification of PCBs
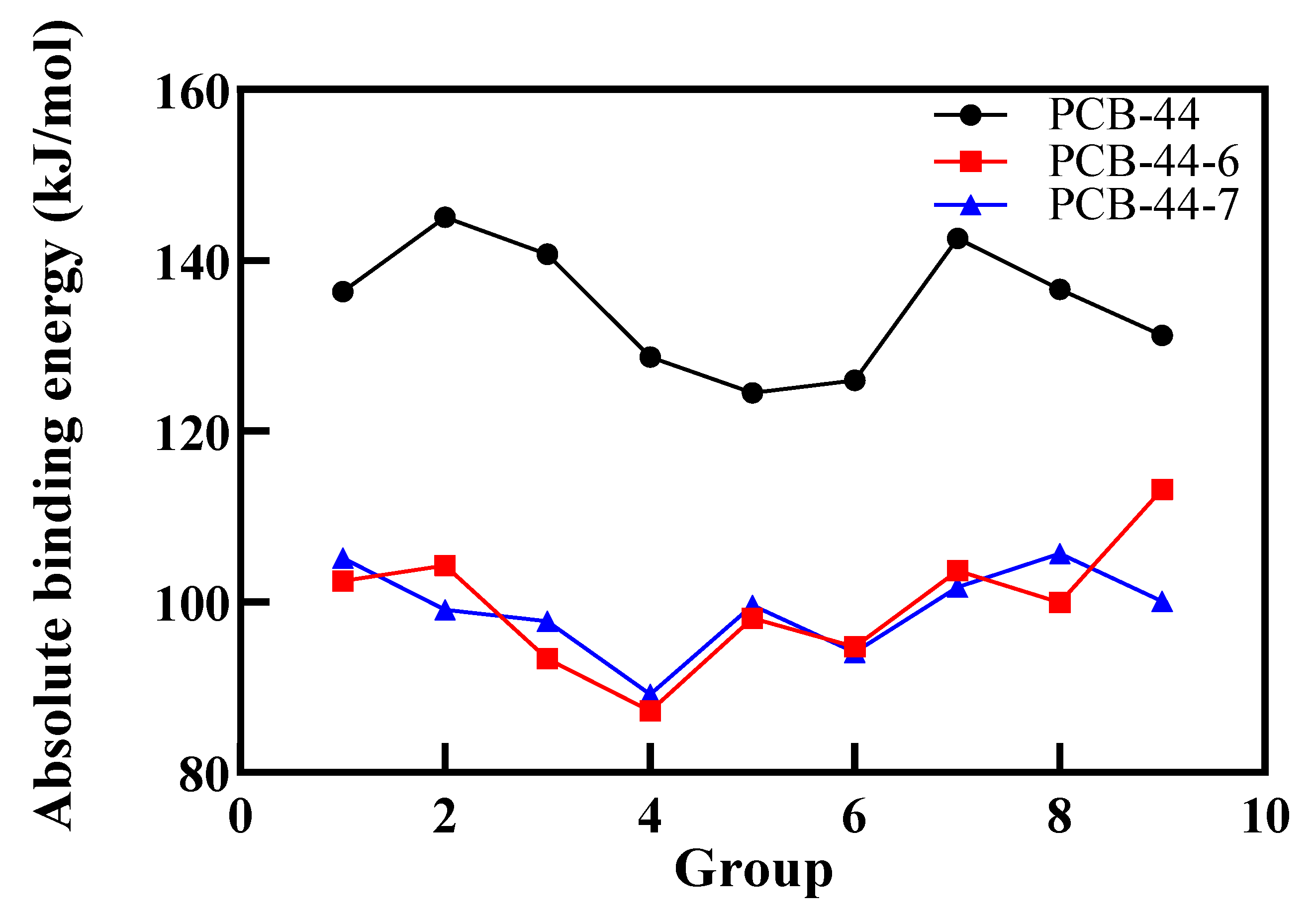
3.3. Validation in Oestrogenic Effect Mitigation of PCBs Based on Molecular Dynamics Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviations in English | Full English Name |
| PCBs | Polychlorinated biphenyls |
| POPs | Persistent organic pollutants |
| E1 | Estrone |
| E2 | 17β-estradiol |
| 3D-QSAR | Three-dimensional quantitative structure-activity relationship |
| hERα | Human oestrogen receptor α |
| REC20 | Estrogen Activity Indicators |
| CoMSIA | Comparative molecular similarity index analysis |
| n | Optimal number of principal components |
| q2 | Cross-validation coefficient |
| r2 | Non-cross-validation coefficient |
| SEE | Standard deviation |
| F | F-test value |
| r2pred | external test coefficient |
| SEP | Standard error of prediction |
| Q2ext | External sample calibration complex correlation coefficient |
| E | Electrostatic field |
| H | Hydrophobic field |
| S | Stereo field |
| D | Hydrogen bond donor field |
| A | Hydrogen bond acceptor field |
| PDB | Protein Data Bank |
| NCBI | National Centre for Biotechnology Information |
| MD | Molecular docking |
| logBCF | Bioconcentration factors |
| logt1/2 | Biological Half-life |
| logKOA | N-octanol/air partitioning coefficients |
References
- Safe, S. Toxicology, structure-function relationship, and human and environmental health impacts of polychlorinated biphenyls: Progress and problems. Environ. Health Perspect. 1993, 100, 259–268. [Google Scholar] [CrossRef]
- Alkhatib, E.; Weigand, C. Parameters affecting partitioning of 6 PCB congeners in natural sediments. Environ. Monit. Assess. 2002, 78, 1–17. [Google Scholar] [CrossRef]
- Fu, J.; Mai, B.; Sheng, G.; Zhang, G.; Wang, X.; Peng, P.; Xiao, X.; Ran, R.; Cheng, F.; Peng, X.; et al. Persistent organic pollutants in environment of the Pearl River Delta, China: An overview. Chemosphere 2003, 52, 1411–1422. [Google Scholar] [CrossRef]
- Nussey, S.; Whitehead, S. Endocrinology: An Integrated Approach. Mayo Clin. Proc. 2001, 77, 1015. [Google Scholar]
- Tilson, A.H.; Kodavanti, P.R.S.; Mundy, W.R.; Bushnell, P.J. Neurotoxicity of environmental chemicals and their mechanism of action. Toxicol. Lett. 1998, 102–103, 631–635. [Google Scholar]
- Zhou, T.; Ross, D.G.; DeVito, M.J.; Crofton, K.M. Effects of short term in vivo exposure to polybrominated diphenyl ethers on thyroid hormones and hepatic enzyme activities in weanling rats. Toxicol. Sci. 2001, 61, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Herbstman, J.B.; Mall, J.K. Developmental exposure to polybrominated diphenyl ethers and neurodevelopment. Curr. Environ. Health Rep. 2014, 1, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Vuong, A.M.; Webster, G.M.; Romano, M.E.; Braun, J.M.; Zoeller, R.T.; Hoofnagle, A.N.; Sjödin, A.; Yolton, K.; Lanphear, B.P.; Chen, A. Maternal polybrominated diphenyl ether (PBDE) exposure and thyroid hormones in Maternal and Cord Sera: The HOME study, Cincinnati, USA. Environ. Health Perspect. 2015, 123, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.; Makela, S.; Treuter, E.; Tujague, M.; Thomsen, J.; Andersson, G.; Enmark, E.; Pettersson, K.; Warner, M.; Gustafsson, J.Å. Mechanisms of estrogen action. Physiol. Rev. 2001, 81, 1535–1565. [Google Scholar] [CrossRef]
- Yang, F.X.; Xu, Y. Hydroxylated metabolites of polychlorinated biphenyls and their endocrine disrupting mechanism. Prog. Chem. 2005, 17, 740–748. [Google Scholar]
- Li, X.L. Quantitative Structure-Activity Relationships and Molecular Simulation Studies on Several Endocrine Disruptors’ Chemicals. Ph.D. Thesis, Nanjing University, Nanjing, China, 2013. [Google Scholar]
- Vakharia, D.D.; Gierthy, J.F. Rapid assay for oestrogen receptor binding to PCB metabolites. Toxicol. In Vitro Int. J. Publ. Assoc. Bibra 1999, 13, 275–282. [Google Scholar] [CrossRef]
- Vakharia, D.D.; Gierthy, J.F. Use of a combined human liver microsome-estrogen receptor binding assay to assess potential estrogen modulating activity of PCB metabolites. Toxicol. Lett. 2000, 114, 55–65. [Google Scholar] [CrossRef]
- Schrader, T.J.; Cooke, G.M. Effects of Aroclors and individual PCB congeners on activation of the human androgen receptor in vitro. Reprod. Toxicol. 2003, 17, 15–23. [Google Scholar] [CrossRef]
- Bonefeld-Jørgensen, E.C.; Andersen, H.R.; Rasmussen, T.H.; Vinggaard, A.M. Effect of highly bioaccumulated polychlorinated biphenyl congeners on estrogen and androgen receptor activity. Toxicology 2001, 158, 141–153. [Google Scholar] [CrossRef]
- Ulbrich, B.; Stahlmann, R. Developmental toxicity of polychlorinated biphenyls (PCBs): A systematic review of experimental data. Arch. Toxicol. 2004, 78, 252–268. [Google Scholar]
- Ross, G. The public health implications of polychlorinated biphenyls (PCBs) in the environment. Ecotoxicol. Environ. Saf. 2004, 59, 275–291. [Google Scholar] [CrossRef]
- Yang, J.; Gu, W.; Li, Y. Biological enrichment prediction of polychlorinated biphenyls and novel molecular design based on 3D-QSAR/HQSAR associated with molecule docking. Biosci. Rep. 2019, 39, 1–20. [Google Scholar] [CrossRef]
- Qu, R.; Liu, H.; Feng, M.; Yang, X.; Wang, Z. Investigation on Intramolecular Hydrogen Bond and Some Thermodynamic Properties of Polyhydroxylated Anthraquinones. J. Chem. Eng. Data 2012, 57, 2442–2445. [Google Scholar] [CrossRef]
- Zeng, X.; Qu, R.; Feng, M.; Chen, J.; Wang, L.; Wang, Z. Photodegradation of Polyfluorinated Dibenzo-p-Dioxins (PFDDs) in Organic Solvents: Experimental and Theoretical Studies. Environ. Sci. Technol. 2016, 50, 8128–8134. [Google Scholar] [CrossRef]
- Meerts, I.A.; Letcher, R.J.; Hoving, S.; Marsh, G.; Bergman, K.E.; Lemmen, J.G.; Burg, B.V.; Brouwer, A. In vitro estrogenicity of polybrominated diphenyl ethers, hydroxylated PDBEs, and polybrominated bisphenol A compounds. Environ. Health Perspect. 2001, 109, 399–407. [Google Scholar] [CrossRef]
- Chen, Y. Research on the Migration and Biodegradation of PCBs Based on 3D-QSAR and Molecular Docking. Ph.D. Thesis, North China Electric Power University, Beijing, China, 2017. [Google Scholar]
- Zhang, Q.; Lu, M.; Wang, C.; Du, J.; Zhou, P.; Zhao, M. Characterization of estrogen receptor alpha activities in polychlorinated biphenyls by in vitro dual-luciferase reporter gene assay. Environ. Pollut. 2014, 189, 169–175. [Google Scholar] [CrossRef]
- Liu, H.X.; Shi, J.Q.; Liu, H.; Wang, Z.Y. Improved 3D-QSPR analysis of the predictive octanol–air partition coefficients of hydroxylated and methoxylated polybrominated diphenyl ethers. Atmos. Environ. 2013, 77, 840–845. [Google Scholar] [CrossRef]
- Hummel, C.W.; Geiser, A.G.; Bryant, H.U.; Cohen, I.R.; Dally, R.D.; Fong, K.C.; Frank, S.A.; Hinklin, R.; Jones, S.A.; Lewis, G.; et al. A selective estrogen receptor modulator designed for the treatment of uterine leiomyoma with unique tissue specificity for uterus and ovaries in rats. Med. Chem. 2005, 48, 6772–6775. [Google Scholar] [CrossRef]
- Richardson, T.I.; Dodge, J.A.; Durst, G.L.; Pfeifer, L.A.; Shah, J.; Yong, W.; Durbin, J.D.; Krishnan, V.; Norman, B.H. Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 3: Synthesis of cyclopentanone and cyclohexanone intermediates for C-ring modification. Bioorg. Med. Chem. Lett. 2007, 17, 4824–4828. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, A.M.; Pike, A.C.; Dauter, Z.; Hubbard, R.E.; Bonn, T.; Engström, O.; Öhman, L.; Greene, G.L.; Gustafsson, J.-Å.; Carlquist, M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 1997, 389, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.B.; Nettles, K.W.; Bruning, J.B.; Kim, Y.; Joachimiak, A.; Sharma, S.; Carlson, K.E.; Stossi, F.; Katzenellenbogen, B.S.; Greene, G.L. Elemental isomerism: A boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands. Cell Chem. Biol. 2007, 14, 659–669. [Google Scholar] [CrossRef]
- Dykstra, K.D.; Guo, L.; Birzin, E.T.; Chan, W.; Yang, Y.T.; Hayes, E.C.; DaSilva, C.A.; Pai, L.Y.; Mosley, R.T.; Kraker, B.; et al. Estrogen receptor ligands. Part 16: 2-aryl indoles as highly subtype selective ligands for ERalpha. Cheminform 2007, 17, 2322–2328. [Google Scholar]
- Halgren, T.A. Merck molecular force field.1. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 1, 490–519. [Google Scholar] [CrossRef]
- Li, X.L.; Ye, L.; Wang, X.X.; Wang, X.; Liu, H.; Zhu, Y.; Yu, H. Combined 3D-QSAR, molecular docking and molecular dynamics study on thyroidhormone activity of hydroxylated polybrominated diphenyl ethers to thyroidreceptors β. Toxicol. Appl. Pharmacol. 2012, 265, 300–307. [Google Scholar] [CrossRef]
- Holt, P.A.; Chairs, J.B.; Trent, J.O. Molecular Docking of Intercalators and Groove-Binders to Nucleic Acids Using Autodock and Surflex. J. Chem. Inf. Model. 2008, 48, 1602–1615. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.Y.; Qian, L.; Miao, T.F.; Lu, H.L.; Zheng, K.C. CoMFA and docking studies of 2-phenylindole derivatives with anticancer activity. Eur. J. Med. Chem. 2009, 44, 2822–2827. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wei, D.; Zhao, H.; Du, Y. Genotoxicity of quinolones: Substituents contribution and transformation products QSAR evaluation using 2D and 3D models. Chemosphere 2013, 95, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Salahinejad, M.; Ghasemi, J.B. 3D-QSAR studies on the toxicity of substituted benzenes to Tetrahymena pyriformis: CoMFA, CoMSIA and VolSurf approaches. Ecotoxicol. Environ. Saf. 2014, 105, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gu, W.; Li, Y. Molecular design of 1,3,5,7-TetraCN derivatives with reduced bioconcentration using 3D-QSAR modeling, full factorial design, and molecular docking. J. Mol. Graph. Model. 2018, 84, 197–214. [Google Scholar] [CrossRef]
- Li, M.H.; Wang, X.L.; Chu, Z.H.; Li, Y. Multiple-Site Molecular Modification of Dioxin-Like PCBs to Eliminate Bioconcentration. Pol. J. Environ. Stud. 2020, 2, 1–21. [Google Scholar]
- Xu, Z.; Chen, Y.; Qiu, Y.L.; Gu, W.W.; Li, Y. Prediction of stability for polychlorinated biphenyls in transformer insulation oil through three-dimensional quantitative structure-activity relationship pharmacophore model and full factor experimental design. Chem. Res. Chin. Univ. 2016, 32, 348–356. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, X.; Jiang, L.; Li, Y. Prediction of octanol-air partition coefficients for polychlorinated biphenyls (PCBs) using 3D-QSAR models. Ecotoxicol. Environ. Saf. 2016, 124, 202–212. [Google Scholar] [CrossRef]
- Li, F.; Li, X.; Liu, X.; Zhang, L.; You, L.; Zhao, J.; Wu, H. Docking and 3D-QSAR studies on the Ah receptor binding affinities of polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs) and dibenzofurans (PCDFs). Environ. Toxicol. Pharmacol. 2011, 32, 478–485. [Google Scholar] [CrossRef]
- Chen, Y.; Chu, Z.H.; Li, Y. Based on molecular docking technology and CoMSIA/HQSAR assisted Molecular modification of dihydroxy polychlorinated biphenyl derivatives. Chem. J. Chin. Univ. 2018, 39, 299–309. [Google Scholar]
- Fang, H.; Tong, W.D.; Shi, L.M.; Blair, R.; Perkins, R.; Branham, W.; Hass, B.S.; Xie, Q.; Dial, S.L.; Moland, C.L. Structure-activity relationships for a large diverse set of natural, synthetic, and environmental estrogens. Chem. Res. Toxicol. 2001, 14, 280–294. [Google Scholar] [CrossRef]
- Fang, H.; Tong, W.D.; Perkins, R.; Soto, A.M.; Prechtl, N.V.; Sheehan, D.M. Quantitative Comparisons of in Vitro Assays for Estrogenic Activities. Environ. Health Perspect. 2000, 108, 723–729. [Google Scholar] [CrossRef]
- Chu, Z.H.; Li, Y. Designing modified polybrominated diphenyl ether BDE-47, BDE-99, BDE-100, BDE-183, and BDE-209 molecules with decreased estrogenic activities using 3D-QSAR, pharmacophore models coupled with resolution V of the 210-3 fractional factorial design and molecular docking. J. Hazard. Mater. 2019, 364, 151–162. [Google Scholar] [PubMed]
- Yang, W.H. Three-Dimensional Structure Activity Relationship of Endocrine Disrupting Activity of New Persistent Organic Pollutants; China University of Mining and Technology Press: Beijing, China, 2011. [Google Scholar]
- Wertz, D.H.; Scheraga, H.A. Influence of Water on Protein Structure. An Analysis of the Preferences of Amino Acid Residues for the Inside or Outside and for Specific Conformations in a Protein Molecule. Macromolecules 1978, 11, 9–15. [Google Scholar] [CrossRef]
- Castorena-Cortés, G.; Roldán-Carrillo, T.; Zapata-Penasco, I.; Reyes-Avila, J.; Quej-Aké, l.; Marín-Cruz, J.; Olguín-Lora, P. Microcosm assays and Taguchi experimental design for treatment of oil sludge containing high concentration of hydrocarbons. Bioresour. Technol. 2009, 100, 5671–5677. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.Q.; Liu, F.; Ma, X.T.; Yang, X.L.; Zhao, H.Y. Hydrophobic Interaction Induced Coassembly of Homopolymer and Protein. Langmuir 2019, 35, 10958–10964. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Yang, X.C.; Liu, H.Y.; Gao, Y.X. Effect of xylitol and mannitol on the structural and functional properties of peanut proteins. Food Mach. 2018, 34, 17–22. [Google Scholar]
- Ferreira, D.; Gross, G.G.; Hagerman, A.E.; Kolodziej, H.; Yoshida, T. Tannins and related polyphenols: Perspectives on their chemistry, biology, ecological effects, and human health protection. Phytochemistry 2008, 69, 3006–3008. [Google Scholar] [CrossRef]
- Yousuf, B.; Gul, K.; Wani, A.A.; Singh, P. Health benefits of anthocyanins and their encapsulation for potential use in food systems: A review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2223–2230. [Google Scholar] [CrossRef]
- Yang, J.W.; Li, Q.; Li, Y. Enhanced biodegradation/photodegradation of organophosphorus fire retardant using an integrated method of modified pharmacophore model with molecular dynamics and polarizable continuum model. Polymers 2020, 12, 1672. [Google Scholar] [CrossRef]
- Gu, W.W.; Zhao, Y.Y.; Li, Q.; Li, Y. Plant-microorganism Combined Remediation of Polychlorinated Naphthalenes Contaminated Soils Based on Molecular Directed Transformation and Taguchi Experimental Design-assisted Dynamics Simulation. J. Hazard. Mater. 2020, 396, 122753. [Google Scholar] [CrossRef]


| No. | Compounds | Observed Value |
|---|---|---|
| 1a | 2,2′,5-Trichlorobiphenyl (PCB-18) | 5.006 |
| 2 a | 2,4,6-Trichlorobiphenyl (PCB-30) | 6.614 |
| 3 a | 2,2′,3,5′-Tetrachlorobiphenyl (PCB-44) | 8.158 |
| 4 a | 2,2′,5,5′-Tetrachlorobiphenyl (PCB-52) | 6.509 |
| 5 a | 2,2′,4,4′,5-Pentachlorobiphenyl (PCB-99) | 6.048 |
| 6 a | 2,2′,4,5,5′-Pentachlorobiphenyl (PCB-101) | 7.189 |
| 7 a | 2,3,3′,4′,6-Pentachlorobiphenyl (PCB-110) | 8.009 |
| 8 b | 2,2′,3,3′,4,4′-Hexachlorobiphenyl (PCB-128) | 7.073 |
| 9 b | 2,2′,4,5′,6-Pentachlorobiphenyl (PCB-103) | 7.770 |
| 10 b | 2,2′,4,5′-Tetrachlorobiphenyl (PCB-49) | 6.672 |
| 11 b | 2,4,4′-Trichlorobiphenyl (PCB-28) | 5.430 |
| Model | n | q2 | r2 | SEE | F | r2pred | SEP | Q2ext | S | E | H | D | A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CoMSIA | 3 | 0.665 | 0.991 | 0.146 | 114.424 | 0.902 | 0.206 | 0.904 | 0.50% | 85.70% | 13.70% | 0 | 0 |
| Compounds | pREC20 | logBCF [33] | logt1/2 [36] | logKOA [37] | pEC50 [38] | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CoMSIA Pred. | Fall Rate (%) | CoMSIA | Change Rate (%) | CoMSIA | Change Rate (%) | CoMSIA | Change Rate (%) | CoMFA | Change Rate (%) | |
| Target molecule PCB-44 | 8.158 | 4.542 | 0.94 | 9.318 | 4.327 | |||||
| 3-carboxyl-PCB-44 | 7.712 | 5.5 | 4.204 | −7.44 | 0.830 | −11.70 | 8.731 | −6.30 | 4.878 | 12.73 |
| 3-amino-PCB-44 | 7.009 | 14.1 | 4.042 | −11.01 | 0.694 | −26.17 | 8.210 | −11.89 | 3.943 | −8.87 |
| 3-vinyl-PCB-44 | 5.201 | 36.2 | 4.258 | −6.25 | 0.695 | −26.06 | 8.459 | −9.22 | 4.774 | 10.33 |
| 3-methyl-PCB-44 | 6.991 | 14.3 | 4.036 | −11.14 | 0.704 | −25.11 | 8.201 | −11.99 | 4.153 | −4.02 |
| 3-ethyl-PCB-44 | 7.200 | 11.7 | 4.040 | −11.05 | 0.709 | −24.57 | 8.291 | −11.02 | 4.423 | 2.22 |
| 3-isopropyl-PCB-44 | 7.353 | 9.9 | 4.143 | −8.78 | 0.710 | −24.47 | 8.370 | −10.17 | 4.401 | 1.71 |
| 5′-carboxyl-PCB-44 | 5.260 | 35.5 | 4.817 | 6.05 | 1.075 | 14.36 | 9.770 | 4.85 | 3.243 | −25.05 |
| 5′-amino-PCB-44 | 6.459 | 20.8 | 3.757 | −17.28 | 0.546 | −41.91 | 7.990 | −14.25 | 4.641 | 7.26 |
| 5′-hydroxyl-PCB-44 | 7.637 | 6.4 | 4.367 | −3.85 | 0.832 | −11.49 | 8.863 | −4.88 | 4.754 | 9.87 |
| 5′-vinyl-PCB-44 | 5.881 | 27.9 | 4.354 | −4.14 | 0.740 | −21.28 | 8.667 | −6.99 | 4.688 | 8.34 |
| 5′-methyl-PCB-44 | 6.957 | 14.7 | 4.936 | 8.67 | 1.206 | 28.30 | 9.181 | −1.47 | 5.171 | 19.51 |
| 5′-ethyl-PCB-44 | 6.616 | 18.9 | 4.230 | −6.87 | 0.601 | −36.06 | 8.030 | −13.82 | 4.728 | 9.27 |
| 5′-isopropyl-PCB-44 | 6.094 | 25.3 | 4.169 | −8.21 | 0.667 | −29.04 | 8.321 | −10.70 | 4.657 | 7.63 |
| 5′-tert-butyl-PCB-44 | 5.000 | 38.7 | 4.998 | 10.04 | 1.046 | 11.28 | 9.826 | 5.45 | 5.235 | 20.98 |
| 3-Br-5′-aldehyde-PCB-44 | 6.838 | 16.2 | 4.845 | 6.67 | 1.177 | 25.21 | 9.096 | −2.38 | 5.171 | 19.51 |
| 3-Br-5′-amino-PCB-44 | 7.057 | 13.5 | 4.314 | −5.02 | 0.819 | −12.87 | 8.790 | −5.67 | 4.869 | 12.53 |
| 3-Br-5′-hydroxyl-PCB-44 | 7.581 | 7.1 | 4.310 | −5.11 | 0.796 | −15.32 | 8.758 | −6.01 | 4.752 | 9.82 |
| 3-methoxy-5′-aldehyde-PCB-44 | 5.698 | 30.2 | 5.563 | 22.48 | 0.901 | −4.15 | 9.292 | −0.28 | 3.186 | −26.37 |
| 3-methoxy-5′-carboxyl-PCB-44 | 5.690 | 30.3 | 5.483 | 20.72 | 0.969 | 3.09 | 9.623 | 3.27 | 4.043 | −6.56 |
| 3-methoxy-5′-amino-PCB-44 | 6.665 | 18.3 | 2.559 | −43.66 | 0.458 | −51.28 | 7.816 | −16.12 | 5.187 | 19.88 |
| 3-methoxy-5′-hydroxyl-PCB-44 | 5.682 | 30.4 | 4.727 | 4.07 | 0.743 | −20.96 | 8.813 | −5.42 | 3.999 | −7.58 |
| 3-ethinyl-5′-aldehyde-PCB-44 | 5.676 | 30.4 | 5.220 | 14.93 | 0.939 | −0.11 | 9.188 | −1.40 | 2.945 | −31.94 |
| 3-ethinyl-5′-carboxyl-PCB-44 | 5.669 | 30.5 | 5.133 | 13.01 | 1.007 | 7.13 | 9.518 | 2.15 | 3.795 | −12.29 |
| 3-ethinyl-5′-amino-PCB-44 | 6.503 | 20.3 | 3.652 | −19.59 | 0.494 | −47.45 | 7.863 | −15.61 | 4.595 | 6.19 |
| 3-ethinyl-5′-hydroxyl-PCB-44 | 6.084 | 25.4 | 2.715 | −40.22 | 0.840 | −10.64 | 7.970 | −14.47 | 4.424 | 2.24 |
| 3-vinyl-5′-aldehyde-PCB-44 | 7.783 | 4.6 | 4.818 | 6.08 | 0.960 | 2.13 | 8.803 | −5.53 | 4.606 | 6.45 |
| 3-vinyl-5′-carboxyl-PCB-44 | 5.290 | 35.2 | 4.929 | 8.52 | 0.877 | −6.70 | 8.983 | −3.60 | 3.901 | −9.85 |
| 3-vinyl-5′-amino-PCB-44 | 5.629 | 31 | 2.585 | −43.09 | 0.548 | −41.70 | 6.910 | −25.84 | 4.070 | −5.94 |
| 3-vinyl-5′-hydroxyl-PCB-44 | 7.167 | 12.1 | 3.911 | −13.89 | 0.667 | −29.04 | 8.265 | −11.30 | 4.596 | 6.22 |
| 3-methyl-5′-aldehyde-PCB-44 | 6.950 | 14.7 | 4.755 | 4.69 | 0.889 | −5.43 | 8.277 | −11.17 | 4.576 | 5.75 |
| 3-methyl-5′-carboxyl-PCB-44 | 7.714 | 5.4 | 4.801 | 5.70 | 1.006 | 7.02 | 8.819 | −5.36 | 3.853 | −10.95 |
| 3-methyl-5′-amino-PCB-44 | 6.202 | 24 | 2.687 | −40.84 | 0.441 | −53.09 | 6.455 | −30.73 | 4.017 | −7.16 |
| 3-methyl-5′-hydroxyl-PCB-44 | 6.343 | 22.2 | 3.859 | −15.04 | 0.596 | −36.60 | 7.747 | −16.86 | 4.584 | 5.94 |
| 3-ethyl-5′-aldehyde-PCB-44 | 7.135 | 12.5 | 4.993 | 9.93 | 0.893 | −5.00 | 8.378 | −10.09 | 4.750 | 9.78 |
| 3-ethyl-5′-carboxyl-PCB-44 | 7.893 | 3.2 | 5.041 | 10.99 | 1.010 | 7.45 | 8.918 | −4.29 | 4.026 | −6.96 |
| 3-ethyl-5′-amino-PCB-44 | 5.341 | 34.5 | 3.491 | −23.14 | 0.313 | −66.70 | 6.967 | −25.23 | 4.625 | 6.89 |
| 3-ethyl-5′-hydroxyl-PCB-44 | 6.524 | 20 | 4.090 | −9.95 | 0.600 | −36.17 | 7.844 | −15.82 | 4.749 | 9.75 |
| 3-isopropyl-5′-aldehyde-PCB-44 | 7.272 | 10.9 | 4.754 | 4.67 | 0.894 | −4.89 | 8.421 | −9.63 | 4.683 | 8.23 |
| 3-isopropyl-5′-carboxyl-PCB-44 | 8.034 | 1.5 | 4.804 | 5.77 | 1.011 | 7.55 | 8.962 | −3.82 | 3.960 | −8.48 |
| 3-isopropyl-5′-amino-PCB-44 | 5.917 | 27.5 | 2.615 | −42.43 | 0.454 | −51.70 | 6.559 | −29.61 | 4.098 | −5.29 |
| 3-isopropyl-5′-hydroxyl-PCB-44 | 6.665 | 18.3 | 3.865 | −14.91 | 0.601 | −36.06 | 7.889 | −15.34 | 4.684 | 8.25 |
| 3-tert-butyl-5′-amino-PCB-44 | 7.539 | 7.6 | 4.409 | −2.93 | 0.617 | −34.36 | 8.620 | −7.49 | 5.426 | 25.40 |
| 3-Br-5′-Br-PCB-44 | 8.106 | 0.6 | 4.406 | −2.99 | 0.863 | −8.19 | 9.081 | −2.54 | 4.347 | 0.46 |
| 3-carboxyl-5′-carboxyl-PCB-44 | 5.726 | 29.8 | 5.193 | 14.33 | 1.277 | 35.85 | 10.435 | 11.99 | 2.538 | −41.35 |
| 3-amino-5′-amino-PCB-44 | 5.182 | 36.5 | 3.276 | −27.87 | 0.300 | −68.09 | 6.882 | −26.14 | 4.246 | −1.87 |
| 3-hydroxyl-5′-hydroxyl-PCB-44 | 5.809 | 28.8 | 4.255 | -6.32 | 0.762 | −18.94 | 8.407 | −9.78 | 3.549 | −17.98 |
| 3-vinyl-5′-vinyl-PCB-44 | 5.333 | 34.6 | 4.032 | −11.23 | 0.570 | −39.36 | 7.996 | −14.19 | 4.543 | 4.99 |
| 3-methyl-5′-methyl-PCB-44 | 5.131 | 37.1 | 3.440 | −24.26 | 0.346 | −63.19 | 6.804 | −26.98 | 4.406 | 1.83 |
| 3-ethyl-5′-ethyl-PCB-44 | 5.530 | 32.2 | 3.544 | −21.97 | 0.370 | −60.64 | 7.003 | −24.84 | 4.801 | 10.95 |
| 3-isopropyl-5′-isopropyl-PCB-44 | 5.834 | 28.5 | 4.542 | 0.00 | 0.394 | −58.09 | 7.194 | −22.79 | 4.327 | 0.00 |
| Compounds | 2AYR | 2Q70 | 1ERR | 2Q6J | 2IOK | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Docking Score | Change Rate (%) | Docking Score | Change Rate (%) | Docking Score | Change Rate (%) | Docking Score | Change Rate (%) | Docking Score | Change Rate (%) | |
| Target molecule PCB-44 | 3.06 | 3.40 | 4.28 | 2.94 | 2.90 | |||||
| 3-amino-PCB-44 | 2.74 | −10.46 | 3.57 | 5.00 | 3.67 | −14.25 | 3.41 | 16.10 | 3.49 | 20.39 |
| 3-methyl-PCB-44 | 4.22 | 37.91 | 3.28 | −3.53 | 4.35 | 1.64 | 2.57 | −12.61 | 2.82 | −2.84 |
| 3-vinyl-5′-amino-PCB-44 | 5.05 | 65.03 | 5.86 | 72.35 | 4.30 | 0.47 | 3.22 | 9.74 | 4.29 | 47.88 |
| 3-methyl-5′-amino-PCB-44 | 4.23 | 38.24 | 5.20 | 52.94 | 5.84 | 36.45 | 3.63 | 23.70 | 4.76 | 64.34 |
| 3-isopropyl-5′-amino-PCB-44 | 5.11 | 66.99 | 5.78 | 70.00 | 4.38 | 2.34 | 4.04 | 37.45 | 4.81 | 65.93 |
| 3-amino-5′-amino-PCB-44 | 4.39 | 43.46 | 5.54 | 62.94 | 3.50 | −18.22 | 4.03 | 37.13 | 5.43 | 87.41 |
| 3-hydroxyl-5′-hydroxyl-PCB-44 | 4.10 | 33.99 | 4.38 | 28.82 | 4.95 | 15.65 | 4.45 | 51.48 | 4.59 | 58.43 |
| Before Modification | After Modification | |||||||
|---|---|---|---|---|---|---|---|---|
| Name of Amino Acid Residue | Target Molecule PCB-44 | 3-methyl-PCB-44 | 3-amino-PCB-44 | 3-hydroxyl-5′-hydroxyl-PCB-44 | 3-vinyl-5′-amino-PCB-44 | 3-methyl-5′-amino-PCB-44 | 3-isopropyl-5′-amino-PCB-44 | 3-amino-5′-amino-PCB-44 |
| Docking score | 2.94 | 2.57 | 3.41 | 4.45 | 3.22 | 3.63 | 4.04 | 4.03 |
| Hydrophobic residues | Leu346 | 3.5 | 3.7 | 5.6 | 3.9 | 5.2 | 4.3 | 3.5 |
| Ale350 | 5.7 | 4.1 | 4.0 | 4.7 | 4.2 | 3.7 | 3.8 | |
| Leu387 | 5.1 | - | 4.6 | 5.3 | 5.0 | 4.8 | 3.8 | |
| Met388 | 6.6 | - | 5.8 | 7.1 | 5.9 | 7.6 | 5.0 | |
| Phe404 | 3.1 | - | 4.6 | 4.0 | 6.0 | 4.6 | 3.6 | |
| Met421 | 7.4 | 4.8 | 4.3 | 5.7 | 3.8 | 5.4 | 6.9 | |
| Average distance (Å) | 5.23 | 4.20 | 4.82 | 5.12 | 5.02 | 5.07 | 4.43 | 5.00 |
| Hydrophilic residues | Thr347 | - | 3.5 | - | - | - | - | - |
| Glu353 | - | - | 3.9 | 3.9 | 4.0 | 3.9 | 4.1 | |
| His524 | - | - | - | - | - | - | - | |
| Combinations | A (mmol/L) | B (mmol/L) | C (mmol/L) | D (mmol/L) |
|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0.96 | 0.94 | 0.96 |
| 3 | 0 | 1.92 | 1.88 | 1.92 |
| 4 | 0.95 | 0 | 0.94 | 1.92 |
| 5 | 0.95 | 0.96 | 1.88 | 0 |
| 6 | 0.95 | 1.92 | 0 | 0.96 |
| 7 | 1.90 | 0 | 0.94 | 0.96 |
| 8 | 1.90 | 0.96 | 0 | 1.92 |
| 9 | 1.90 | 1.92 | 1.88 | 0 |
| PCB-44 | 3-methyl-PCB-44 | 3-amino-PCB-44 | 3-hydroxyl-5′-hydroxyl-PCB-44 | 3-vinyl-5′-amino-PCB-44 | 3-methyl-5′-amino-PCB-44 | 3-isopropyl-5′-amino-PCB-44 | 3-amino-5′-amino-PCB-44 | |
|---|---|---|---|---|---|---|---|---|
| Combination 1 | −136.317 | −133.293 | −138.483 | −128.696 | −135.967 | −140.651 | −102.431 | −105.153 |
| Change rate (%) | −2.22 | 1.59 | −5.59 | −0.26 | 3.18 | −24.86 | −22.86 | |
| Combination 2 | −145.044 | −139.843 | −107.763 | −115.925 | −128.898 | −129.797 | −104.278 | −99.087 |
| Change rate (%) | −3.59 | −25.7 | −20.08 | −11.13 | −10.51 | −28.11 | −31.68 | |
| Combination 3 | −140.755 | −134.72 | −131.681 | −127.886 | −124.711 | −151.602 | −93.373 | −97.705 |
| Change rate (%) | −4.29 | −6.45 | −9.14 | −11.4 | 7.71 | −33.66 | −30.59 | |
| Combination 4 | −128.654 | −135.583 | −136.997 | −112.013 | −132.642 | −136.842 | −87.189 | −89.155 |
| Change rate (%) | 5.39 | 6.48 | −12.93 | 3.1 | 6.36 | −32.23 | −30.7 | |
| Combination 5 | −124.468 | −126.849 | −129.241 | −124.585 | −121.917 | −133.491 | −98.075 | −99.583 |
| Change rate (%) | 1.91 | 3.83 | 0.09 | −2.05 | 7.25 | −21.2 | −19.99 | |
| Combination 6 | −125.964 | −137.308 | −119.652 | −129.574 | −131.267 | −140.547 | −94.746 | −94.105 |
| Change rate (%) | 9.01 | −5.01 | 2.87 | 4.21 | 11.58 | −24.78 | −25.29 | |
| Combination 7 | −142.61 | −127.914 | −120.375 | −116.144 | −129.26 | −137.648 | −103.637 | −101.748 |
| Change rate (%) | −10.31 | −15.59 | −18.56 | −9.36 | −3.48 | −27.33 | −28.65 | |
| Combination 8 | −136.574 | −127.681 | −119.583 | −126.673 | −106.3 | −126.598 | −99.966 | −105.671 |
| Change rate (%) | −6.51 | −12.44 | −7.25 | −22.17 | −7.3 | −26.8 | −22.63 | |
| Combination 9 | −131.23 | −107.546 | −126.97 | −123.287 | −132.44 | −138.777 | −113.162 | −100.038 |
| Change rate (%) | −18.05 | −3.25 | −6.05 | 0.92 | 5.75 | −13.77 | −23.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.; Zhang, W.; Chu, Z.; Li, Y. Mitigating the Adverse Effects of Polychlorinated Biphenyl Derivatives on Estrogenic Activity via Molecular Modification Techniques. Int. J. Environ. Res. Public Health 2021, 18, 4999. https://doi.org/10.3390/ijerph18094999
He W, Zhang W, Chu Z, Li Y. Mitigating the Adverse Effects of Polychlorinated Biphenyl Derivatives on Estrogenic Activity via Molecular Modification Techniques. International Journal of Environmental Research and Public Health. 2021; 18(9):4999. https://doi.org/10.3390/ijerph18094999
Chicago/Turabian StyleHe, Wei, Wenhui Zhang, Zhenhua Chu, and Yu Li. 2021. "Mitigating the Adverse Effects of Polychlorinated Biphenyl Derivatives on Estrogenic Activity via Molecular Modification Techniques" International Journal of Environmental Research and Public Health 18, no. 9: 4999. https://doi.org/10.3390/ijerph18094999
APA StyleHe, W., Zhang, W., Chu, Z., & Li, Y. (2021). Mitigating the Adverse Effects of Polychlorinated Biphenyl Derivatives on Estrogenic Activity via Molecular Modification Techniques. International Journal of Environmental Research and Public Health, 18(9), 4999. https://doi.org/10.3390/ijerph18094999







