In Silico Insights on the Pro-Inflammatory Potential of Polycyclic Aromatic Hydrocarbons and the Prospective Anti-Inflammatory Capacity of Andrographis paniculata Phytocompounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Retrieval and Preparation of PAH Ligand Structures
2.2. Retrieval and Preparation of AP Ligand Structures
2.3. Retrieval of Protein Structures and Validation
2.4. Molecular Docking
2.5. Molecular Dynamics Simulation (MDS)
3. Results
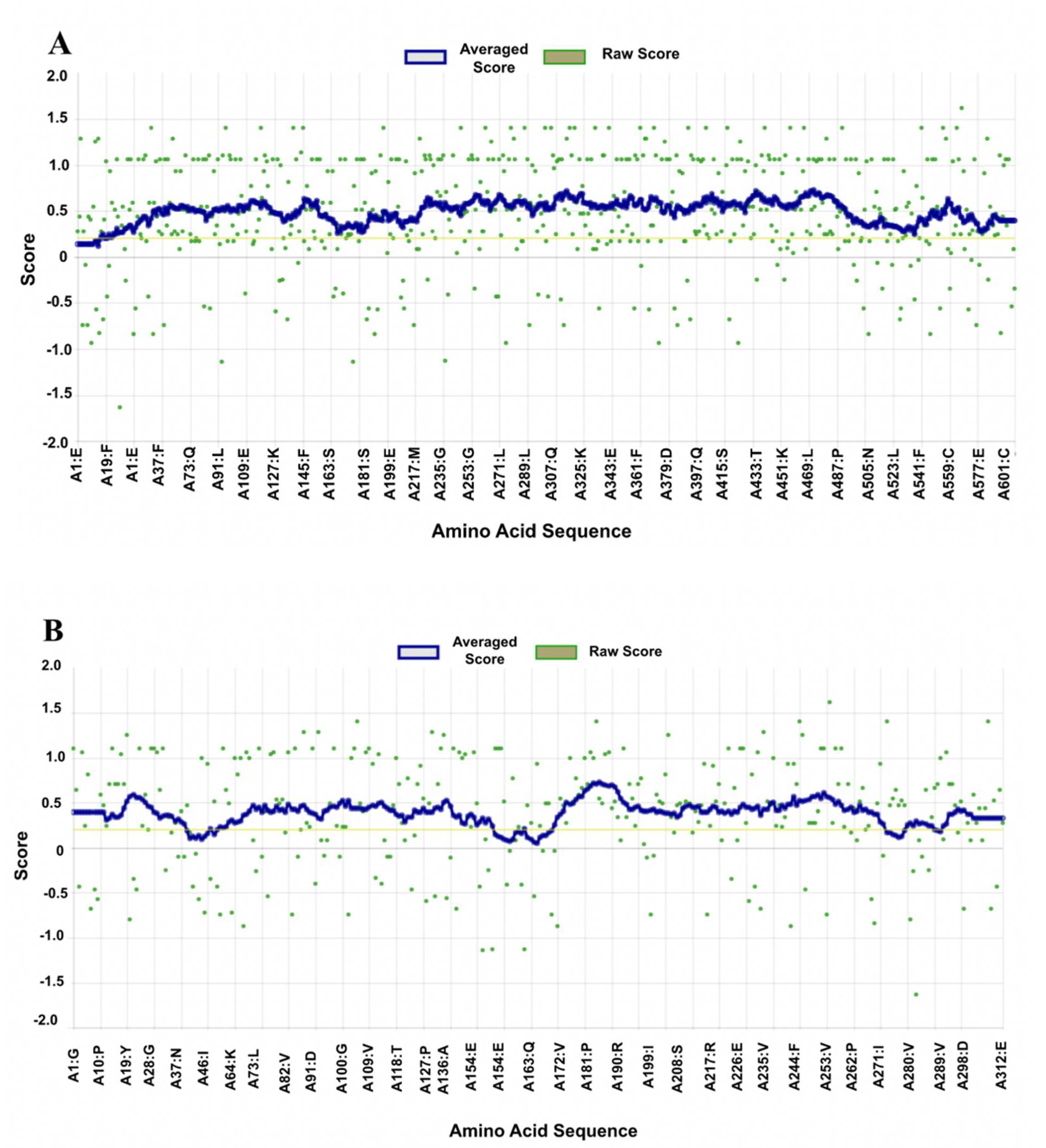
3.1. Structural Stability of Prepared Proteins
3.2. PAHs and Human TLR4 Interaction
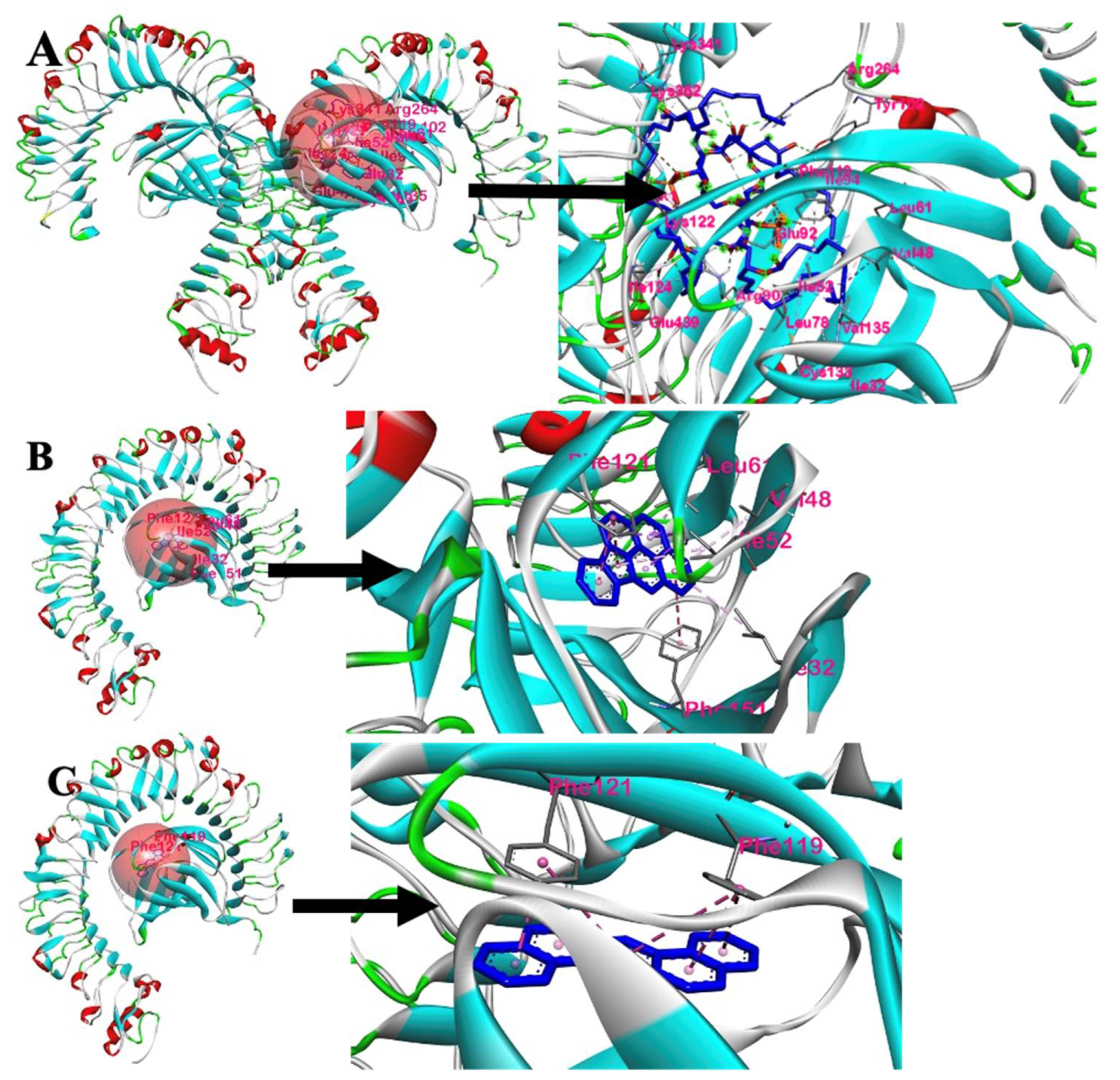
3.2.1. Analysis of Docking Scores and Amino Acid Interactions
3.2.2. Intermolecular Forces of Attraction
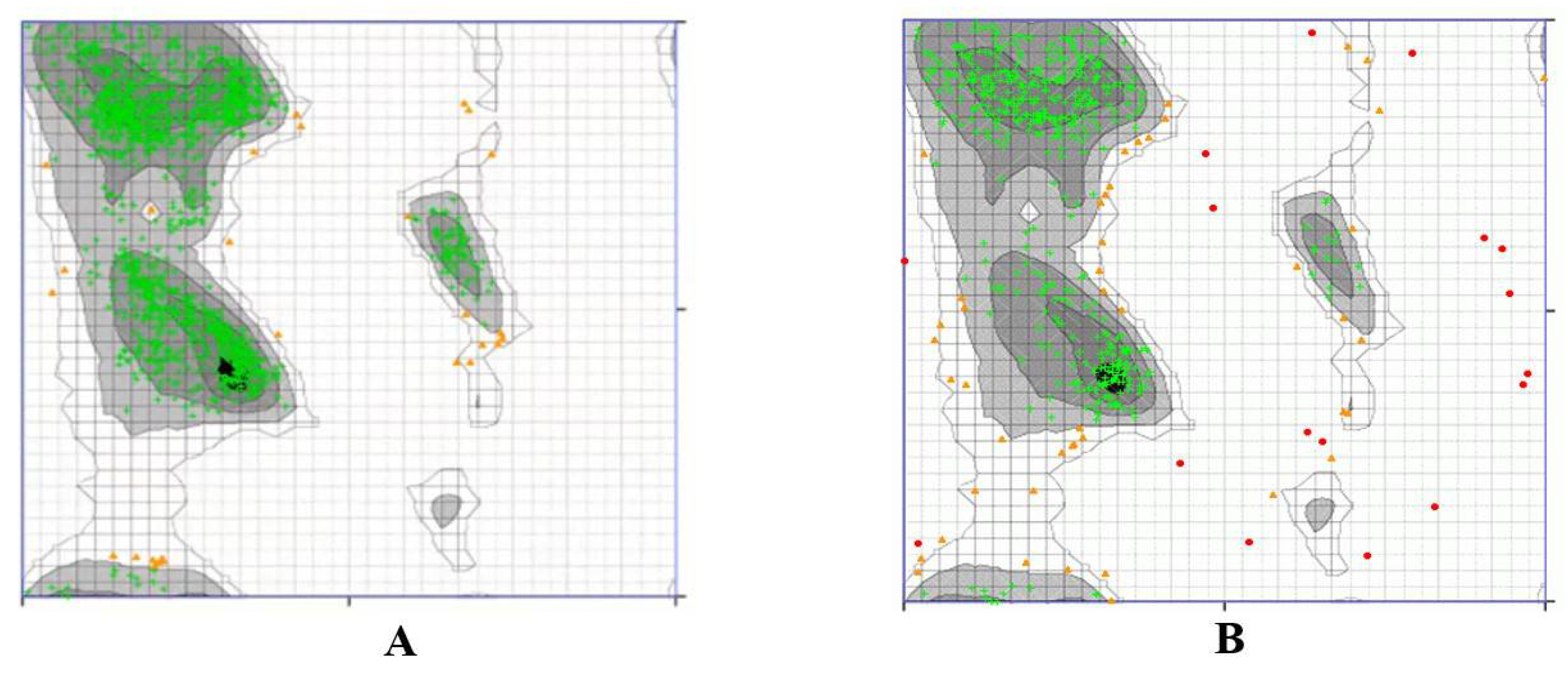
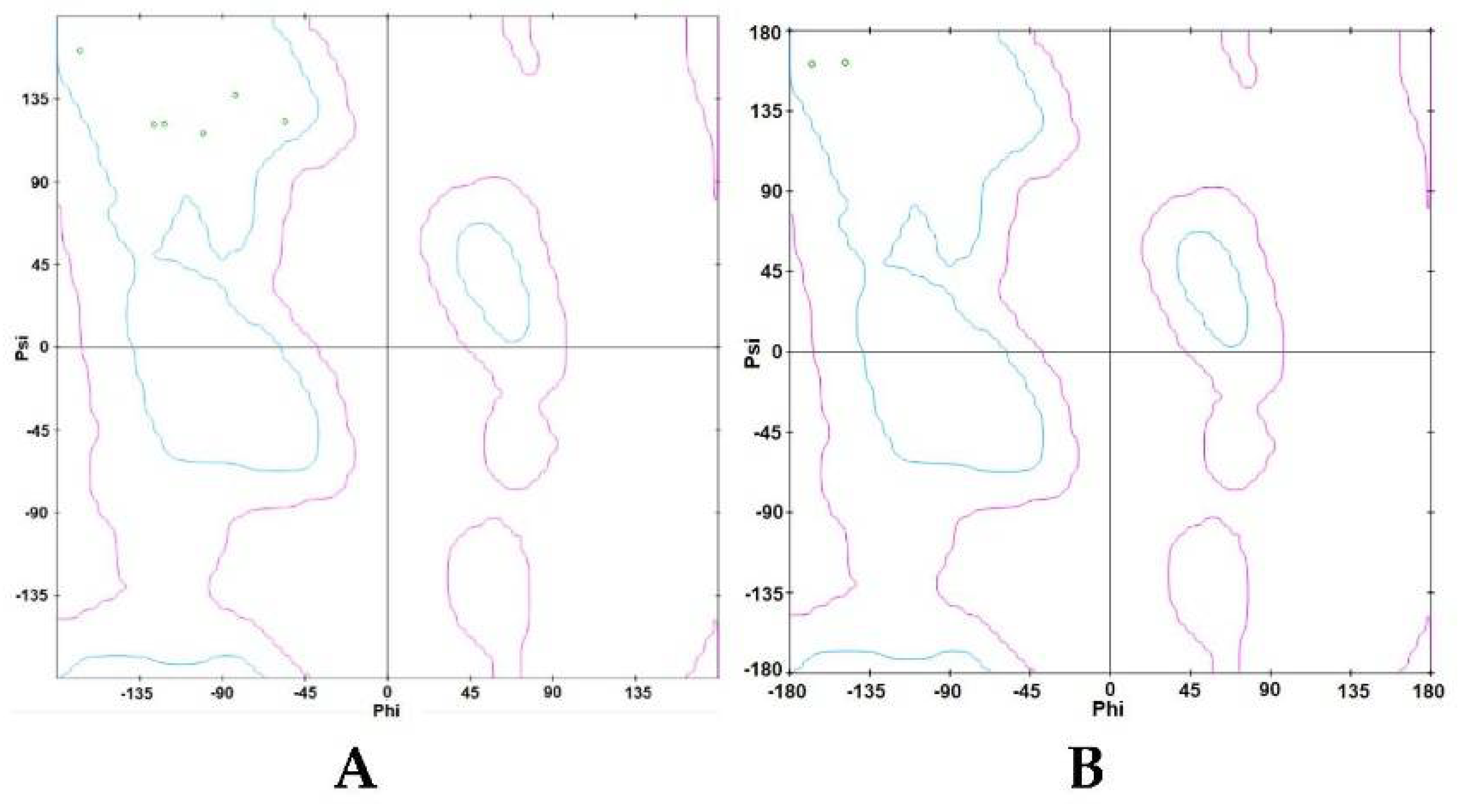
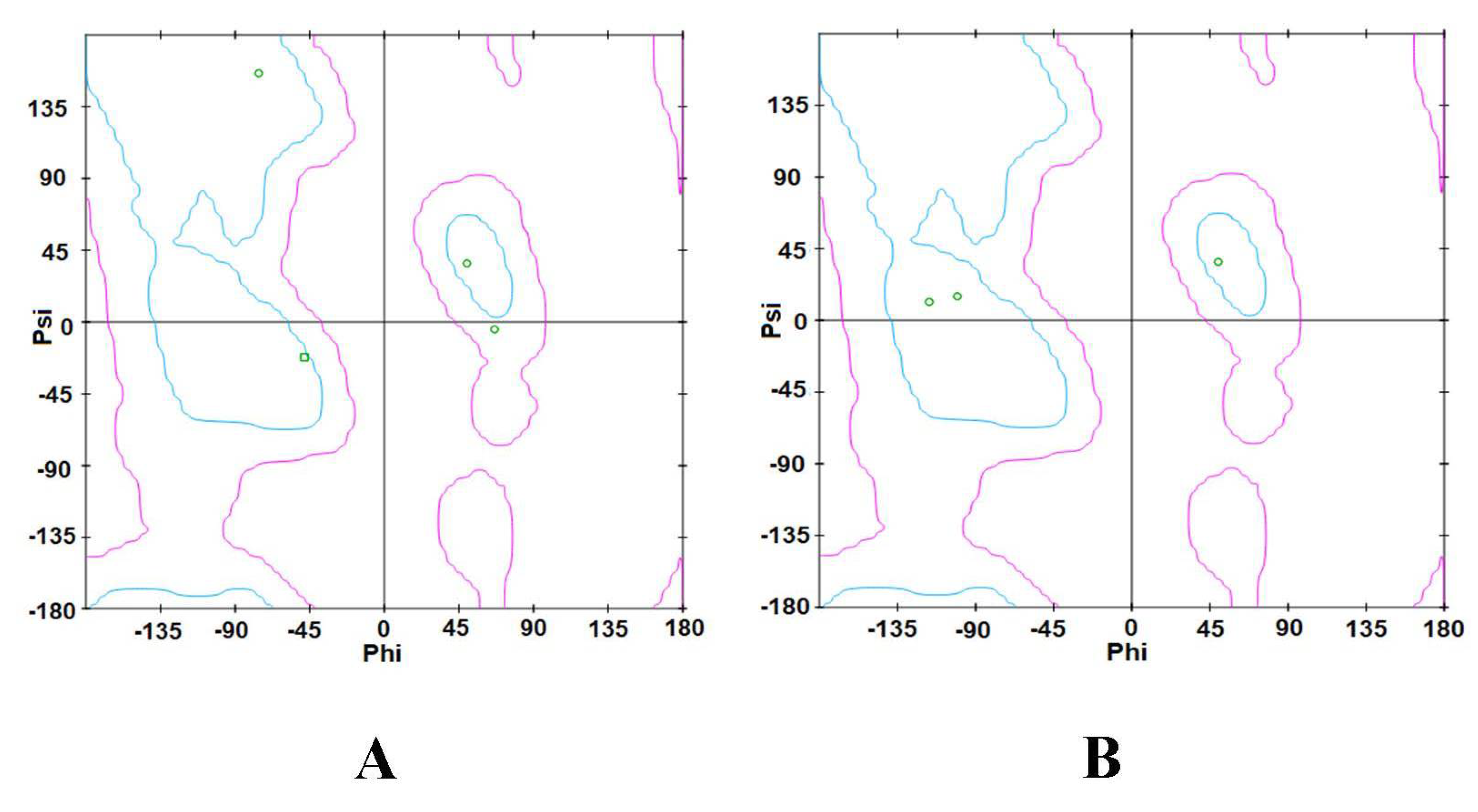
3.2.3. Ramachandran Plot
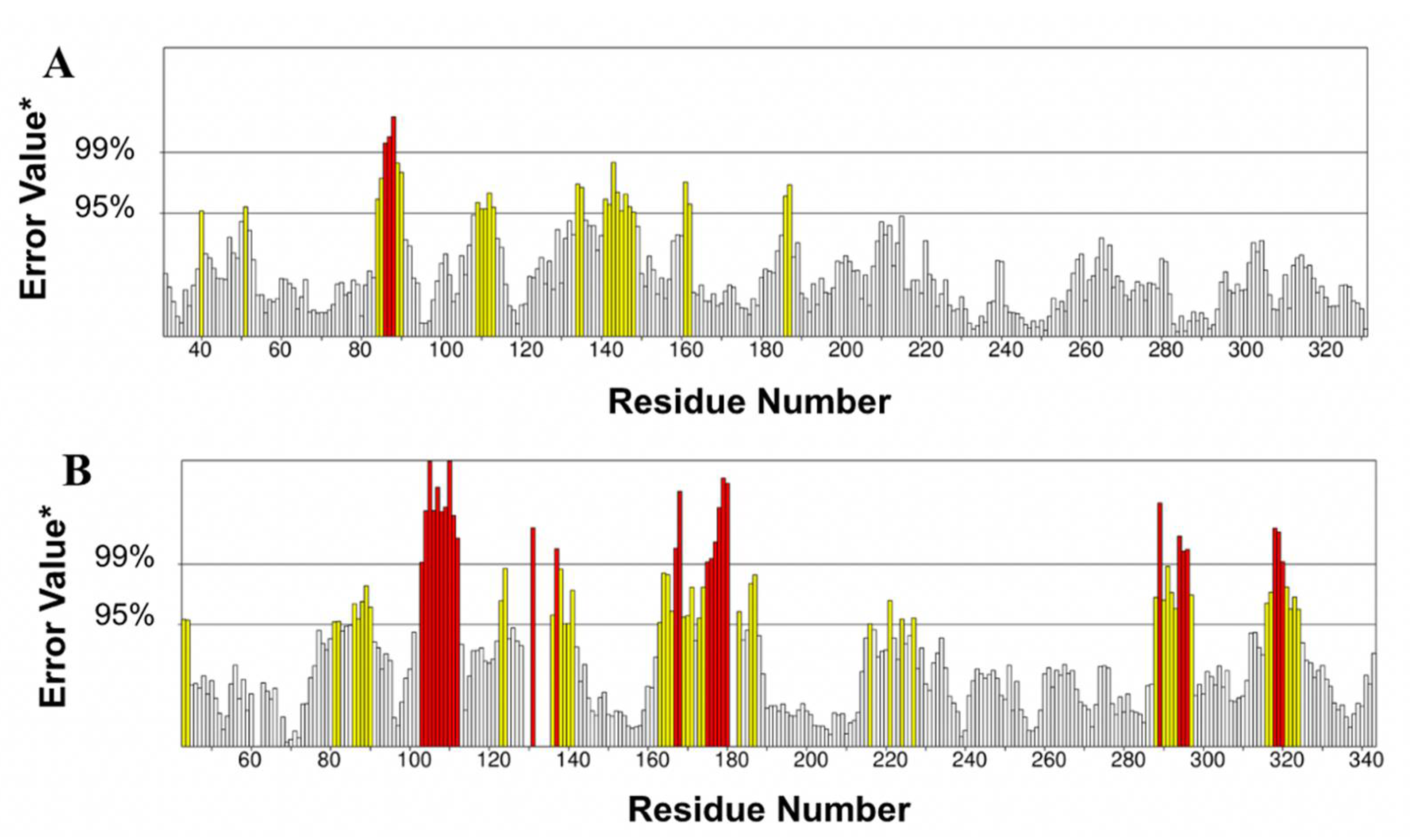
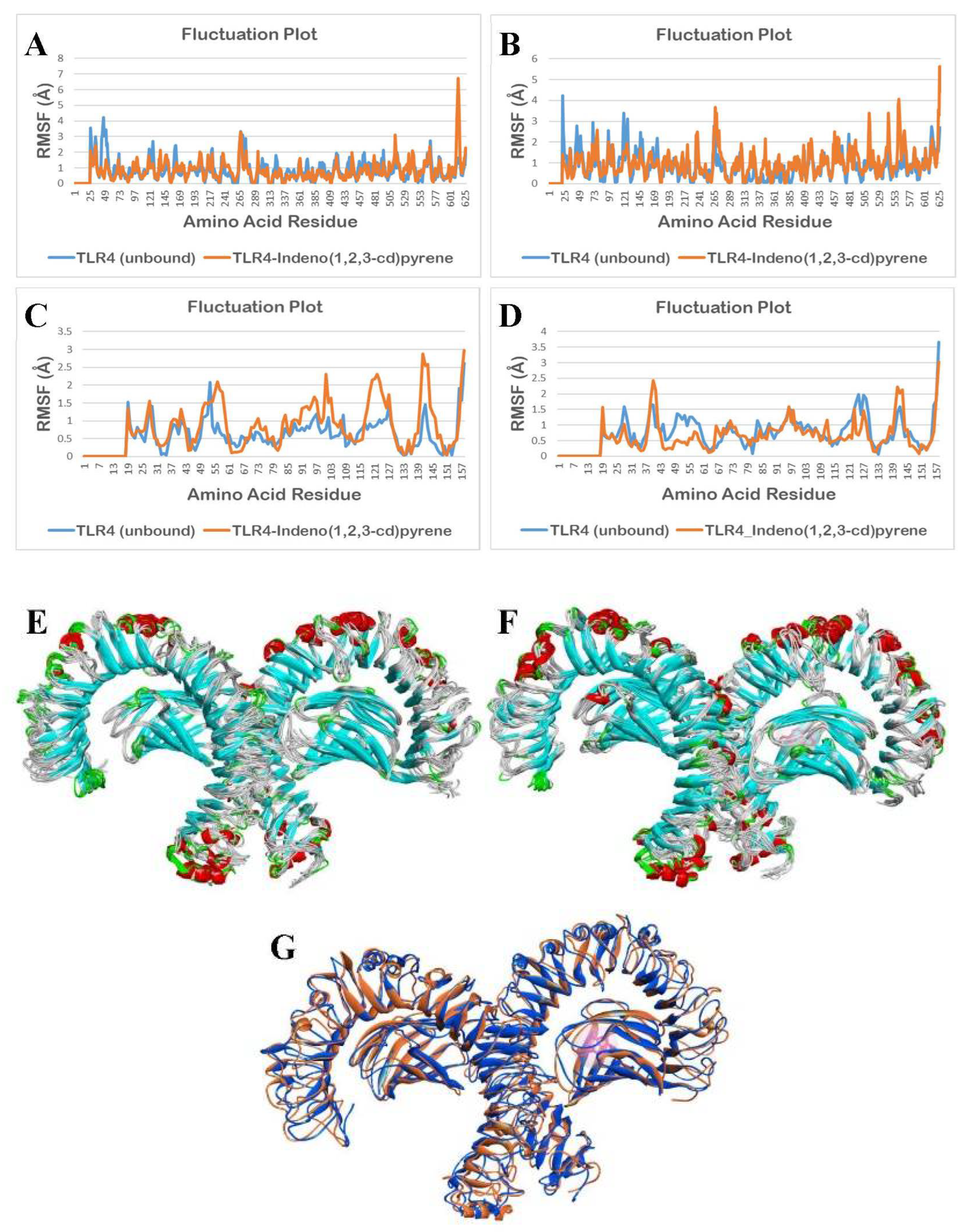
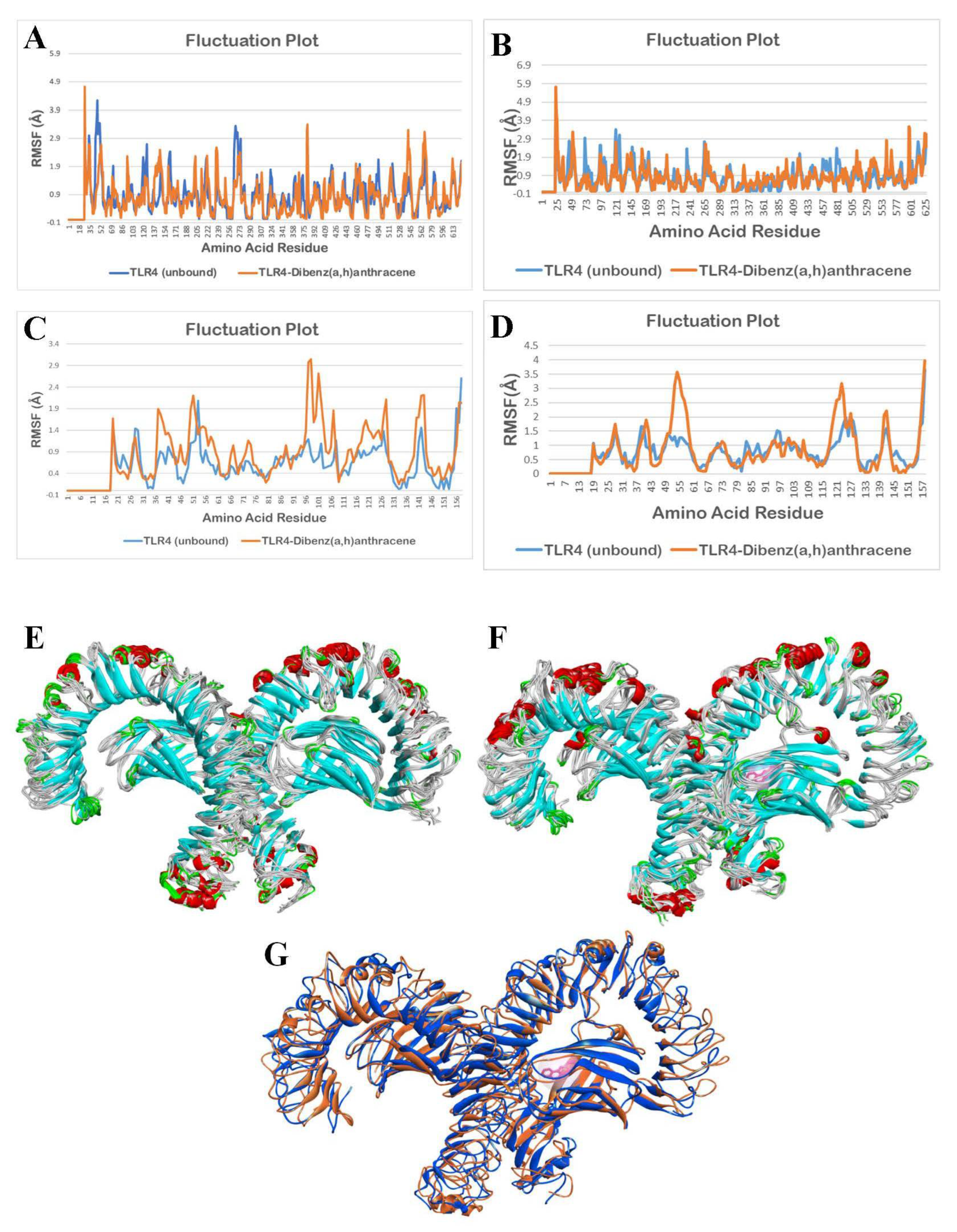
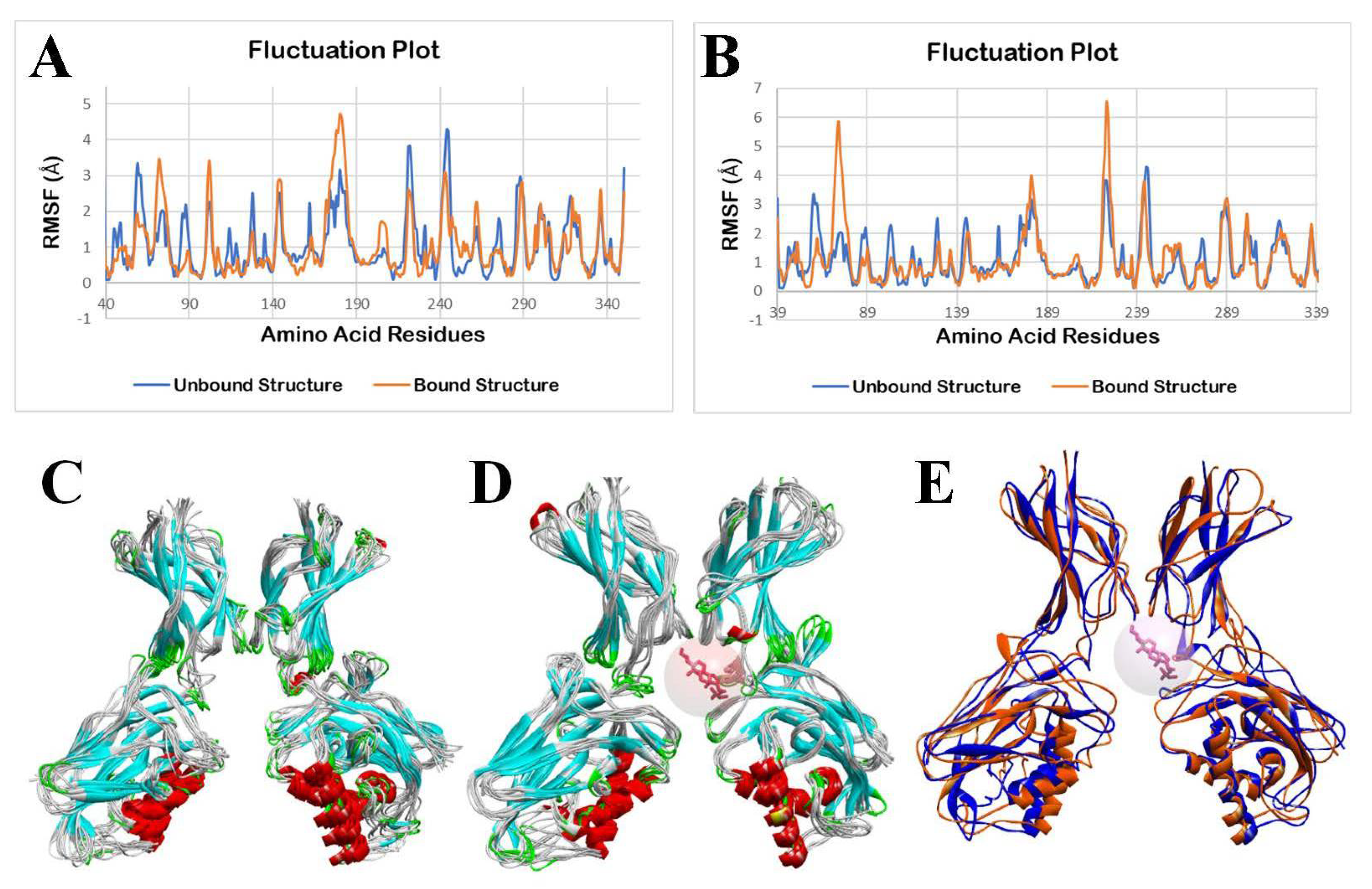
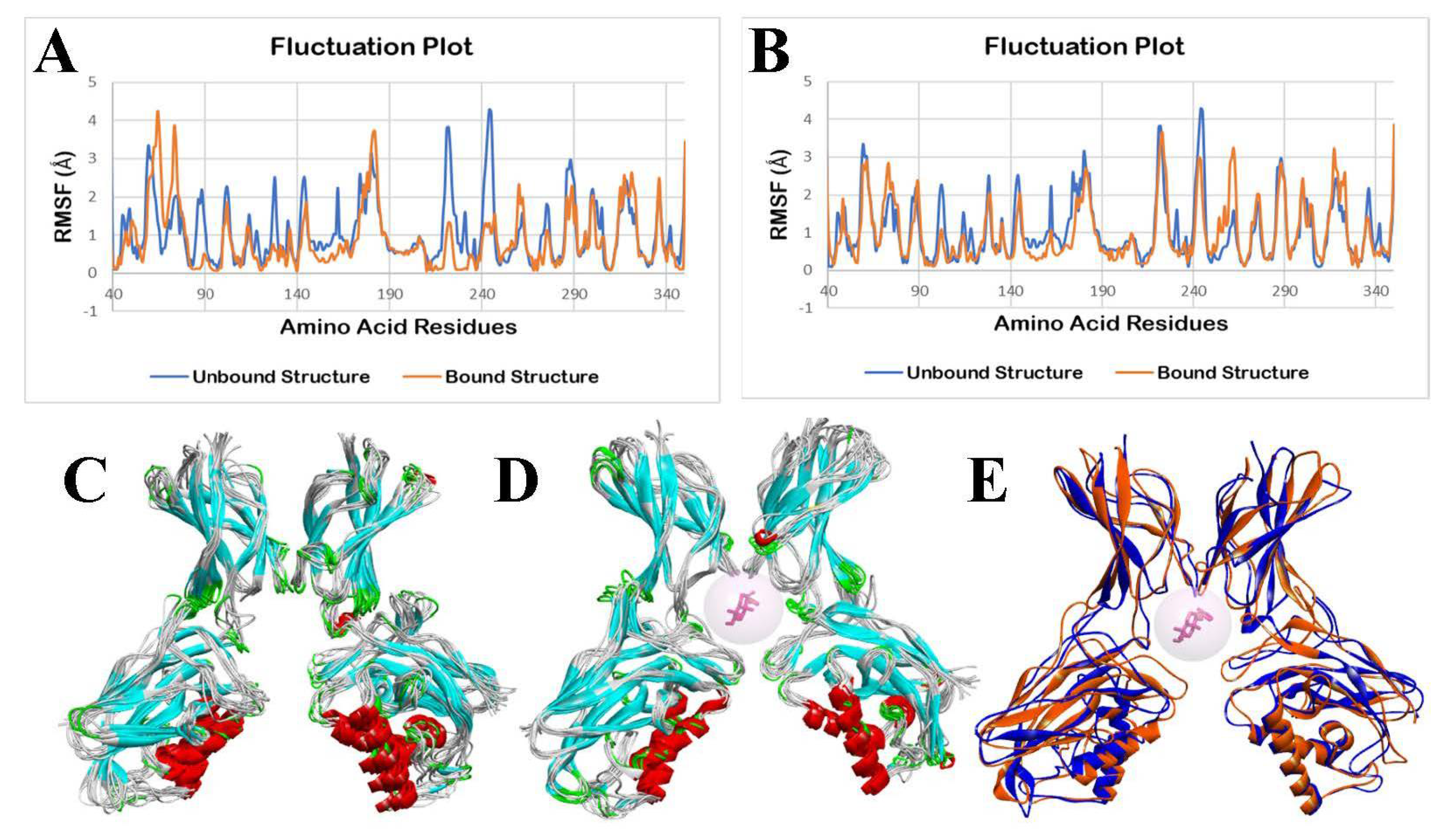
3.2.4. Molecular Dynamics Simulation
3.3. AP Phytocompounds and NF-κB p50 Interaction
3.3.1. Analysis of Docking Scores and Amino Acid Interactions
3.3.2. Intermolecular Forces of Attraction
3.3.3. Ramachandran Plot
3.3.4. Molecular Dynamics Simulation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Han, Y.; Qiu, X.; Wang, Y.; Li, W.; Liu, J.; Chen, X.; Li, R.; Xu, F.; Chen, W.; et al. Association of Internal Exposure to Polycyclic Aromatic Hydrocarbons with Inflammation and Oxidative Stress in Prediabetic and Healthy Individuals. Chemosphere 2020, 253, 126748. [Google Scholar] [CrossRef] [PubMed]
- Santiago, E.C.; Cayetano, M.G. Polycyclic Aromatic Hydrocarbons in Ambient Air in the Philippines Derived from Passive Sampler with Polyurethane Foam Disk. Atmos. Environ. 2007, 41, 4138–4147. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, D.; Zhang, L.; Zhou, S. Spatial Distribution of Polycyclic Aromatic Hydrocarbons in the Philippine Sea, Western Pacific and the Impact Factors Analysis. Mar. Pollut. Bull. 2021, 173, 113083. [Google Scholar] [CrossRef] [PubMed]
- Chavez, L. Philippines Declares No New Coal Plants—But Lets Approved Projects Through. Mongabay. 5 November 2020. Available online: https://news.mongabay.com/2020/11/philippines-declares-no-new-coal-plants-but-lets-approved-projects-through/ (accessed on 10 July 2022).
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic Inflammation in the Etiology of Disease across the Life Span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.S.D.; Liao, W.; Zhou, S.; Wong, W.S.F. Is There a Future for Andrographolide to Be an Anti-Inflammatory Drug? Deciphering Its Major Mechanisms of Action. Biochem. Pharmacol. 2017, 139, 71–81. [Google Scholar] [CrossRef]
- Tarachand, S.P.; Thirumoorthy, G.; Lakshmaiah, V.V.; Nagella, P. In Silico Molecular Docking Study of Andrographis Paniculata Phytochemicals against TNF-α as a Potent Anti-Rheumatoid Drug. J. Biomol. Struct. Dyn. 2022, 1–11. [Google Scholar] [CrossRef]
- Okhuarobo, A.; Ehizogie Falodun, J.; Erharuyi, O.; Imieje, V.; Falodun, A.; Langer, P. Harnessing the Medicinal Properties of Andrographis Paniculata for Diseases and beyond: A Review of Its Phytochemistry and Pharmacology. Asian Pac. J. Trop. Dis. 2014, 4, 213–222. [Google Scholar] [CrossRef]
- Yan, Y.; Fang, L.-H.; Du, G.-H. Andrographolide. In Natural Small Molecule Drugs from Plants; Springer: Singapore, 2018; pp. 357–362. ISBN 978-981-10-8021-0. [Google Scholar]
- Richard, E.J.; Murugan, S.; Bethapudi, B.; Illuri, R.; Mundkinajeddu, D.; Velusami, C.C. Is Andrographis Paniculata Extract and Andrographolide Anaphylactic? Toxicol. Rep. 2017, 4, 431–437. [Google Scholar] [CrossRef]
- Chao, W.-W.; Kuo, Y.-H.; Hsieh, S.-L.; Lin, B.-F. Inhibitory Effects of Ethyl Acetate Extract of Andrographis Paniculata on NF-κ B Trans-Activation Activity and LPS-Induced Acute Inflammation in Mice. Evid. Based Complement. Alternat. Med. 2011, 2011, 254531. [Google Scholar] [CrossRef] [Green Version]
- Kobori, M.; Yoshida, M.; Ohnishi-Kameyama, M.; Shinmoto, H. Ergosterol Peroxide from an Edible Mushroom Suppresses Inflammatory Responses in RAW264.7 Macrophages and Growth of HT29 Colon Adenocarcinoma Cells: Antitumour, Anti-Inflammation of Ergosterol Peroxide. Br. J. Pharmacol. 2007, 150, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Merdivan, S.; Lindequist, U. Ergosterol Peroxide: A Mushroom-Derived Compound with Promising Biological Activities—A Review. Int. J. Med. Mushrooms 2017, 19, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.-W.; Lin, B.-F. Isolation and Identification of Bioactive Compounds in Andrographis Paniculata (Chuanxinlian). Chin. Med. 2010, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Chao, W.-W.; Kuo, Y.-H.; Lin, B.-F. Anti-Inflammatory Activity of New Compounds from Andrographis Paniculata by NF-ΚB Transactivation Inhibition. J. Agric. Food Chem. 2010, 58, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Xiao, Z.; Wen, X.; Luo, J.; Chen, S.; Cheng, Z.; Xiang, D.; Hu, J.; He, J. The Anti-Inflammatory Effect of Andrographis Paniculata (Burm. f.) Nees on Pelvic Inflammatory Disease in Rats through down-Regulation of the NF-ΚB Pathway. BMC Complement. Altern. Med. 2016, 16, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevelin, S.C.; Sag, C.M.; Zhang, M.; Alves-Filho, J.C.; Cunha, T.M.; Dos Santos, C.X.; Sawyer, G.; Murray, T.; Brewer, A.; Laurindo, F.R.M.; et al. Endothelial Nox2 Limits Systemic Inflammation and Hypotension in Endotoxemia by Controlling Expression of Toll-Like Receptor 4. Shock 2021, 56, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Neupane, R.; McAmis, W.; Singh, U.; Chatterjee, S.; Raychoudhury, S. Toxicity of Polycyclic Aromatic Hydrocarbons Involves NOX2 Activation. Toxicol. Rep. 2019, 6, 1176–1181. [Google Scholar] [CrossRef]
- Shoenfelt, J.; Mitkus, R.J.; Zeisler, R.; Spatz, R.O.; Powell, J.; Fenton, M.J.; Squibb, K.A.; Medvedev, A.E. Involvement of TLR2 and TLR4 in Inflammatory Immune Responses Induced by Fine and Coarse Ambient Air Particulate Matter. J. Leukoc. Biol. 2009, 86, 303–312. [Google Scholar] [CrossRef] [Green Version]
- Jarukamjorn, K. Andrographis paniculata: A review of aspects of regulatory mechanisms of hepatic CYP1A enzymes. Bol. Latinoam. Caribe Plantas Med. Aromáticas 2008, 7, 100–107. [Google Scholar]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-ΚB Signaling in Inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [Green Version]
- Christian, F.; Smith, E.; Carmody, R. The Regulation of NF-ΚB Subunits by Phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Oeckinghaus, A.; Ghosh, S. The NF- B Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Research and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational Protein–Ligand Docking and Virtual Drug Screening with the AutoDock Suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Advisory Note—Classification of Polycyclic Aromatic Hydrocarbons 2012. Available online: https://epa.tas.gov.au/Documents/Advisory%20Note%20for%20Classification%20of%20PAHs.pdf (accessed on 10 July 2022).
- Berry, M.; Fielding, B.; Gamieldien, J. Practical Considerations in Virtual Screening and Molecular Docking. In Emerging Trends in Computational Biology, Bioinformatics, and Systems Biology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 487–502. ISBN 978-0-12-802508-6. [Google Scholar]
- Sreeja, T.G.; Raghunandan, R. Insilico Insight into the Association between Polycyclic Aromatic Hydrocarbons and Human Toll like Receptor in Progression of Esophageal Carcinogenesis. Polycycl. Aromat. Compd. 2021, 1–16. [Google Scholar] [CrossRef]
- Cuadrado, I.; Amesty, Á.; Cedrón, J.; Oberti, J.; Estévez-Braun, A.; Hortelano, S.; de las Heras, B. Semisynthesis and Inhibitory Effects of Solidagenone Derivatives on TLR-Mediated Inflammatory Responses. Molecules 2018, 23, 3197. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Sohn, K.H.; Rhee, S.H.; Hwang, D. Saturated Fatty Acids, but Not Unsaturated Fatty Acids, Induce the Expression of Cyclooxygenase-2 Mediated through Toll-like Receptor 4. J. Biol. Chem. 2001, 276, 16683–16689. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Wang, S.; Zhang, C.; Zhao, Y. Coordinated Modulation of Energy Metabolism and Inflammation by Branched-Chain Amino Acids and Fatty Acids. Front. Endocrinol. 2020, 11, 617. [Google Scholar] [CrossRef]
- Guo, H.; Ahn, S.; Zhang, L. Benzene-Associated Immunosuppression and Chronic Inflammation in Humans: A Systematic Review. Occup. Environ. Med. 2021, 78, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Polycyclic Aromatic Hydrocarbons (PAHs); Government of South Australia SA Health: Adelaide, Australia, 2022.
- Kubik, S.; Mungalpara, D. Amino Acid-Based Receptors. In Comprehensive Supramolecular Chemistry II; Elsevier: Amsterdam, The Netherlands, 2017; pp. 293–310. ISBN 978-0-12-803199-5. [Google Scholar]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-ΚB Pathway for the Therapy of Diseases: Mechanism and Clinical Study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xie, F.; Chen, R.; Zhang, Z.; Dai, R.; Zhao, N.; Wang, R.; Sun, Y.; Chen, Y. Transcription Factor NF-ΚB Promotes Acute Lung Injury via MicroRNA-99b-Mediated PRDM1 down-Regulation. J. Biol. Chem. 2020, 295, 18638–18648. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liang, X.; Feng, Q.; Li, J.; Pan, X.; Xie, P.; Jiang, Z.; Yang, Z. Ergosterol Peroxide Suppresses Influenza A Virus-Induced pro-Inflammatory Response and Apoptosis by Blocking RIG-I Signaling. Eur. J. Pharmacol. 2019, 860, 172543. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.X.; Tran, H.; Schwaiger, S.; Stuppner, H.; Marzocco, S. Effect of Non-Volatile Constituents of Elsholtzia Ciliata (Thunb.) Hyl. from Southern Vietnam on Reactive Oxygen Species and Nitric Oxide Release in Macrophages. Chem. Biodivers. 2021, 18, e2000577. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.-P.; Kong, L.-R.; Cheng, C.; Lim, J.C.W.; Wong, W.S.F. Protective Role of 14-Deoxy-11,12-Didehydroandrographolide, a Noncytotoxic Analogue of Andrographolide, in Allergic Airway Inflammation. J. Nat. Prod. 2011, 74, 1484–1490. [Google Scholar] [CrossRef]
- Cai, W.; Chen, S.; Li, Y.; Zhang, A.; Zhou, H.; Chen, H.; Jin, M. 14-Deoxy-11,12-Didehydroandrographolide Attenuates Excessive Inflammatory Responses and Protects Mice Lethally Challenged with Highly Pathogenic A(H5N1) Influenza Viruses. Antivir. Res. 2016, 133, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Gao, J.; Liu, W.; Jiang, C.; Yang, X.; Sun, Y.; Guo, W.; Xu, Q. Andrographolide Ameliorates OVA-Induced Lung Injury in Mice by Suppressing ROS-Mediated NF-ΚB Signaling and NLRP3 Inflammasome Activation. Oncotarget 2016, 7, 80262–80274. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Sarwar, A.H.M.G.; Rahat, R.; Ahmed, R.S.; Umar, S. Stigmasterol Protects Rats from Collagen Induced Arthritis by Inhibiting Proinflammatory Cytokines. Int. Immunopharmacol. 2020, 85, 106642. [Google Scholar] [CrossRef]
- Loizou, S.; Lekakis, I.; Chrousos, G.P.; Moutsatsou, P. β-Sitosterol Exhibits Anti-Inflammatory Activity in Human Aortic Endothelial Cells. Mol. Nutr. Food Res. 2010, 54, 551–558. [Google Scholar] [CrossRef]
- Lim, J.C.W.; Chan, T.K.; Ng, D.S.; Sagineedu, S.R.; Stanslas, J.; Wong, W.F. Andrographolide and Its Analogues: Versatile Bioactive Molecules for Combating Inflammation and Cancer: Andrographolide for Inflammation and Cancer. Clin. Exp. Pharmacol. Physiol. 2012, 39, 300–310. [Google Scholar] [CrossRef] [PubMed]




| Ligand Reference | Binding Energy (kcal/mol) | Interacting Amino Acids | Interaction | Type | Location of Interaction |
|---|---|---|---|---|---|
| LPS | −4.1 | LYS122 | Attractive charge | Hydrogen Bond | Chain D of TLR4 |
| ARG264 | Conventional hydrogen bond | Hydrogen Bond | Chain B of TLR4 | ||
| LYS362 | Conventional hydrogen bond | Hydrogen Bond | Chain B of TLR4 | ||
| ARG90 | Conventional hydrogen bond | Hydrogen Bond | Chain D of TLR4 | ||
| TYR102 | Conventional hydrogen bond | Hydrogen Bond | Chain D of TLR4 | ||
| LYS122 | Conventional hydrogen bond | Hydrogen Bond | Chain D of TLR4 | ||
| GLU439 | Conventional hydrogen bond | Hydrogen Bond | Chain A of TLR4 | ||
| LYS362 | Alkyl | Hydrophobic | Chain B of TLR4 | ||
| VAL48 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| ILE52 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| LEU61 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| LEU78 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| ILE124 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| CYS133 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| VAL135 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| ILE32 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
| ILE94 | Alkyl | Hydrophobic | Chain D of TLR4 | ||
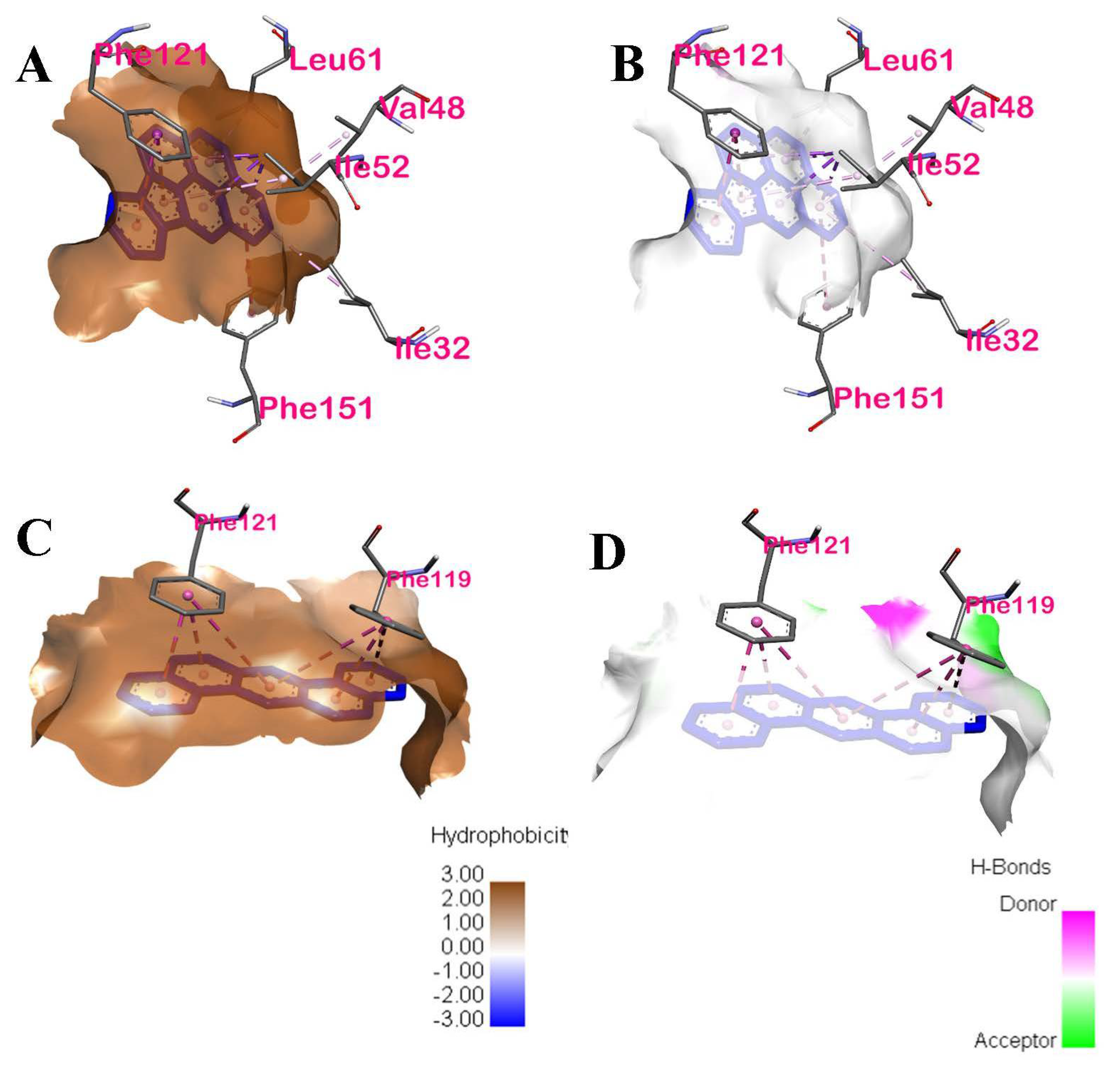
| PHE119 | Pi-alkyl | Hydrophobic | Chain D of TLR4 |
| Ligand | Binding Energy (kcal/mol) | Interacting Amino Acids | Interaction | Type | Location of Interaction | Common Amino Acid LPS and Ligands Interacted with |
|---|---|---|---|---|---|---|
| IP | −10 | ILE52, PHE121, PHE151, LEU61, ILE32, VAL48 | Hydrophobic | Pi-sigma and Pi-alkyl, Pi-Pi stacked, Pi-Pi T-shaped, Pi-alkyl, Pi-alkyl, Pi-alkyl | Chain D of TLR4 | 4: Val48, ILE52, ILE32 and LEU61 |
| DahA | −9.2 | PHE121, PHE119 | Hydrophobic | Pi-Pi stacked, Pi-Pi stacked | Chain D of TLR4 | 1: PHE119 |
| BkF | −8.9 | PHE119, PHE121, ILE52, LEU61 | Hydrophobic | Pi-Pi stacked, Pi-Pi stacked, Pi-Alkyl, Pi-Alkyl respectively | Chain D of TLR-4 | 3: PHE119, ILE52, and LEU61 |
| BaP | −8.9 | PHE119, PHE121, ILE52, LEU61 | Hydrophobic | Pi-Pi stacked, Pi-Pi stacked, Pi-Alkyl, Pi-Alkyl respectively | Chain D of TLR-4 | 3: PHE119, ILE52, and LEU61 |
| Ligand | PubChem ID | Binding Energy (kcal/mol) | TEF | Carcinogenic Classification |
|---|---|---|---|---|
| IP | 9131 | −10 | 0.1 | Probable human carcinogen |
| DahA | 5889 | −9.2 | 1 | Probable human carcinogen |
| BkF | 9158 | −8.9 | 0.1 | Probable human carcinogen |
| BaP | 2336 | −8.9 | 1 | Probable human carcinogen |
| BbF | 9153 | −8.8 | 0.1 | Probable human carcinogen |
| Chrysene | 9171 | −8.2 | 0.01 | Probable human carcinogen |
| BaA | 5954 | −8.1 | 0.1 | Probable human carcinogen |
| Ligand | Amino Acid | Interaction | Type | Distance |
|---|---|---|---|---|
| IP | ILE52 | Hydrophobic | Pi-sigma, Pi-sigma, Pi-sigma, Pi-Alkyl | 3.3891, 3.89425, 3.44148, 4.62034 |
| PHE121 | Hydrophobic | Pi-Pi stacked, Pi-Pi stacked | 4.14965, 4.64929 | |
| PHE151 | Hydrophobic | Pi-Pi T-shaped | 5.84498 | |
| LEU61 | Hydrophobic | Pi-Alkyl | 5.18222 | |
| ILE32 | Hydrophobic | Pi-Alkyl | 5.47123 | |
| VAL48 | Hydrophobic | Pi-Alkyl | 5.16885 | |
| DahA | PHE119 | Hydrophobic | Pi-Pi stacked, Pi-Pi stacked, Pi-Pi stacked | 5.52106, 4.47965, 5.42439 |
| PHE121 | Hydrophobic | Pi-Pi stacked, Pi-Pi stacked, Pi-Pi stacked | 5.11694, 3.86586, 3.74824 |
| Ligand Reference | Binding Energy (kcal/mol) | Interacting Amino Acids | Interaction | Type | Location of Interaction |
|---|---|---|---|---|---|
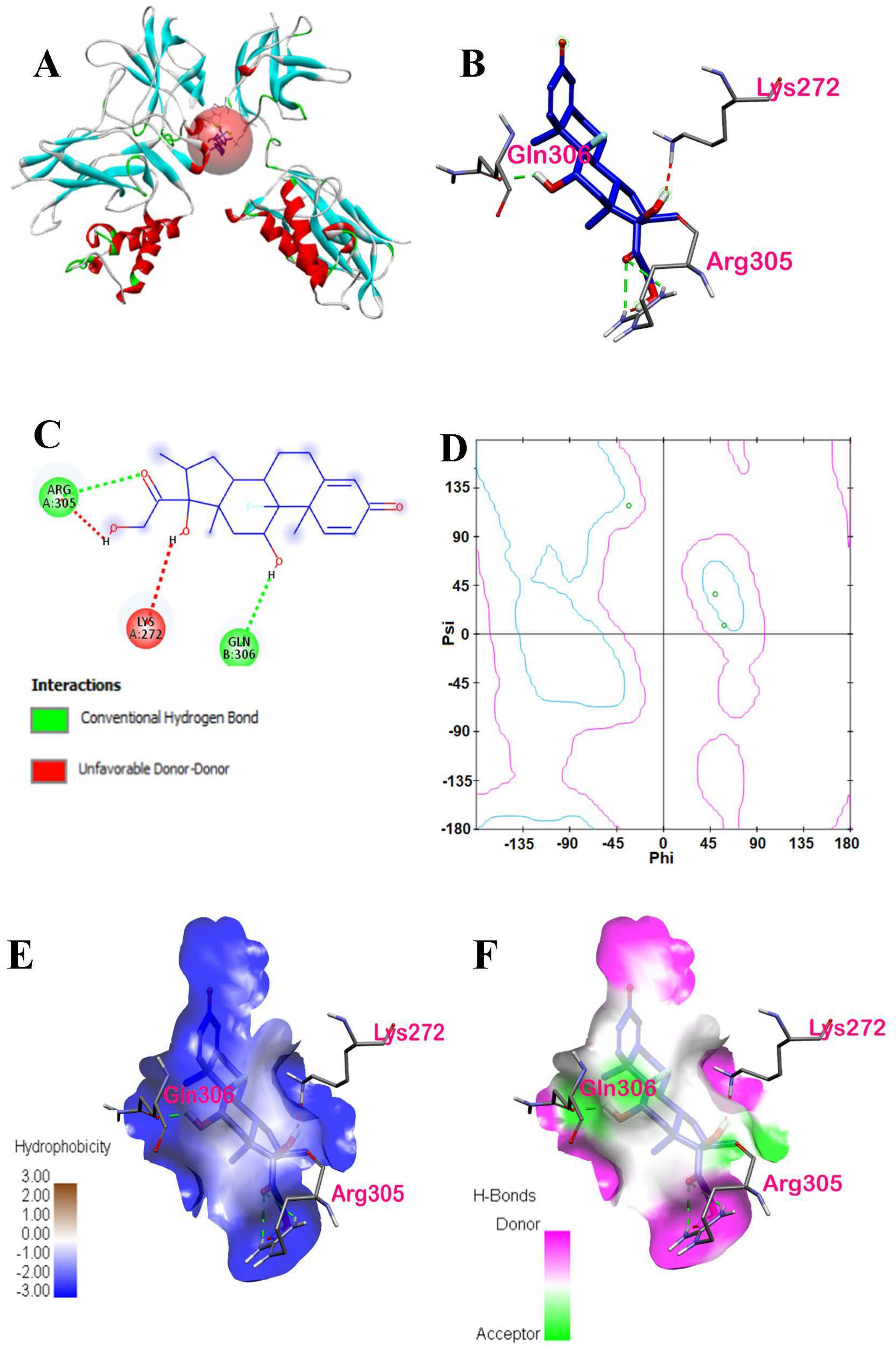
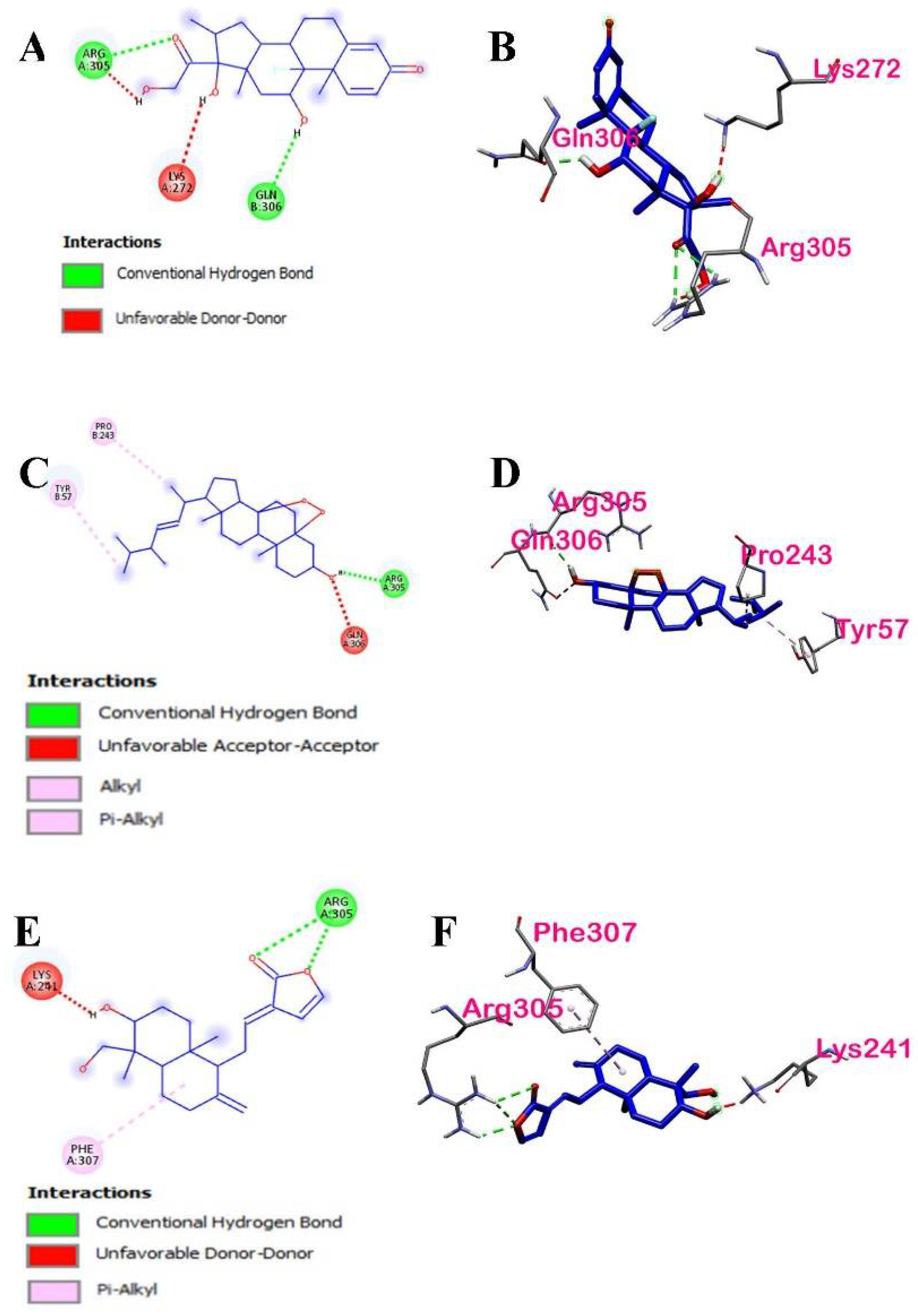
| Dexamethasone | −5.4 | ARG305 | Conventional Hydrogen Bond | Hydrogen Bond | Chain A of p50 |
| GLN306 | Conventional Hydrogen Bond | Hydrogen Bond | Chain B of p50 | ||
| LYS272 | Conventional Hydrogen Bond | Hydrogen Bond | Chain A of p50 |
| AP Phytochemical | PubChem ID | Binding Energy (kcal/mol) |
|---|---|---|
| Dexamethasone * | 5743 | −5.4 |
| ergosterol peroxide | 5351516 | −5.6 |
| 14-deoxy-14,15-dehydroandrographolide | 5351516 | −5.3 |
| 5-hydroxy-7,8-dimethoxyflavanone | 13963770 | −5.3 |
| 5-hydroxy-7,8-dimethoxyflavone | 188316 | −5.2 |
| 14-deoxy-11,12-didehydroandrographolide | 5708351 | −5.2 |
| andrographolide | 5318517 | −5.2 |
| stigmasterol | 5280794 | −5.1 |
| β-sitosterol | 222284 | −5.0 |
| 19-O-acetyl-14-deoxy-11,12-didehydroandrographolide | 46179874 | −4.7 |
| Ligand | Binding Energy (kcal/mol) | Interacting Amino Acids | Interactions | Type | Location of Interaction | Common Amino Acid Dexamethasone and Ligands Interacted with |
|---|---|---|---|---|---|---|
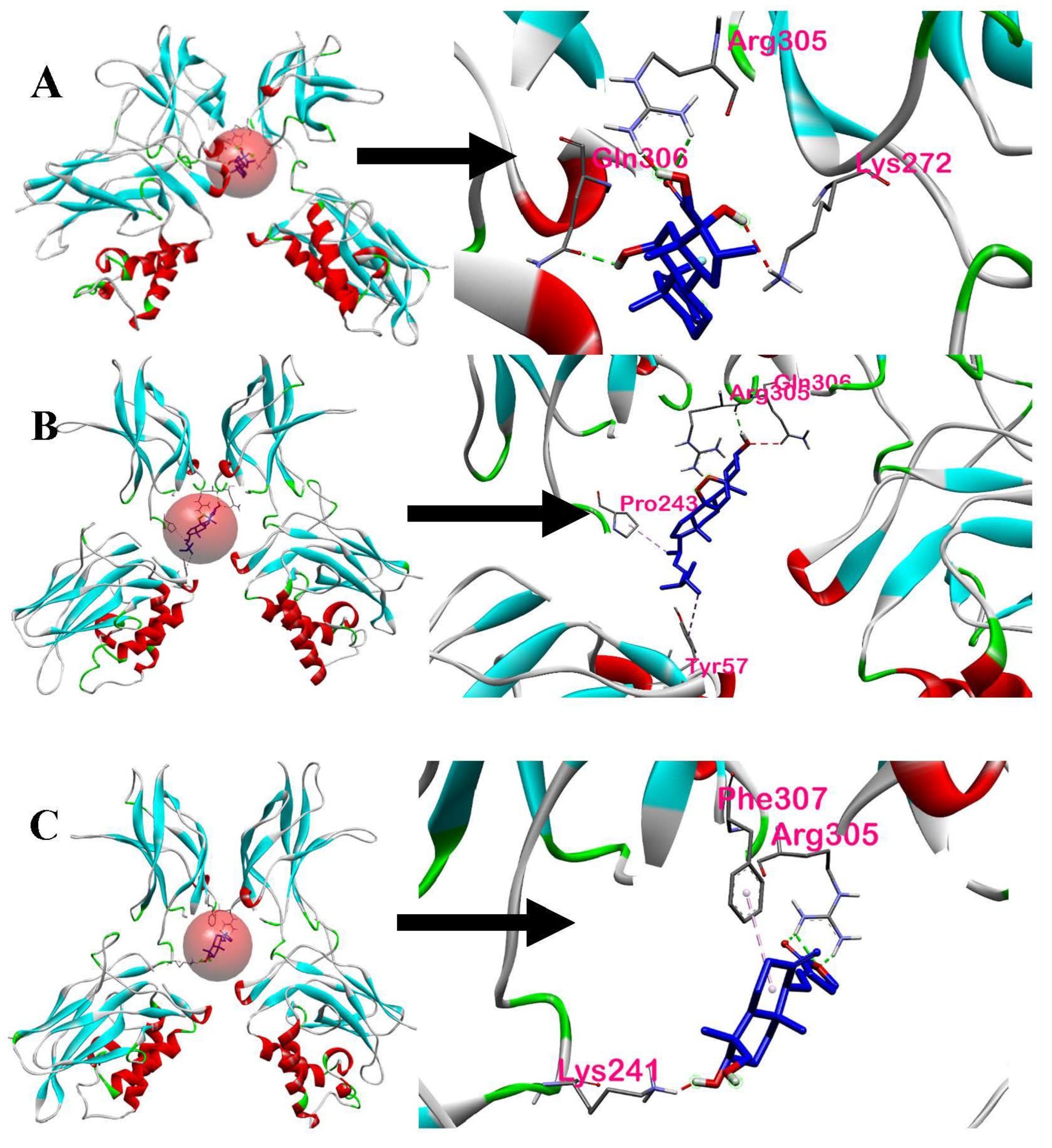
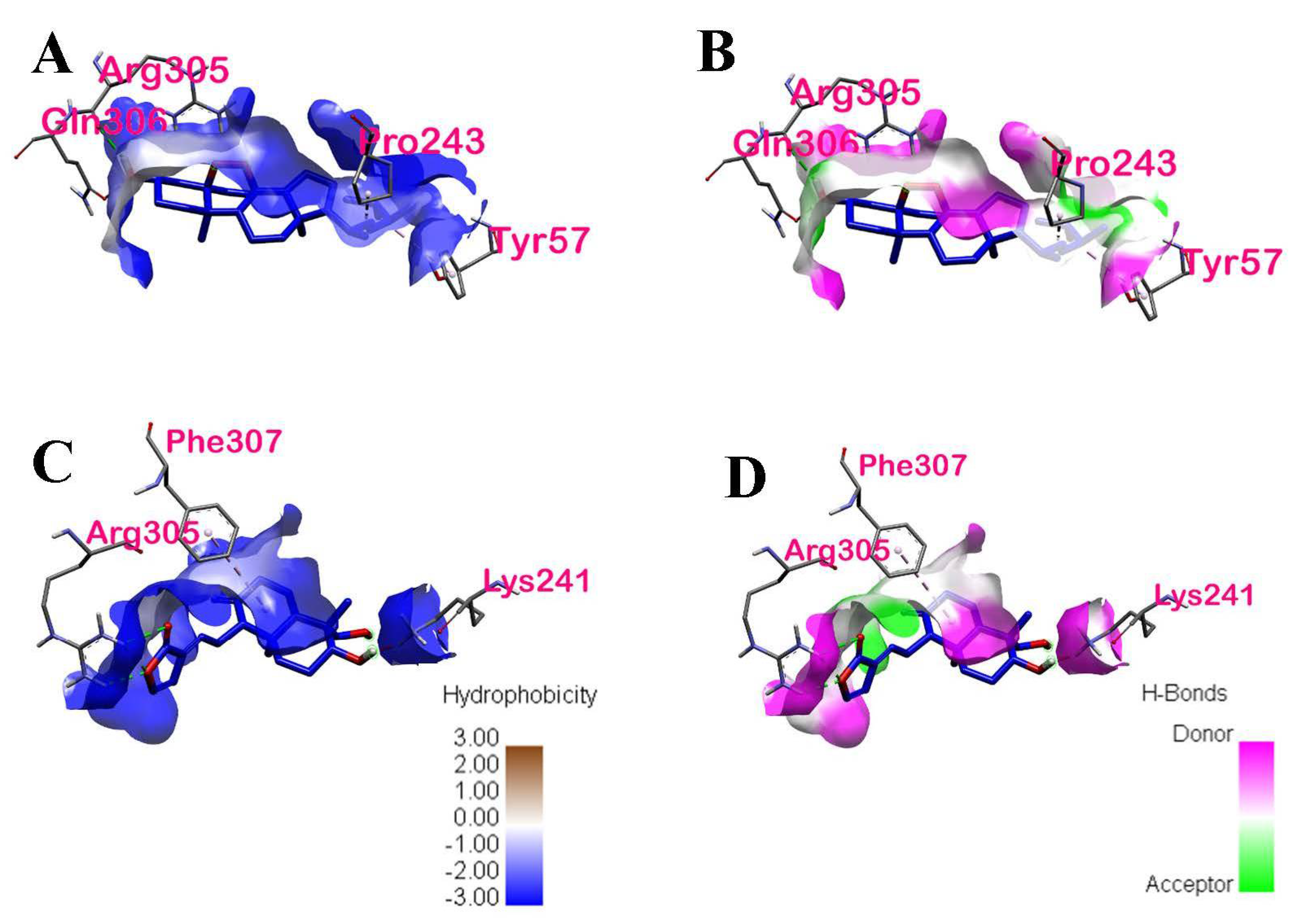
| Ergosterol peroxide | −5.6 | ARG305, PRO243, TYR57 | Hydrogen Bond, Hydrophobic | Conventional Hydrogen Bond, Alkyl, Pi-Alkyl | Chain A and Chain B of p50 | 1: ARG305 |
| 14-deoxy-14,15-dehydroandrographolide | −5.3 | ARG305, PHE307, LYS241, LYS272 | Hydrogen Bond, Hydrophobic | Conventional Hydrogen Bond, Pi-Alkyl | Chain A of p50 | 2: ARG305, LYS272 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Julaton, T.; Taclendo, A.; Oyong, G.; Rempillo, O.; Galvez, M.C.; Vallar, E. In Silico Insights on the Pro-Inflammatory Potential of Polycyclic Aromatic Hydrocarbons and the Prospective Anti-Inflammatory Capacity of Andrographis paniculata Phytocompounds. Int. J. Environ. Res. Public Health 2022, 19, 8588. https://doi.org/10.3390/ijerph19148588
Julaton T, Taclendo A, Oyong G, Rempillo O, Galvez MC, Vallar E. In Silico Insights on the Pro-Inflammatory Potential of Polycyclic Aromatic Hydrocarbons and the Prospective Anti-Inflammatory Capacity of Andrographis paniculata Phytocompounds. International Journal of Environmental Research and Public Health. 2022; 19(14):8588. https://doi.org/10.3390/ijerph19148588
Chicago/Turabian StyleJulaton, Trixia, Aibelou Taclendo, Glenn Oyong, Ofelia Rempillo, Maria Cecilia Galvez, and Edgar Vallar. 2022. "In Silico Insights on the Pro-Inflammatory Potential of Polycyclic Aromatic Hydrocarbons and the Prospective Anti-Inflammatory Capacity of Andrographis paniculata Phytocompounds" International Journal of Environmental Research and Public Health 19, no. 14: 8588. https://doi.org/10.3390/ijerph19148588
APA StyleJulaton, T., Taclendo, A., Oyong, G., Rempillo, O., Galvez, M. C., & Vallar, E. (2022). In Silico Insights on the Pro-Inflammatory Potential of Polycyclic Aromatic Hydrocarbons and the Prospective Anti-Inflammatory Capacity of Andrographis paniculata Phytocompounds. International Journal of Environmental Research and Public Health, 19(14), 8588. https://doi.org/10.3390/ijerph19148588








