Clinical Network for Big Data and Personalized Health: Study Protocol and Preliminary Results
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
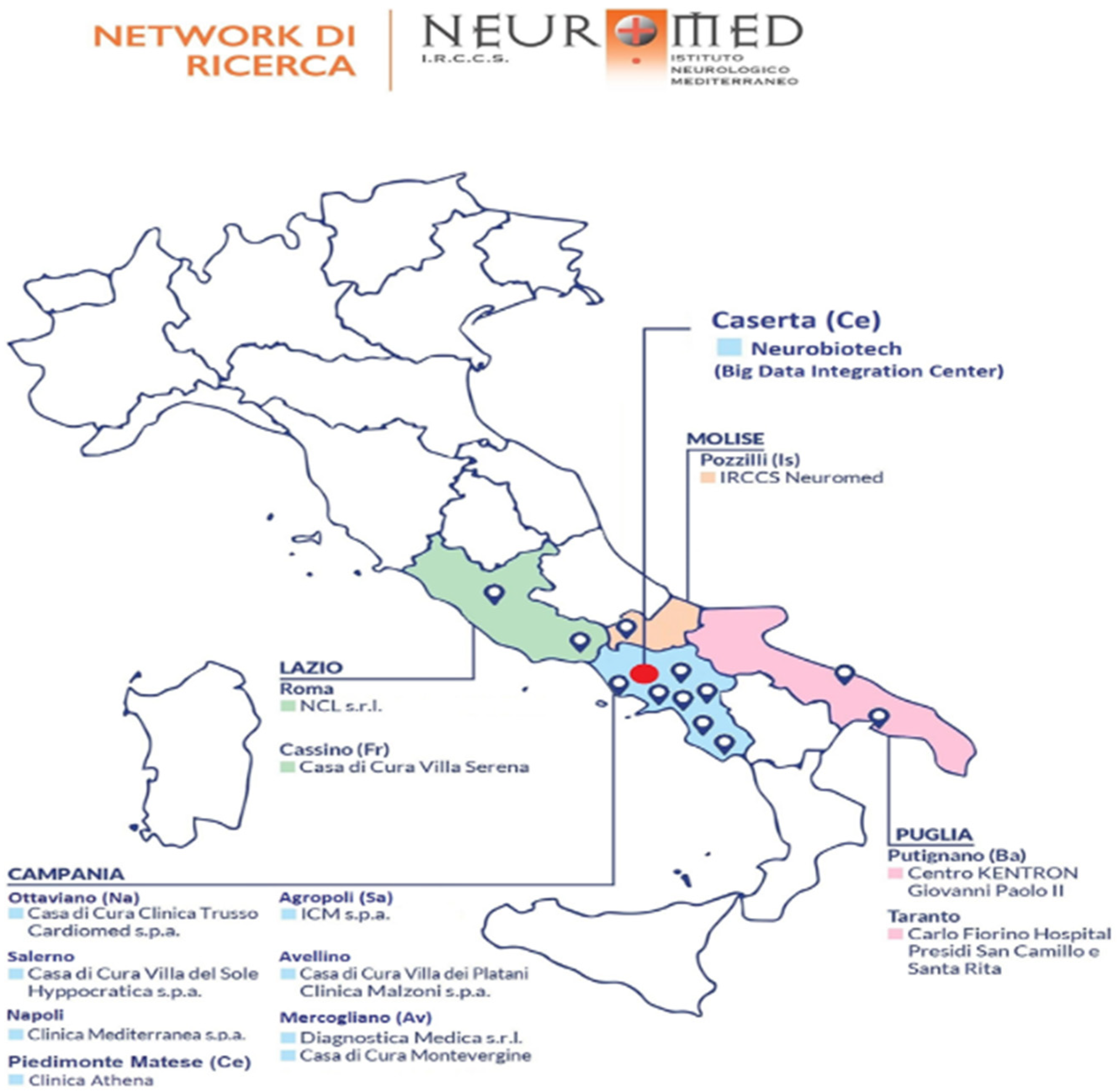
- being hospitalized for at least 24 h in one of the involved clinics of the Neuromed group, which cover almost all regions of Central-Southern Italy: IRCCS Neuromed (Pozzilli, Isernia), Clinica Mediterranea (Napoli), Istituto Clinico Mediterraneo (Agropoli, Salerno), Villa del Sole (Salerno), Diagnostica Medica (Avellino), Clinica Malzoni (Avellino), Casa di cura Trusso (Ottaviano, Napoli), Neurological Centre of Latium (Roma), Villa Serena (Cassino, Frosinone), Carlo Fiorino Hospital (Taranto), Centro Giovanni Paolo II (Putignano, Bari), Clinica Athena (Piedimonte Matese, Caserta) (Figure 1).
- being hospitalized for day surgery
- accessing an intensive care unit
- being under 18 years of age.
2.2. Questionnaires
2.3. Biological Samples and Biobanking
2.4. Alternative Strategies during COVID-19 Pandemic
3. Results
4. Discussion
4.1. Strengths of the Project
4.2. Potential Limitations
5. Conclusions
Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- Clinical Network Big Data and Personalised Health Project Study Investigators
- Principal Investigators: Licia Iacoviello, MD, PhD, (IRCCS Neuromed, Pozzilli and Università dell’Insubria, Varese, Italy)
- Steering Committee: Giovanni de Gaetano, Maria Benedetta Donati, Chiara Cerletti, Alessandro Gialluisi, Amalia De Curtis, Simona Costanzo, Marialaura Bonaccio (Department of Epidemiology and Prevention, IRCCS Neuromed, Pozzilli), Augusto Di Castelnuovo (Mediterranea Cardiocentro, Napoli).
- Recruitment coordinator: Simona Esposito (Department of Epidemiology and Prevention, IRCCS Neuromed).
- Neuromed Research Network:
- I.R.C.C.S. Neuromed, Pozzilli (Simona Esposito, Sabatino Orlandi)Clinica Malzoni, Avellino (Elena Bonanno, Maria Bianco, Annarita Vinciguerra)Diagnostica Medica, Avellino (Paola Bruni, Maria Bianco)N.C.L., Roma (Anna Campanella, Ida D’Anselmo, Edoardo Romoli, Pasquale Scognamiglio)Villa del Sole, Salerno (Maria Ceglia, Maria Grazia Caputo, Michelina Contangelo, Maria Rosaria Pandolfi, Giovanni Ricco)Clinica Athena, Piedimonte Matese (Maria Addolorata D’Abbraccio)Casa di Cura Trusso, Ottaviano (Alessandro Del Giudice, Camilla Esposito)Clinica Mediterranea, Napoli (Francesca De Micco)Carlo Fiorino Hospital, Taranto (Giovanni Pulito)I.C.M., Agropoli (Paola De Domenico, Aniello Formisano, Mariafiorella Tomasino)Centro Giovanni Paolo II, Putignano (Angela Vinci)Villa Serena, Cassino (Anna Izzo, Edoardo Romoli).
- Data analysis: Simona Costanzo (Department of Epidemiology and Prevention, IRCCS Neuromed), Augusto Di Castelnuovo (Mediterranea Cardiocentro, Napoli), Alessandro Gialluisi (Department of Epidemiology and Prevention, IRCCS Neuromed), Sabatino Orlandi (Department of Epidemiology and Prevention, IRCCS Neuromed).
- Informatics: Sabatino Orlandi (Department of Epidemiology and Prevention, IRCCS Neuromed)
- Biobanking: Amalia De Curtis, Simona Esposito (Department of Epidemiology and Prevention, IRCCS Neuromed), Sara Magnacca (Mediterranea Cardiocentro, Napoli).
- Communication and Press Office: Americo Bonanni (Department of Epidemiology and Prevention, IRCCS Neuromed).
References
- GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [Green Version]
- GBD 2017 Risk Factor Collaborators. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1923–1994. [Google Scholar]
- Donati, M.B. The “common soil hypothesis”: Evidence from population studies? Thromb. Res. 2010, 125 (Suppl. S2), S92–S95. [Google Scholar] [CrossRef]
- Nationaler Ethikrat. Biobanks for Research: Opinion; German National Ethics Council Publisher Nationaler Ethikrat: Berlin, Germany, 2004. [Google Scholar]
- Hansson, M.G.; Levin, M. (Eds.) Biobanks as Resources for Health; Uppsala University: Uppsala, Sweden, 2003. [Google Scholar]
- Iacoviello, L.; De Curtis, A.; Donati, M.B.; de Gaetano, G. Biobanks for cardiovascular epidemiology and prevention. Future Cardiol. 2014, 10, 243–254. [Google Scholar] [CrossRef]
- Yuille, M.; van Ommen, G.-J.; Bréchot, C.; Cambon-Thomsen, A.; Dagher, G.; Landegren, U.; Litton, J.-E.; Pasterk, M.; Peltonen, L.; Taussig, M.; et al. Biobanking for Europe. Brief. Bioinform. 2008, 9, 14–24. [Google Scholar] [CrossRef]
- Melville, S.; Byrd, J.B. Personalized Medicine and the Treatment of Hypertension. Curr. Hypertens. Rep. 2019, 21, 13. [Google Scholar] [CrossRef]
- Li, B.; He, X.; Jia, W.; Li, H. Novel Applications of Metabolomics in Personalized Medicine: A Mini-Review. Molecules 2017, 22, 1173. [Google Scholar] [CrossRef]
- Margolis, R.; Derr, L.; Dunn, M.; Huerta, M.; Larkin, J.; Sheehan, J.; Guyer, M.; Green, E.D. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: Capitalizing on biomedical big data. J. Am. Med. Inform. Assoc. 2014, 21, 957–958. [Google Scholar] [CrossRef] [Green Version]
- Cox, M.; Ellsworth, D. Application-controlled demand paging for out-of-core visualization. In Proceedings of the Visualization ‘97 (Cat. No. 97CB36155), Phoenix, AZ, USA, 19–24 October 1997; pp. 235–244. [Google Scholar]
- Rehm, H.L. Evolving health care through personal genomics. Nat. Rev. Genet. 2017, 18, 259–267. [Google Scholar] [CrossRef]
- Khoury, M.J.; Ioannidis, J.P. Big data meets public health. Science 2014, 346, 1054–1055. [Google Scholar] [CrossRef] [Green Version]
- Gunter, T.D.; Terry, N.P. The emergence of national electronic health record architectures in the United States and Australia: Models, costs, and questions. J. Med. Internet Res. 2005, 7, e3. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinh-Le, C.; Chuang, R.; Chokshi, S.; Mann, D. Wearable Health Technology and Electronic Health Record Integration: Scoping Review and Future Directions. JMIR mHealth uHealth 2019, 7, e12861. [Google Scholar] [CrossRef] [PubMed]
- Birkhead, G.S.; Klompas, M.; Shah, N.R. Uses of electronic health records for public health surveillance to advance public health. Annu. Rev. Public Health 2015, 36, 345–359. [Google Scholar] [CrossRef] [Green Version]
- Kruse, C.S.; Kristof, C.; Jones, B.; Mitchell, E.; Martinez, A. Barriers to Electronic Health Record Adoption: A Systematic Literature Review. J. Med. Syst. 2016, 40, 252. [Google Scholar] [CrossRef] [Green Version]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis. IEEE J. Biomed. Health Inform. 2018, 22, 1589–1604. [Google Scholar] [CrossRef]
- Huber, M.T.; Highland, J.D.; Krishnamoorthi, V.R.; Tang, J.W. Utilizing the Electronic Health Record to Improve Advance Care Planning: A Systematic Review. Am. J. Hosp. Palliat. Care 2018, 35, 532–541. [Google Scholar] [CrossRef]
- Zhou, L.; Sordo, M. Chapter 5—Expert systems in medicine. In Artificial Intelligence in Medicine; Xing, L., Giger, M.L., Min, J.K., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 75–100. [Google Scholar] [CrossRef]
- Sofi, F.; Dinu, M.; Pagliai, G.; Marcucci, R.; Casini, A. Validation of a literature-based adherence score to Mediterranean diet: The MEDI-LITE score. Int. J. Food Sci. Nutr. 2017, 68, 757–762. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripathi, T. One year update on the COVID-19 pandemic: Where are we now? Acta Trop. 2021, 214, 105778. [Google Scholar] [CrossRef]
- Wyatt, D.; Lampon, S.; McKevitt, C. Delivering healthcare’s ‘triple aim’: Electronic health records and the health research participant in the UK National Health Service. Sociol. Health Illn. 2020, 42, 1312–1327. [Google Scholar] [CrossRef]
- Wang, M.; Pantell, M.S.; Gottlieb, L.M.; Adler-Milstein, J. Documentation and review of social determinants of health data in the EHR: Measures and associated insights. J. Am. Med. Inform. Assoc. 2021, 28, 2608–2616. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Di Castelnuovo, A.; Costanzo, S.; Persichillo, M.; Bracone, F.; Cerletti, C.; Donati, M.B.; De Gaetano, G.; Iacoviello, L.; Bonaccio, M.; et al. Socioeconomic and psychosocial determinants of adherence to the Mediterranean diet in a general adult Italian population. Eur. J. Public Health 2018, 29, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Pounis, G.; Bonanni, A.; Ruggiero, E.; Di Castelnuovo, A.; Costanzo, S.; Persichillo, M.; Bonaccio, M.; Cerletti, C.; Riccardi, G.; Donati, M.; et al. Food group consumption in an Italian population using the updated food classification system FoodEx2: Results from the Italian Nutrition & HEalth Survey (INHES) study. Nutr. Metab. Cardiovasc. Dis. 2017, 27, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Fiorito, G.; Di Gaetano, C.; Guarrera, S.; Rosa, F.; Feldman, M.W.; Piazza, A.; Matullo, G. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur. J. Hum. Genet. 2016, 24, 1056–1062. [Google Scholar] [CrossRef] [Green Version]
- Colonna, V.; Nutile, T.; Astore, M.; Guardiola, O.; Antoniol, G.; Ciullo, M.; Persico, M.G. Campora: A young genetic isolate in South Italy. Hum. Hered. 2007, 64, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Sarno, S.; Petrilli, R.; Abondio, P.; De Giovanni, A.; Boattini, A.; Sazzini, M.; De Fanti, S.; Cilli, E.; Ciani, G.; Gentilini, D.; et al. Genetic history of Calabrian Greeks reveals ancient events and long term isolation in the Aspromonte area of Southern Italy. Sci. Rep. 2021, 11, 3045. [Google Scholar] [CrossRef]
- Babalini, C.; Martínez-Labarga, C.; Tolk, H.-V.; Kivisild, T.; Giampaolo, R.; Tarsi, T.; Contini, I.; Barać, L.; Janicijevic, B.; Klarić, I.M.; et al. The population history of the Croatian linguistic minority of Molise (southern Italy): A maternal view. Eur. J. Hum. Genet. 2005, 13, 902–912. [Google Scholar] [CrossRef] [Green Version]
- Sarno, S.; Boattini, A.; Pagani, L.; Sazzini, M.; De Fanti, S.; Quagliariello, A.; Ruscone, G.A.G.; Guichard, E.; Ciani, G.; Bortolini, E.; et al. Ancient and recent admixture layers in Sicily and Southern Italy trace multiple migration routes along the Mediterranean. Sci. Rep. 2017, 7, 1984. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, L.; Chen, R.; Zhao, Y.; Lv, L.; Xu, Z.; Xu, P. Electronic healthcare records and external outcome data for hospitalized patients with heart failure. Sci. Data 2021, 8, 46. [Google Scholar] [CrossRef]
- Pisesky, A.; Benchimol, E.I.; Wong, C.A.; Hui, C.; Crowe, M.; Bélair, M.-A.; Pojsupap, S.; Karnauchow, T.; O’Hearn, K.; Yasseen, A.S., 3rd; et al. Incidence of Hospitalization for Respiratory Syncytial Virus Infection amongst Children in Ontario, Canada: A Population-Based Study Using Validated Health Administrative Data. PLoS ONE 2016, 11, e0150416. [Google Scholar] [CrossRef]
- Xiao, C.; Bruner, D.W.; Dai, T.; Guo, Y.; Hanlon, A. A Comparison of Missing-Data Imputation Techniques in Exploratory Factor Analysis. J. Nurs. Meas. 2019, 27, 313–334. [Google Scholar] [CrossRef] [PubMed]

| Variables | N of Subjects (%) |
|---|---|
| Age groups (%) | |
| 18–30 | 759 (12.6%) |
| 31–50 | 2108 (34.9%) |
| 51–70 | 2027 (33.6%) |
| 71–90 | 1114 (18.5%) |
| 90+ | 28 (0.5%) |
| Women (%) | 3874 (64.2%) |
| Educational level (%) | |
| Up to lower school | 929 (15.4%) |
| Upper secondary | 3833 (63.5%) |
| Postsecondary education | 1093 (18.1%) |
| Missing | 181 (3.0%) |
| Occupation (%) | |
| Student | 134 (2.2%) |
| Manual | 1374 (22.8%) |
| Non-manual | 779 (12.9%) |
| Specialized/management | 398 (6.6%) |
| Housewife | 1287 (21.3%) |
| Retired | 1536 (25.4%) |
| Unemployed | 391 (6.5%) |
| Do not wish to answer | 66 (1.1%) |
| Missing | 71 (1.2%) |
| Prevalent occupation (%) | |
| Agri-food | 498 (8.2%) |
| Textile | 175 (2.9%) |
| Engineering | 167 (2.8%) |
| Chemical/pharmaceutical | 122 (2.0%) |
| Extractive | 6 (0.1%) |
| Electronics | 56 (0.9%) |
| Construction | 157 (2.6%) |
| Metallurgic | 39 (0.6%) |
| Other | 3405 (56.4%) |
| Missing | 1411 (23.4%) |
| Marital status (%) | |
| Married/living in a couple or de facto relationship | 4514 (74.8%) |
| Separated/divorced | 300 (5.0%) |
| Single | 756 (12.5%) |
| Widowed | 422 (7.0%) |
| Missing | 44 (0.7%) |
| Variables | N of Subjects (%) |
|---|---|
| Mediterranean diet (%) | |
| Low adherence (2 to 10) | 2150 (35.6%) |
| Average adherence (11) | 1389 (23.0%) |
| High adherence (12 to 18) | 2105 (34.9%) |
| Missing | 392 (6.5%) |
| Type of water (%) | |
| Plastic bottles | 4828 (80.0%) |
| Glass bottles | 280 (4.6%) |
| Tap water | 698 (11.5%) |
| Missing | 233 (3.9%) |
| Smoking status (%) | |
| Yes | 1824 (30.2%) |
| No | 2836 (47.0%) |
| Former | 1356 (22.5%) |
| Missing | 20 (0.3%) |
| Hours spent with mobile phone (%) | |
| <2 h | 2596 (43.0%) |
| 2–4 h | 2284 (37.8%) |
| 5–14 h | 815 (13.5%) |
| >15 h | 92 (1.5%) |
| Missing | 249 (4.1%) |
| Hours spent with cordless phone (%) | |
| <2 h | 2417 (40.0%) |
| 2–4 h | 149 (2.5%) |
| 5–14 h | 27 (0.4%) |
| >15 h | 12 (0.2%) |
| Missing | 3431 (56.8%) |
| Sleeping with phone nearby (%) | |
| Yes | 2705 (44.8%) |
| No | 3197 (53.0%) |
| Missing | 134 (2.2%) |
| Physically active lifestyle (%) | |
| Yes | 3451 (57.2%) |
| No | 2461 (40.8%) |
| Missing | 124 (2.0%) |
| Body mass index (%) | |
| Under/normal weight (<25 kg/m²) | 2415 (40.0%) |
| Overweight (≥25, <30 kg/m²) | 2164 (35.8%) |
| Obese (≥30 kg/m²) | 1327 (22.0%) |
| Missing | 132 (2.2%) |
| Quality of sleep (%) | |
| < 4 h | 251 (4.1%) |
| 5–6 h | 1813 (30.0%) |
| 6–7 h | 2449 (40.6%) |
| 7–8 h | 1278 (21.2%) |
| > 8 h | 188 (3.1%) |
| Missing | 10 (0.2%) |
| Variables | N of Subjects (%) |
|---|---|
| Number of pregnancies (median; SD) | (2; 1.6) |
| Menopausal status (%) | |
| Yes | 1686 (43.5%) |
| No | 2128 (54.9%) |
| Missing | 60 (1.5%) |
| Hypertension (%) | |
| Yes | 2195 (36.4%) |
| No | 3779 (62.6%) |
| Do not wish to answer | 11 (0.2%) |
| Missing | 51 (0.8%) |
| Diabetes (%) | |
| Yes | 671 (11.1%) |
| No | 5287 (87.6%) |
| Do not wish to answer | 19 (0.3%) |
| Missing | 59 (1.0%) |
| Hyperlipidaemia (%) | |
| Yes | 1183 (19.6%) |
| No | 4773 (79.1%) |
| Do not wish to answer | 19 (0.3%) |
| Missing | 61 (1.0%) |
| Systolic blood pressure (mmHg) (median; SD) | (121.8; 13.1) |
| Min | 60 |
| Max | 225 |
| Diastolic blood pressure (mmHg) (median; SD) | (74.0; 9.0) |
| Min | 20 |
| Max | 160 |
| Heart rate (bpm) (median; SD) | (73.2; 7.8) |
| Min | 34 |
| Max | 180 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, S.; Orlandi, S.; Magnacca, S.; De Curtis, A.; Gialluisi, A.; Iacoviello, L.; on behalf of The Neuromed Clinical Network Big Data and Personalised Health Investigators. Clinical Network for Big Data and Personalized Health: Study Protocol and Preliminary Results. Int. J. Environ. Res. Public Health 2022, 19, 6365. https://doi.org/10.3390/ijerph19116365
Esposito S, Orlandi S, Magnacca S, De Curtis A, Gialluisi A, Iacoviello L, on behalf of The Neuromed Clinical Network Big Data and Personalised Health Investigators. Clinical Network for Big Data and Personalized Health: Study Protocol and Preliminary Results. International Journal of Environmental Research and Public Health. 2022; 19(11):6365. https://doi.org/10.3390/ijerph19116365
Chicago/Turabian StyleEsposito, Simona, Sabatino Orlandi, Sara Magnacca, Amalia De Curtis, Alessandro Gialluisi, Licia Iacoviello, and on behalf of The Neuromed Clinical Network Big Data and Personalised Health Investigators. 2022. "Clinical Network for Big Data and Personalized Health: Study Protocol and Preliminary Results" International Journal of Environmental Research and Public Health 19, no. 11: 6365. https://doi.org/10.3390/ijerph19116365






