Discovering Thematically Coherent Biomedical Documents Using Contextualized Bidirectional Encoder Representations from Transformers-Based Clustering
Abstract
:1. Introduction
- We developed a GMM-based efficient clustering framework that incorporates heavily pre-trained BioBERT domain-specific language representations to improve clustering accuracy for biomedical document analysis;
- We compared six distinct kinds of representative feature that highlight different aspects and have a significant impact on clustering effort when used in combination with different clustering techniques. The findings are useful for investigating comparable articles based on their inherent characteristics;
- The empirical comparison analysis demonstrates that the suggested proposed framework outperforms a variety of baseline models for biomedical-specific document analysis;
- The research findings are likely to contribute not only to biomedical document analysis but also to a wide range of applications in the healthcare area such as trend analysis and recommendation systems, as well as drug and gene expression identification.
2. Related Work
3. Proposed BioBERT-Based Clustering Framework for Biomedical Document Analysis Materials and Methods
3.1. Proposed BioBERT-Based Clustering Framework for Biomedical Document Analysis
3.1.1. Data Preprocessing
3.1.2. Bidirectional Encoder Representations from Transformers for Biomedical Text Mining
3.1.3. Gaussian Mixture Model
3.2. Experimental Setup
3.2.1. Experimental Environment
3.2.2. Baseline Models
- Embedding Techniques
- Clustering Techniques
3.2.3. Evaluation Metrics
4. Experimental Result and Analysis
4.1. Text Data Preprocessing
4.2. Comparison Results of Clustering Models
| Representations | Fowlkes–Mallows Score | Silhouette Coefficient | Adjusted Rand Index | Davies–Bouldin Score |
|---|---|---|---|---|
| BioBERT | 0.6798 | 0.2762 | 0.3258 | 2.2121 |
| Word2Vec | 0.5339 | 0.2112 | 0.0875 | 4.8135 |
| Glove | 0.5573 | 0.2355 | 0.2216 | 3.8381 |
| TF–IDF with PCA | 0.4545 | 0.0752 | 0.0626 | 5.7447 |
| TF–IDF with AE | 0.5102 | 0.1118 | 0.0684 | 4.8371 |
| BioWordVec | 0.6356 | 0.2995 | 0.3055 | 1.9482 |
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 25 April 2022).
- Batbaatar, E.; Pham, V.H.; Ryu, K.H. Multi-Task Topic Analysis Framework for Hallmarks of Cancer with Weak Supervision. Appl. Sci. 2020, 10, 834. [Google Scholar] [CrossRef] [Green Version]
- Prabhakar Kaila, D.; Prasad, D.A. Informational flow on Twitter–Corona virus outbreak–topic modelling approach. Int. J. Adv. Res. Eng. Technol. (IJARET) 2020, 11, 128–134. [Google Scholar]
- Zhu, Y.; Jung, W.; Wang, F.; Che, C. Drug repurposing against Parkinson’s disease by text mining the scientific literature. Libr. Hi Tech 2020, 38, 741–750. [Google Scholar] [CrossRef]
- Hansson, L.K.; Hansen, R.B.; Pletscher-Frankild, S.; Berzins, R.; Hansen, D.H.; Madsen, D.; Christensen, S.B.; Christiansen, M.R.; Boulund, U.; Wolf, X.A.; et al. Semantic text mining in early drug discovery for type 2 diabetes. PLoS ONE 2020, 15, e0233956. [Google Scholar] [CrossRef]
- Ju, C.; Zhang, S. Doctor Recommendation Model Based on Ontology Characteristics and Disease Text Mining Perspective. BioMed Res. Int. 2021, 7431199. [Google Scholar] [CrossRef] [PubMed]
- Basiri, M.E.; Abdar, M.; Cifci, M.A.; Nemati, S.; Acharya, U.R. A novel method for sentiment classification of drug reviews using fusion of deep and machine learning techniques. Knowl. Based Syst. 2020, 198, 105949. [Google Scholar] [CrossRef]
- Santibáñez, R.; Garrido, D.; Martin, A.J. Atlas: Automatic modeling of regulation of bacterial gene expression and metabolism using rule-based languages. Bioinformatics 2020, 36, 5473–5480. [Google Scholar] [CrossRef]
- Păduraru, O.; Moroșanu, A.; Păduraru, C.Ș.; Cărăușu, E.M. Healthcare Management: A Bibliometric Analysis Based on the Citations of Research Articles Published between 1967 and 2020. Healthcare 2022, 10, 555. [Google Scholar] [CrossRef]
- Franco, P.; Segelov, E.; Johnsson, A.; Riechelmann, R.; Guren, M.G.; Das, P.; Rao, S.; Arnold, D.; Spindler, K.G.; Deutsch, E. A Machine-Learning-Based Bibliometric Analysis of the Scientific Literature on Anal Cancer. Cancers 2022, 14, 1697. [Google Scholar] [CrossRef]
- Ahadi, A.; Singh, A.; Bower, M.; Garrett, M. Text Mining in Education—A Bibliometrics-Based Systematic Review. Educ. Sci. 2022, 12, 210. [Google Scholar] [CrossRef]
- Berardi, M.; Santamaria Amato, L.; Cigna, F.; Tapete, D.; Siciliani de Cumis, M. Text Mining from Free Unstructured Text: An Experiment of Time Series Retrieval for Volcano Monitoring. Appl. Sci. 2022, 12, 3503. [Google Scholar] [CrossRef]
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A survey of clustering with deep learning: From the perspective of network architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- Kushwaha, N.; Pant, M. Textual data dimensionality reduction-a deep learning approach. Multimed. Tools Appl. 2020, 79, 11039–11050. [Google Scholar] [CrossRef]
- Karim, M.R.; Beyan, O.; Zappa, A.; Costa, I.G.; Rebholz-Schuhmann, D.; Cochez, M.; Decker, S. Deep learning-based clustering approaches for bioinformatics. Brief. Bioinform. 2021, 22, 393–415. [Google Scholar] [CrossRef] [Green Version]
- Pinto da Costa, J.F.; Cabral, M. Statistical Methods with Applications in Data Mining: A Review of the Most Recent Works. Mathematics 2022, 10, 993. [Google Scholar] [CrossRef]
- Davagdorj, K.; Park, K.H.; Amarbayasgalan, T.; Munkhdalai, L.; Wang, L.; Li, M.; Ryu, K.H. BioBERT Based Efficient Clustering Framework for Biomedical Document Analysis. In Proceedings of the International Conference on Genetic and Evolutionary Computing, Jilin, China, 21–23 October 2021; Springer: Singapore, 2021. [Google Scholar]
- Chuluunsaikhan, T.; Ryu, G.; Yoo, K.H.; Rah, H.; Nasridinov, A. Incorporating Deep Learning and News Topic Modeling for Forecasting Pork Prices: The Case of South Korea. Agriculture 2020, 10, 513. [Google Scholar] [CrossRef]
- Amin, S.; Uddin, M.I.; Hassan, S.; Khan, A.; Nasser, N.; Alharbi, A.; Alyami, H. Recurrent neural networks with TF-IDF embedding technique for detection and classification in tweets of dengue disease. IEEE Access 2020, 8, 131522–131533. [Google Scholar] [CrossRef]
- Park, J.; Park, C.; Kim, J.; Cho, M.; Park, S. ADC: Advanced document clustering using contextualized representations. Expert Syst. Appl. 2019, 137, 157–166. [Google Scholar] [CrossRef]
- Yang, B.; Fu, X.; Sidiropoulos, N.D.; Hong, M. Towards k-means-friendly spaces: Simultaneous deep learning and clustering. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 3861–3870. [Google Scholar]
- Agarwal, N.; Sikka, G.; Awasthi, L.K. Enhancing web service clustering using Length Feature Weight Method for service description document vector space representation. Expert Syst. Appl. 2020, 161, 113682. [Google Scholar] [CrossRef]
- Omar, A.A. Feature selection in text clustering applications of literary texts: A hybrid of term weighting methods. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 99–107. [Google Scholar] [CrossRef]
- Alkhatib, W.; Rensing, C.; Silberbauer, J. Multi-label text classification using semantic features and dimensionality reduction with autoencoders. In Proceedings of the International Conference on Language, Data and Knowledge, Galway, Ireland, 19–20 June 2017; Springer: Cham, Germany, 2017; pp. 380–394. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1802.05365. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Kong, X.; Mao, M.; Wang, W.; Liu, J.; Xu, B. VOPRec: Vector representation learning of papers with text information and structural identity for recommendation. IEEE Trans. Emerg. Top. Comput. 2018, 9, 226–237. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Reynolds, D.A. Gaussian mixture models. Encycl. Biom. 2009, 741, 659–663. [Google Scholar]
- Zhang, Y.; Ghaoui, L.E. Large-scale sparse principal component analysis with application to text data. arXiv 2012, arXiv:1210.7054. [Google Scholar]
- Liou, C.Y.; Cheng, W.C.; Liou, J.W.; Liou, D.R. Autoencoder for words. Neurocomputing 2014, 139, 84–96. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Q.; Yang, Z.; Lin, H.; Lu, Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci. Data 2019, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The global k-means clustering algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Do, C.B.; Batzoglou, S. What is the expectation maximization algorithm? Nat. Biotechnol. 2008, 26, 897–899. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.J.; Meira, W., Jr.; Meira, W. Data Mining and Analysis: Fundamental Concepts and Algorithms; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- Karatzas, E.; Baltoumas, F.A.; Kasionis, I.; Sanoudou, D.; Eliopoulos, A.G.; Theodosiou, T.; Iliopoulos, I.; Pavlopoulos, G.A. Darling: A Web Application for Detecting Disease-Related Biomedical Entity Associations with Literature Mining. Biomolecules 2022, 12, 520. [Google Scholar] [CrossRef] [PubMed]
- Perera, N.; Nguyen, T.T.L.; Dehmer, M.; Emmert-Streib, F. Comparison of Text Mining Models for Food and Dietary Constituent Named-Entity Recognition. Mach. Learn. Knowl. Extr. 2022, 4, 254–275. [Google Scholar] [CrossRef]
- Bonilla, D.A.; Moreno, Y.; Petro, J.L.; Forero, D.A.; Vargas-Molina, S.; Odriozola-Martínez, A.; Orozco, C.A.; Stout, J.R.; Rawson, E.C.; Kreider, R.B. A Bioinformatics-Assisted Review on Iron Metabolism and Immune System to Identify Potential Biomarkers of Exercise Stress-Induced Immunosuppression. Biomedicines 2022, 10, 724. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Shah, S. Concept embedding-based weighting scheme for biomedical text clustering and visualization. In Applied Informatics; SpringerOpen: Berlin/Heidelberg, Germany, 2018; Volume 5, pp. 1–19. [Google Scholar]
- Kavvadias, S.; Drosatos, G.; Kaldoudi, E. Supporting topic modeling and trends analysis in biomedical literature. J. Biomed. Inform. 2020, 110, 103574. [Google Scholar] [CrossRef]
- Muchene, L.; Safari, W. Two-stage topic modelling of scientific publications: A case study of University of Nairobi, Kenya. PLoS ONE 2021, 16, e0243208. [Google Scholar] [CrossRef] [PubMed]
- Karami, A.; Lundy, M.; Webb, F.; Dwivedi, Y.K. Twitter and research: A systematic literature review through text mining. IEEE Access 2020, 8, 67698–67717. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, M.; Zhang, Y. Topic-informed neural approach for biomedical event extraction. Artif. Intell. Med. 2020, 103, 101783. [Google Scholar] [CrossRef]
- Liang, L.; Lu, X.; Lu, S. New Gene Embedding Learned from Biomedical Literature and Its Application in Identifying Cancer Drivers. bioRxiv 2021. [Google Scholar] [CrossRef]
- Boukhari, K.; Omri, M.N. Approximate matching-based unsupervised document indexing approach: Application to biomedical domain. Scientometrics 2020, 124, 903–924. [Google Scholar] [CrossRef]
- Curiskis, S.A.; Drake, B.; Osborn, T.R.; Kennedy, P.J. An evaluation of document clustering and topic modelling in two online social networks: Twitter and Reddit. Inf. Process. Manag. 2020, 57, 102034. [Google Scholar] [CrossRef]
- Koutsomitropoulos, D.A.; Andriopoulos, A.D. Automated MeSH indexing of biomedical literature using contextualized word representations. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, Neos Marmaras, Greece, 5–7 June 2020; Springer: Cham, Germany, 2020; pp. 343–354. [Google Scholar]
- Luo, X.; Gandhi, P.; Storey, S.; Zhang, Z.; Han, Z.; Huang, K. A Computational Framework to Analyze the Associations between Symptoms and Cancer Patient Attributes Post Chemotherapy using EHR data. IEEE J. Biomed. Health Inform. 2021, 25, 4098–4109. [Google Scholar] [CrossRef] [PubMed]
- Batbaatar, E.; Ryu, K.H. Ontology-based healthcare named entity recognition from twitter messages using a recurrent neural network approach. Int. J. Environ. Res. Public Health 2019, 16, 3628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Hu, J.; Ryu, K.H. An Efficient Tool for Semantic Biomedical Document Analysis. In Advances in Intelligent Information Hiding and Multimedia Signal Processing. Smart Innovation, Systems and Technologies; Springer: Singapore, 2021. [Google Scholar] [CrossRef]
- Batbaatar, E.; Li, M.; Ryu, K.H. Semantic-emotion neural network for emotion recognition from text. IEEE Access 2019, 7, 111866–111878. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaise, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; Annual Conference on Neural Information Processing Systems: Long Beach, CA, USA, 2017; pp. 5998–6008. [Google Scholar]
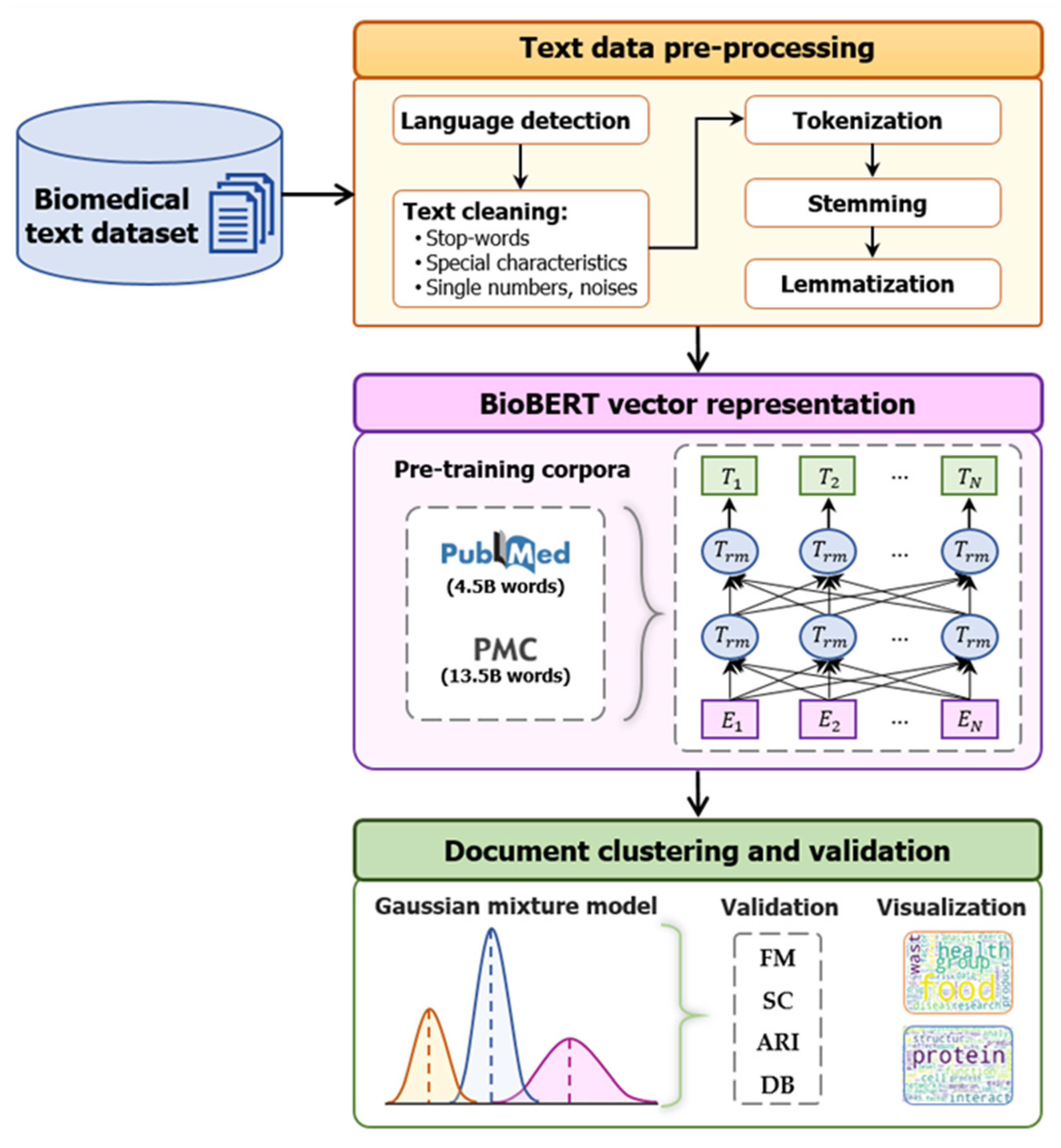







| Corpus | Number of Words (by Billion) | Domain |
|---|---|---|
| BooksCorpus | 2.5 | General |
| English Wikipedia | 0.8 | General |
| PubMed abstracts | 4.5 | Biomedical |
| PMC full-text articles | 13.5 | Biomedical |
| Name | Unit | Level |
|---|---|---|
| BioBERT | Contextual string embedding | Sentences |
| Word2Vec | Words | Local context |
| TF–IDF | Words | Corpus |
| GloVe | Words | Corpus |
| BioWordVec | Contextual sub-word | Local context |
| Representations | Fowlkes–Mallows Score | Silhouette Coefficient | Adjusted Rand Index | Davies–Bouldin Score |
|---|---|---|---|---|
| BioBERT | 0.7817 | 0.3765 | 0.4478 | 1.6849 |
| Word2Vec | 0.5919 | 0.3162 | 0.1143 | 3.1435 |
| GloVe | 0.6994 | 0.2175 | 0.3375 | 2.2419 |
| TF–IDF with PCA | 0.5308 | 0.0969 | 0.0863 | 4.5658 |
| TF–IDF with AE | 0.5659 | 0.0493 | 0.0751 | 3.7854 |
| BioWordVec | 0.7621 | 0.3854 | 0.4095 | 1.7309 |
| Representations | Fowlkes–Mallows Score | Silhouette Coefficient | Adjusted Rand Index | Davies–Bouldin Score |
|---|---|---|---|---|
| BioBERT | 0.7712 | 0.3041 | 0.4369 | 1.8507 |
| Word2Vec | 0.5794 | 0.2395 | 0.1025 | 2.7911 |
| GloVe | 0.5929 | 0.2658 | 0.2904 | 2.8612 |
| TF–IDF with PCA | 0.4672 | 0.0623 | 0.0719 | 3.8127 |
| TF–IDF with AE | 0.5531 | 0.0867 | 0.2758 | 3.4395 |
| BioWordVec | 0.7283 | 0.2624 | 0.4294 | 1.9204 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davagdorj, K.; Wang, L.; Li, M.; Pham, V.-H.; Ryu, K.H.; Theera-Umpon, N. Discovering Thematically Coherent Biomedical Documents Using Contextualized Bidirectional Encoder Representations from Transformers-Based Clustering. Int. J. Environ. Res. Public Health 2022, 19, 5893. https://doi.org/10.3390/ijerph19105893
Davagdorj K, Wang L, Li M, Pham V-H, Ryu KH, Theera-Umpon N. Discovering Thematically Coherent Biomedical Documents Using Contextualized Bidirectional Encoder Representations from Transformers-Based Clustering. International Journal of Environmental Research and Public Health. 2022; 19(10):5893. https://doi.org/10.3390/ijerph19105893
Chicago/Turabian StyleDavagdorj, Khishigsuren, Ling Wang, Meijing Li, Van-Huy Pham, Keun Ho Ryu, and Nipon Theera-Umpon. 2022. "Discovering Thematically Coherent Biomedical Documents Using Contextualized Bidirectional Encoder Representations from Transformers-Based Clustering" International Journal of Environmental Research and Public Health 19, no. 10: 5893. https://doi.org/10.3390/ijerph19105893
APA StyleDavagdorj, K., Wang, L., Li, M., Pham, V.-H., Ryu, K. H., & Theera-Umpon, N. (2022). Discovering Thematically Coherent Biomedical Documents Using Contextualized Bidirectional Encoder Representations from Transformers-Based Clustering. International Journal of Environmental Research and Public Health, 19(10), 5893. https://doi.org/10.3390/ijerph19105893








