Measuring the Usability and Quality of Explanations of a Machine Learning Web-Based Tool for Oral Tongue Cancer Prognostication
Abstract
:1. Introduction
2. Material and Methods
2.1. A Web-Based Prognostic Tool
2.2. Questionnaire
2.2.1. Design and Participants
2.2.2. Questionnaire Development
2.2.3. Questionnaire Validation
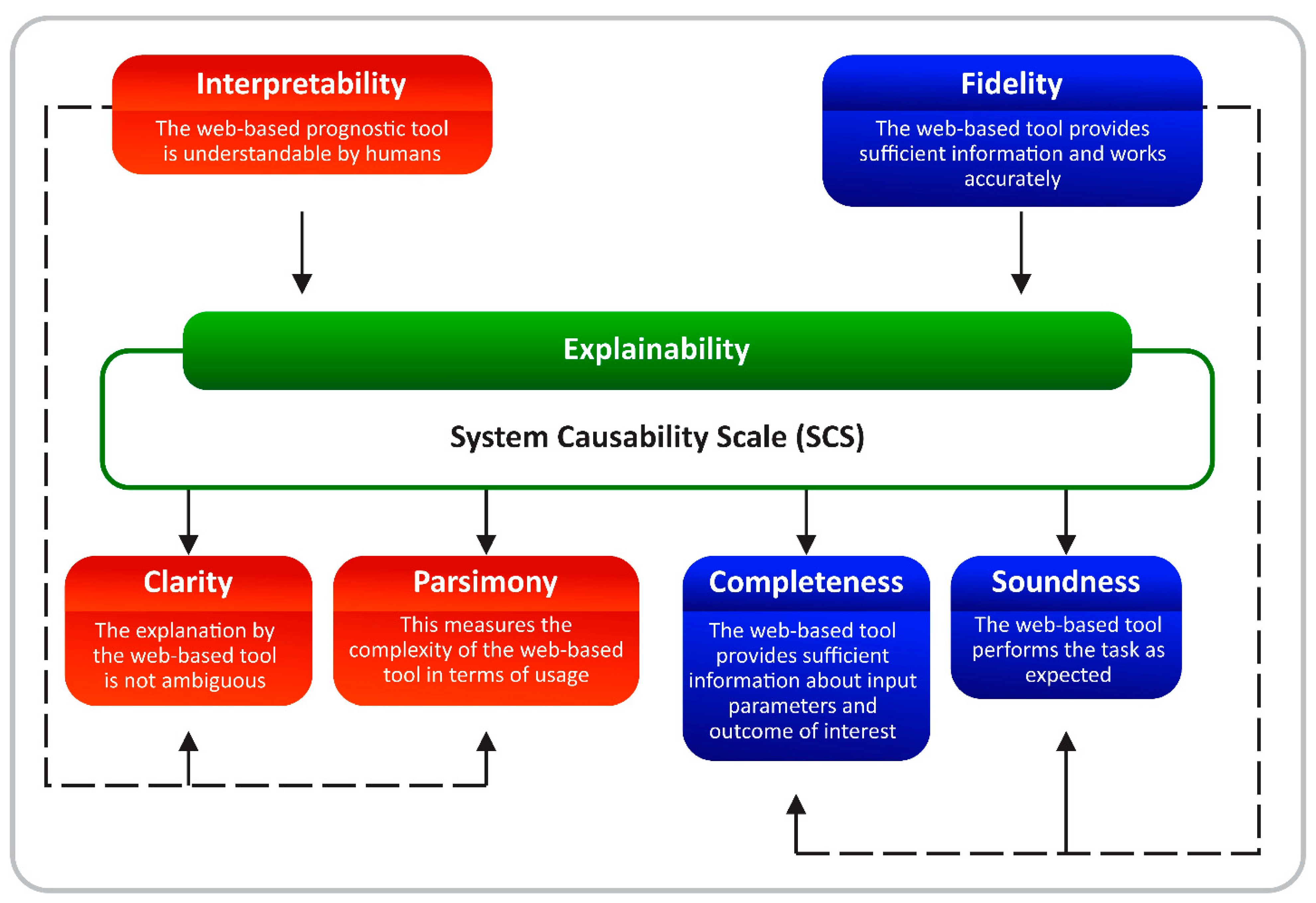
2.3. Measuring the Quality of Explanations
3. Results
3.1. Participants Description
3.2. Usability and Explainability
3.3. Fidelity and Interpretability
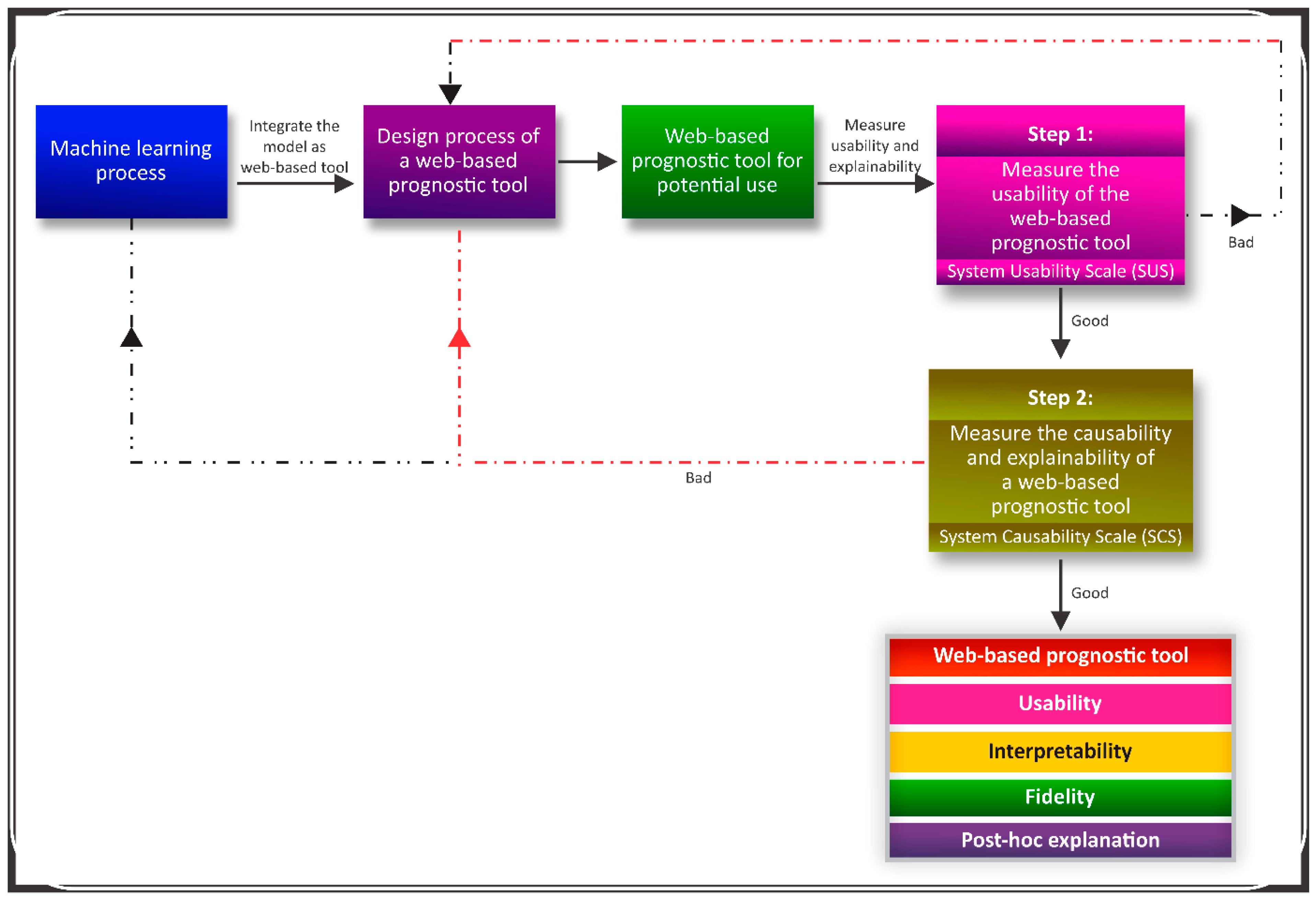
3.4. Framework for an Explainable Web-Based Tool
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alabi, R.O.; Elmusrati, M.; Sawazaki-Calone, I.; Kowalski, L.P.; Haglund, C.; Coletta, R.D.; Mäkitie, A.A.; Salo, T.; Leivo, I.; Almangush, A. Machine Learning Application for Prediction of Locoregional Recurrences in Early Oral Tongue Cancer: A Web-Based Prognostic Tool. Virchows Arch. 2019, 475, 489–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alabi, R.O.; Elmusrati, M.; Sawazaki-Calone, I.; Kowalski, L.P.; Haglund, C.; Coletta, R.D.; Mäkitie, A.A.; Salo, T.; Almangush, A.; Leivo, I. Comparison of Supervised Machine Learning Classification Techniques in Prediction of Locoregional Recurrences in Early Oral Tongue Cancer. Int. J. Med. Inform. 2019, 136, 104068. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Chen, Y.; Li, Z.; Jing, Q.; Hu, C.; Liu, H.; Bao, J.; Hong, Y.; Shi, T.; Li, K.; et al. A Deep Learning Algorithm for Detection of Oral Cavity Squamous Cell Carcinoma from Photographic Images: A Retrospective Study. EClinicalMedicine 2020, 27, 100558. [Google Scholar] [CrossRef]
- Jubair, F.; Al-karadsheh, O.; Malamos, D.; Al Mahdi, S.; Saad, Y.; Hassona, Y. A Novel Lightweight Deep Convolutional Neural Network for Early Detection of Oral Cancer. Oral Diseases 2021, 28, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Sultan, A.S.; Elgharib, M.A.; Tavares, T.; Jessri, M.; Basile, J.R. The Use of Artificial Intelligence, Machine Learning and Deep Learning in Oncologic Histopathology. J. Oral Pathol. Med. 2020, 49, 849–856. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Ghafoorian, M.; Karssemeijer, N.; Heskes, T.; van Uden, I.W.M.; Sanchez, C.I.; Litjens, G.; de Leeuw, F.-E.; van Ginneken, B.; Marchiori, E.; Platel, B. Location Sensitive Deep Convolutional Neural Networks for Segmentation of White Matter Hyperintensities. Sci. Rep. 2017, 7, 5110. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.N.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, Comparison, and Combination of Algorithms for Automatic Detection of Pulmonary Nodules in Computed Tomography Images: The LUNA16 Challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Alabi, R.O.; Tero, V.; Mohammed, E. Machine Learning for Prognosis of Oral Cancer: What Are the Ethical Challenges? CEUR-Workshop Proc. 2020, 2737, 1–22. [Google Scholar]
- Alabi, R.O.; Youssef, O.; Pirinen, M.; Elmusrati, M.; Mäkitie, A.A.; Leivo, I.; Almangush, A. Machine Learning in Oral Squamous Cell Carcinoma: Current Status, Clinical Concerns and Prospects for Future—A Systematic Review. Artif. Intell. Med. 2021, 115, 102060. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Carrington, A.; Müller, H. Measuring the Quality of Explanations: The System Causability Scale (SCS): Comparing Human and Machine Explanations. KI—Künstliche Intell. 2020, 34, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrington, A.M. Kernel Methods and Measures for Classifcation with Transparency, Interpretability and Accuracy in Health Care. Ph.D. Theis, University of Waterloo, Waterloo, ON, Canada, 2018. [Google Scholar]
- Markus, A.F.; Kors, J.A.; Rijnbeek, P.R. The Role of Explainability in Creating Trustworthy Artificial Intelligence for Health Care: A Comprehensive Survey of the Terminology, Design Choices, and Evaluation Strategies. J. Biomed. Inform. 2021, 113, 103655. [Google Scholar] [CrossRef]
- Chander, A.; Srinivasan, R. Evaluating Explanations by Cognitive Value. In Machine Learning and Knowledge Extraction; Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11015, pp. 314–328. ISBN 978-3-319-99739-1. [Google Scholar]
- Lou, Y.; Caruana, R.; Gehrke, J. Intelligible Models for Classification and Regression. In Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining—KDD ’12, Beijing, China, 12–16 August 2012; p. 150. [Google Scholar]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Lundberg, M.; Leivo, I.; Saarilahti, K.; Mäkitie, A.A.; Mattila, P.S. Increased Incidence of Oropharyngeal Cancer and P16 Expression. Acta Oto-Laryngol. 2011, 131, 1008–1011. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. arXiv 2016, arXiv:1602.04938. [Google Scholar]
- Narayanan, M.; Chen, E.; He, J.; Kim, B.; Gershman, S.; Doshi-Velez, F. How Do Humans Understand Explanations from Machine Learning Systems? An Evaluation of the Human-Interpretability of Explanation. arXiv 2018, arXiv:1802.00682. [Google Scholar]
- Almangush, A.; Alabi, R.O.; Mäkitie, A.A.; Leivo, I. Machine Learning in Head and Neck Cancer: Importance of a Web-Based Prognostic Tool for Improved Decision Making. Oral Oncol. 2021, 124, 105452. [Google Scholar] [CrossRef]
- Usability.Gov System Usability Scale (SUS). 2020. Available online: https://www.usability.gov/how-to-and-tools/methods/system-usability-scale.html (accessed on 18 May 2022).
- Brooke, J. SUS: A Quick and Dirty Usability Scale. In Usability Evaluation in Industry; Jordan, P.W., Thomas, B., Weerdmeester, B.A., McClelland, I.L., Eds.; Taylor and Francis: London, UK, 1996. [Google Scholar]
- Miller, T. Explanation in Artificial Intelligence: Insights from the Social Sciences. Artif. Intell. 2019, 267, 1–38. [Google Scholar] [CrossRef]
- Cabitza, F.; Campagner, A.; Ciucci, D. New Frontiers in Explainable AI: Understanding the GI to Interpret the GO. In Machine Learning and Knowledge Extraction; Holzinger, A., Kieseberg, P., Tjoa, A.M., Weippl, E., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11713, pp. 27–47. ISBN 978-3-030-29725-1. [Google Scholar]
- Kulesza, T.; Stumpf, S.; Burnett, M.; Yang, S.; Kwan, I.; Wong, W.-K. Too Much, Too Little, or Just Right? Ways Explanations Impact End Users’ Mental Models. In Proceedings of the 2013 IEEE Symposium on Visual Languages and Human Centric Computing; IEEE, San Jose, CA, USA, 15–19 September 2013; pp. 3–10. [Google Scholar]
- Ras, G.; van Gerven, M.; Haselager, P. Explanation Methods in Deep Learning: Users, Values, Concerns and Challenges. In Explainable and Interpretable Models in Computer Vision and Machine Learning; Escalante, H.J., Escalera, S., Guyon, I., Baró, X., Güçlütürk, Y., Güçlü, U., van Gerven, M., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 19–36. ISBN 978-3-319-98130-7. [Google Scholar]
- Lipton, Z.C. The Mythos of Model Interpretability: In Machine Learning, the Concept of Interpretability Is Both Important and Slippery. Queue 2018, 16, 31–57. [Google Scholar] [CrossRef]
- Gilpin, L.H.; Bau, D.; Yuan, B.Z.; Bajwa, A.; Specter, M.; Kagal, L. Explaining Explanations: An Overview of Interpretability of Machine Learning. In Proceedings of the 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA); IEEE, Turin, Italy, 1–3 October 2018; pp. 80–89. [Google Scholar]
- Heuillet, A.; Couthouis, F.; Díaz-Rodríguez, N. Explainability in Deep Reinforcement Learning. Knowl.-Based Syst. 2021, 214, 106685. [Google Scholar] [CrossRef]
- Langer, M.; Oster, D.; Speith, T.; Hermanns, H.; Kästner, L.; Schmidt, E.; Sesing, A.; Baum, K. What Do We Want from Explainable Artificial Intelligence (XAI)?—A Stakeholder Perspective on XAI and a Conceptual Model Guiding Interdisciplinary XAI Research. Artif. Intell. 2021, 296, 103473. [Google Scholar] [CrossRef]
- Doshi-Velez, F.; Kim, B. Towards A Rigorous Science of Interpretable Machine Learning. arXiv 2017, arXiv:1702.08608. [Google Scholar]
- Hoffman, R.R.; Mueller, S.T.; Klein, G.; Litman, J. Metrics for Explainable AI: Challenges and Prospects. arXiv 2019, arXiv:1812.04608. [Google Scholar]
- Guidotti, R.; Monreale, A.; Ruggieri, S.; Turini, F.; Giannotti, F.; Pedreschi, D. A Survey of Methods for Explaining Black Box Models. ACM Comput. Surv. 2019, 51, 1–42. [Google Scholar] [CrossRef] [Green Version]
- Arambula, A.M.; Bur, A.M. Ethical Considerations in the Advent of Artificial Intelligence in Otolaryngology. Otolaryngol. Head Neck Surg. 2020, 162, 38–39. [Google Scholar] [CrossRef]
- European Commission. High-Level Expert Group on Artificial Intelligence. In Ethics Guidelines for Tustrworthy AI; European Commission: Brussels, Belgium, 2019. [Google Scholar]
- Keskinbora, K.H. Medical Ethics Considerations on Artificial Intelligence. J. Clin. Neurosci. 2019, 64, 277–282. [Google Scholar] [CrossRef]
- Select Committee on Artificial Intelligence. The National Artificial Intelligence Research and Development Strategic Plan: 2019 Update; National Science and Technology Council: Washington, DC, USA, 2019.
- Jamieson, S. Likert Scales: How to (Ab)Use Them. Med. Educ. 2004, 38, 1217–1218. [Google Scholar] [CrossRef]


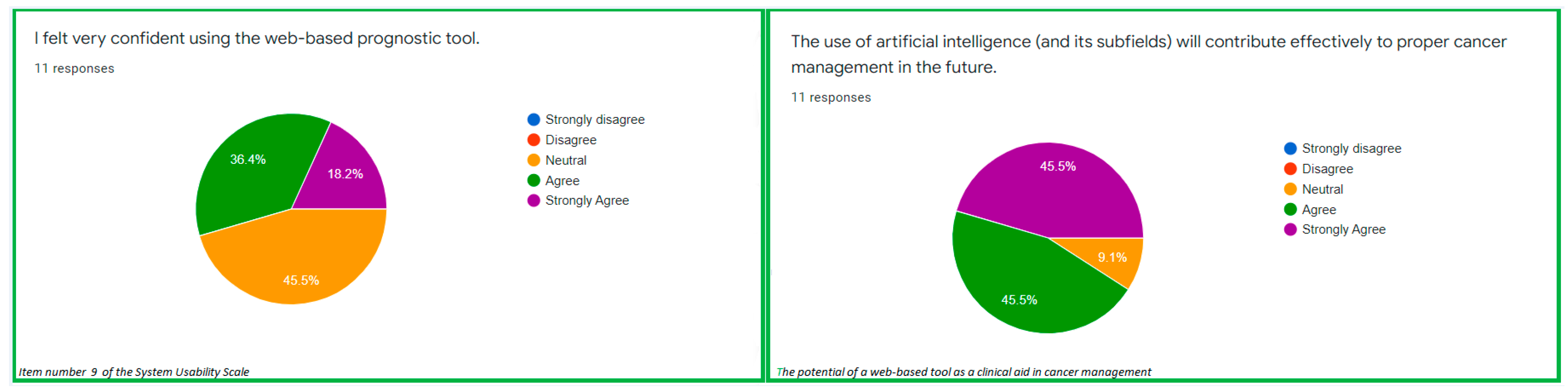

| S/N | System Usability Scale (SUS) | Theme from the SUS | Ratings (%) |
|---|---|---|---|
| 1 | I think that I would like to use this web-based prognostic tool in cancer management. | Readiness to use | Strongly agree: 27.3% Agree: 27.3% Neutral: 27.3% Disagree: 9.1% Strongly disagree: 9.1% |
| 2 | I found the web-based tool unnecessarily complex. | Web-based tool simplicity | Neutral: 18.2% Disagree: 45.5% Strongly disagree: 36.4% |
| 3 | I thought the web-based tool was easy to use. | Ease of use of the web-based tool | Strongly agree: 27.3% Agree: 63.6% Disagree: 9.1% |
| 4 | I think that I would need the support of a technical person to be able to use this web-based prognostic tool. | Need teacher/support to use the web-based tool | Agree: 9.1% Neutral: 9.1% Disagree: 45.5% Strongly disagree: 36.4% |
| 5 | I found the various functions in this web-based prognostic tool to be well integrated. | Understanding the input parameters | Strongly agree: 18.2% Agree: 63.6% Neutral: 9.1% Disagree: 9.1% |
| 6 | I thought there was too much inconsistency in this web-based prognostic tool. | Clarity of the web-based tool | Neutral: 27.3% Disagree: 54.5% Strongly disagree: 18.2% |
| 7 | I would imagine that most people would learn to use this web-based prognostic tool very quickly. | No technicality required | Strongly agree: 36.4% Agree: 45.5% Neutral: 9.1% Disagree: 9.1% |
| 8 | I found the web-based prognostic tool very cumbersome to use. | Less cumbersome tool | Agree: 9.1% Disagree: 81.8% Strongly disagree: 9.1% |
| 9 | I felt very confident using the web-based prognostic tool. | Usability confidence | Strongly agree: 18.2% Agree: 36.4% Neutral: 45.5% |
| 10 | I needed to learn a lot of things before I could get going with this web-based prognostic tool. | Easy to use for everyone | Strongly agree: 18.2% Neutral: 45.5% Disagree: 36.4% |
| S/N | System Causability Scale (SCS) | Theme from the SCS | Ratings Rating (Participants, n = 11) | Score |
|---|---|---|---|---|
| 1 | I found that the prognostic parameters included all relevant known causal factors with sufficient precision and detailed information regarding locoregional recurrence of oral cancer | Causality factors in the data | Agree: 4(6) = 24 Neutral: 3(3) = 9 Disagree: 2(2) = 4 | 37 |
| 2 | I understood the explanations within the context of my work. | Understood the explanations of the web-based tool | Agree: 4(9) = 36 Neutral: 3(1) = 3 Disagree: 2(1) = 2 | 41 |
| 3 | I could change the level of values for each of the prognostic parameters on the web-based tool. | Change in detail level of income parameters | Strongly agree: 5(2) = 10 Agree: 4(5) = 20 Neutral: 3(2) = 6 Disagree: 2(2) = 4 | 40 |
| 4 | I did not need support to understand the explanations provided in the prediction given by the web-based tool. | Need teacher/support to use the web-based tool | Strongly agree: 5(3) = 15 Agree: 4(6) = 24 Neutral: 3(1) = 3 Disagree: 2(1) = 2 | 44 |
| 5 | I found the predictions by the web-based tool helped me to understand causality (cause and effect regarding recurrence of cancer) | Understanding causality based on the inputs and predicted outcome | Strongly agree: 5(1) = 5 Agree: 4(4) = 16 Neutral: 3(4) = 12 Disagree: 2(2) = 4 | 37 |
| 6 | I was able to use the explanations of the prediction by the web-based tool with my knowledge base. | Usage of the tool with knowledge with knowledge | Strongly agree: 5(2) = 10 Agree: 4(6) = 24 Neutral: 3(3) = 9 | 43 |
| 7 | I did not find inconsistencies between explanations provided by the web-based prognostic tool. | No inconsistencies from the web-based tool | Strongly agree: 5(1) = 5 Agree: 4(6) = 24 Neutral: 3(3) = 9 Disagree: 2(1) = 2 | 40 |
| 8 | I think that most people would learn to understand the predictions/explanations provided by the web-based tool very quickly. | Learn to understand | Strongly agree: 5(1) = 5 Agree: 4(9) = 36 Neutral: 3(1) = 3 | 44 |
| 9 | I did not need more references in the explanations: e.g., medical guidelines, regulations to understand the web-based prognostic tool. | Needs references to use the web-based tool | Strongly agree: 5(2) = 10 Agree: 4(4) = 16 Neutral: 3(2) = 6 Disagree: 2(2) = 4 Strongly disagree: 1(1) = 1 | 37 |
| 10 | I received the explanations and prediction of locoregional recurrence in a timely and efficient manner. | Timely and efficient prediction of outcome | Strongly agree: 5(1) = 5 Agree: 4(6) = 24 Neutral: 3(4) = 12 | 41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alabi, R.O.; Almangush, A.; Elmusrati, M.; Leivo, I.; Mäkitie, A. Measuring the Usability and Quality of Explanations of a Machine Learning Web-Based Tool for Oral Tongue Cancer Prognostication. Int. J. Environ. Res. Public Health 2022, 19, 8366. https://doi.org/10.3390/ijerph19148366
Alabi RO, Almangush A, Elmusrati M, Leivo I, Mäkitie A. Measuring the Usability and Quality of Explanations of a Machine Learning Web-Based Tool for Oral Tongue Cancer Prognostication. International Journal of Environmental Research and Public Health. 2022; 19(14):8366. https://doi.org/10.3390/ijerph19148366
Chicago/Turabian StyleAlabi, Rasheed Omobolaji, Alhadi Almangush, Mohammed Elmusrati, Ilmo Leivo, and Antti Mäkitie. 2022. "Measuring the Usability and Quality of Explanations of a Machine Learning Web-Based Tool for Oral Tongue Cancer Prognostication" International Journal of Environmental Research and Public Health 19, no. 14: 8366. https://doi.org/10.3390/ijerph19148366
APA StyleAlabi, R. O., Almangush, A., Elmusrati, M., Leivo, I., & Mäkitie, A. (2022). Measuring the Usability and Quality of Explanations of a Machine Learning Web-Based Tool for Oral Tongue Cancer Prognostication. International Journal of Environmental Research and Public Health, 19(14), 8366. https://doi.org/10.3390/ijerph19148366






