Validation and Improvement of a Convolutional Neural Network to Predict the Involved Pathology in a Head and Neck Surgery Cohort
Abstract
1. Introduction
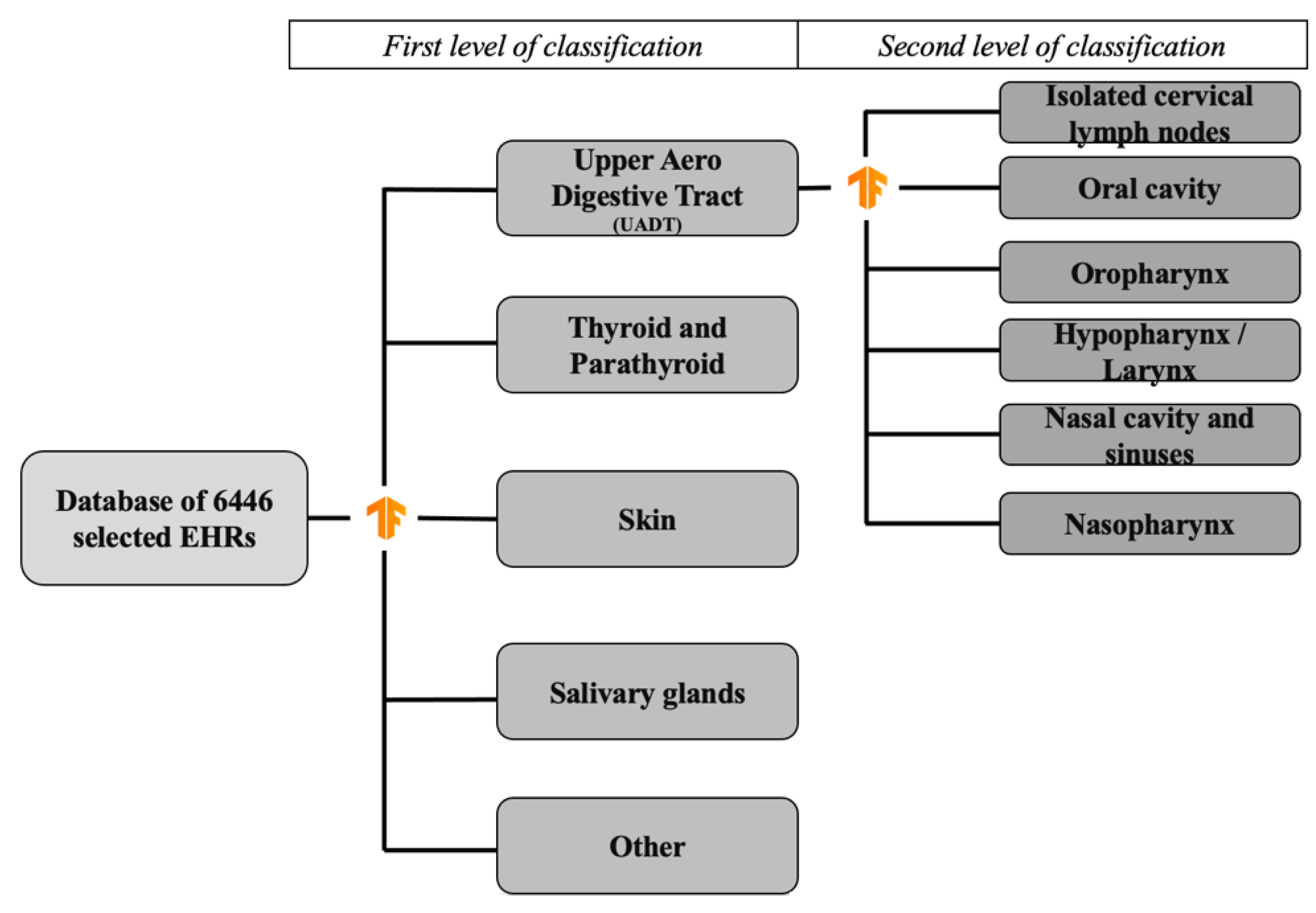
2. Materials and Methods
- -
- “Thyroid and parathyroid pathology”: thyroid nodules, thyroid pathology (Basedow and Hashimoto’s disease), parathyroid dysfunction;
- -
- “Salivary gland pathology”: major salivary gland or accessory salivary gland tumor;
- -
- “Head and neck skin pathology”: squamous cell carcinoma, basocellular carcinoma or melanoma located in the head and neck area;
- -
- “Oral cavity”: oral cavity cancer;
- -
- “Hypopharynx/larynx”: hypopharyngeal cancer, laryngeal cancer;
- -
- “Oropharynx”: oropharyngeal cancer;
- -
- “Nasopharynx”: nasopharyngeal cancer;
- -
- “Isolated cervical lymph nodes”: cervical nodes with unknown primary;
- -
- “Nasal cavity and sinuses”: nasal cavity and paranasal sinuses;
- -
- “Other”: all the other reasons for the consultation.
3. Results
3.1. EHR Extraction and Manual Classification
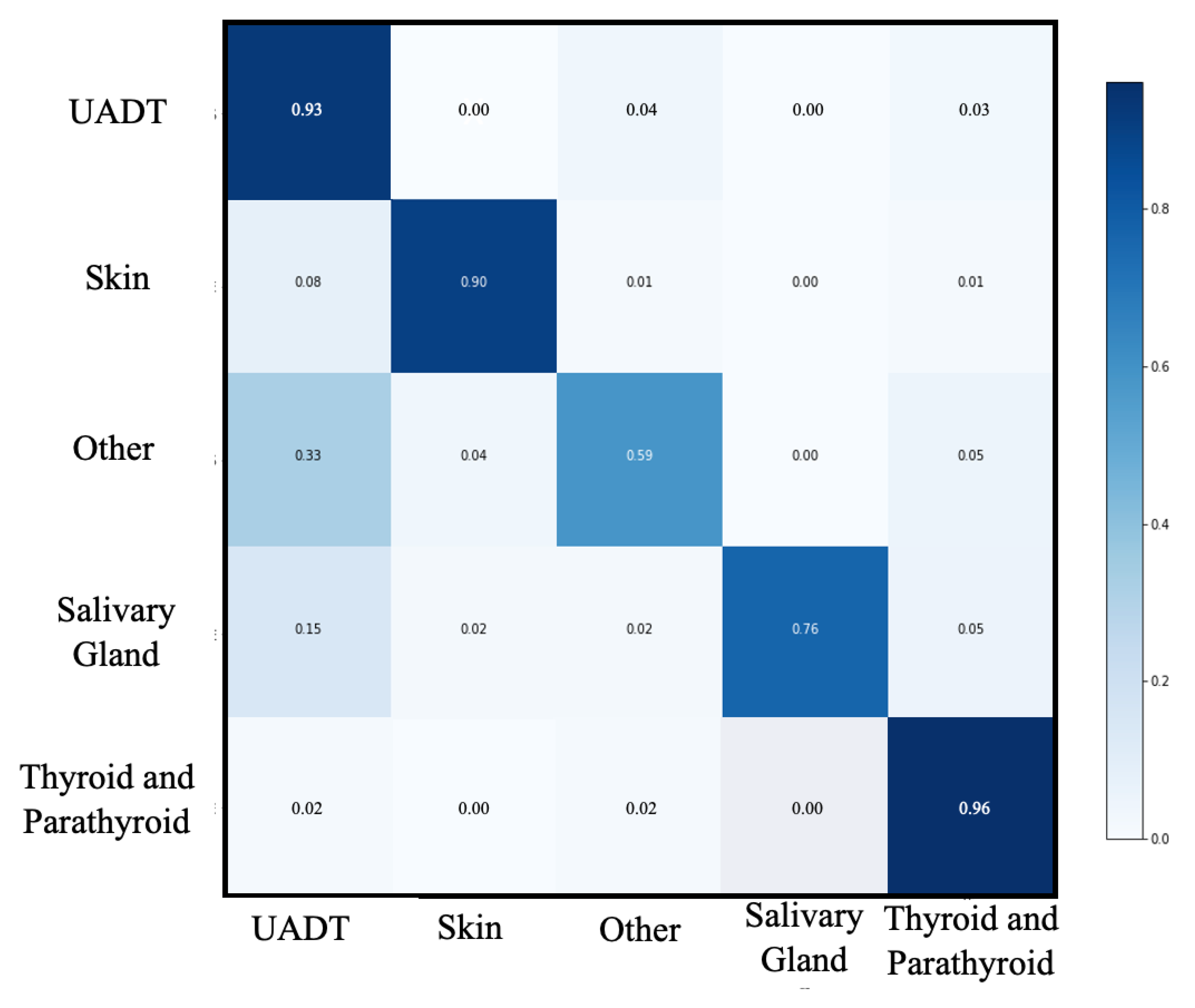
3.2. First Classification Level
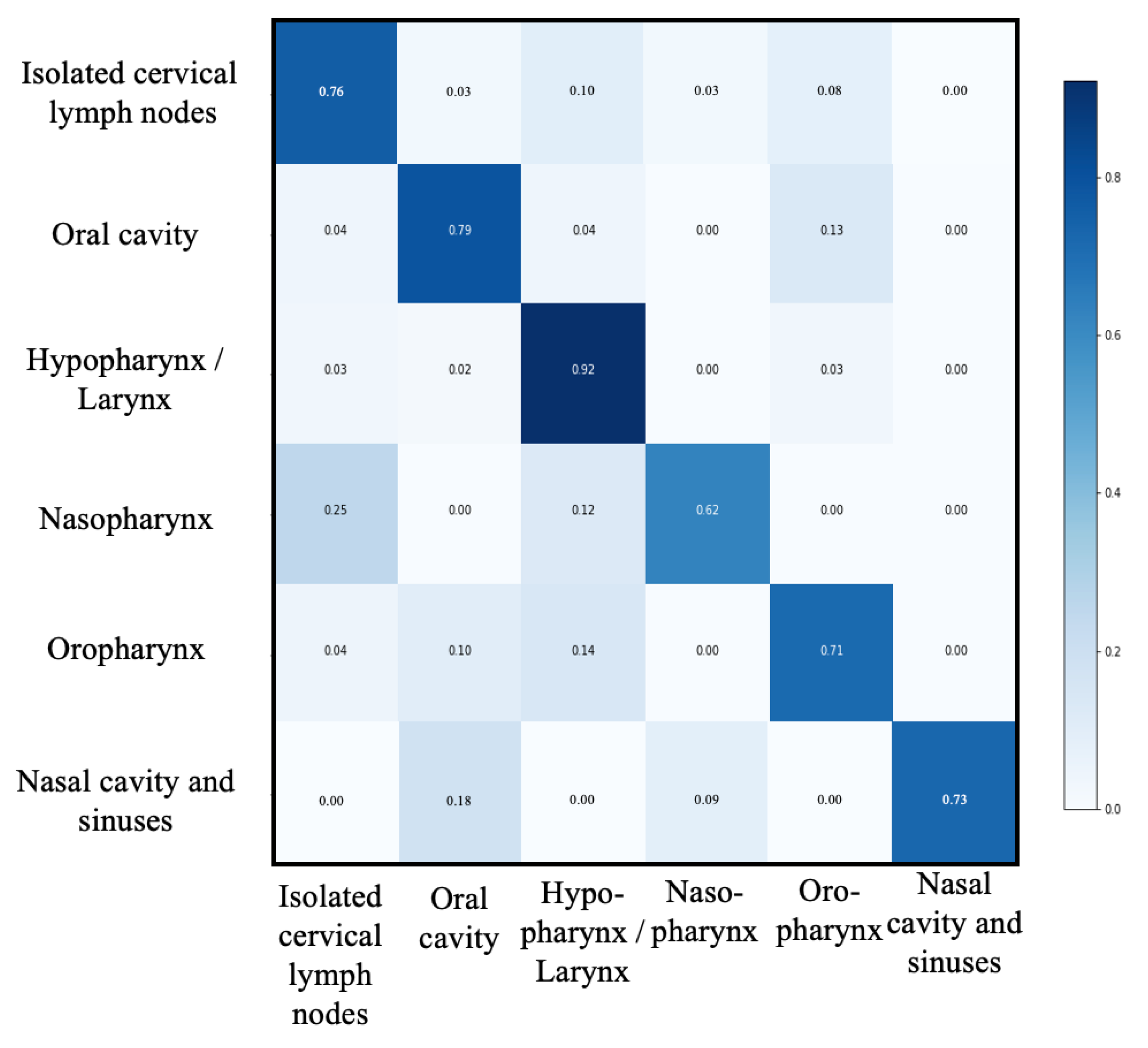
3.3. Second Classification Level
3.4. Extension of the Process
- -
- “Thyroid and parathyroid pathology”: 6798 patients (37.79%)
- -
- “Salivary gland pathology”: 525 patients (2.91%)
- -
- “Head and neck skin pathology”: 2448 patients (13.60%)
- -
- “other”: 1230 patients (6.84%)
- -
- “UADT”: 6987 patients (38.84%)
- ○
- “Oral cavity”: 2083 patients (29.81%)
- ○
- “hypopharynx/larynx”: 2311 patients (33.05%)
- ○
- “oropharynx”: 1379 patients (19.73%)
- ○
- “nasopharynx”: 48 patients (0.69%)
- ○
- “Isolated cervical lymph nodes”: 1062 patients (15.19%)
- ○
- “Nasal cavity and sinuses”: 104 patients (1.48%)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Li, J.; Wu, J.; Zhao, Z.; Zhang, Q.; Shao, J.; Wang, C.; Qiu, Z.; Li, W. Artificial intelligence-assisted decision making for prognosis and drug efficacy prediction in lung cancer patients: A narrative review. J. Thorac. Dis. 2021, 13, 7021–7033. [Google Scholar] [CrossRef]
- Li, D.; Pehrson, L.M.; Lauridsen, C.A.; Tøttrup, L.; Fraccaro, M.; Elliott, D.; Zając, H.D.; Darkner, S.; Carlsen, J.F.; Nielsen, N.B. The added effect of artificial intelligence on physicians’ performance in detecting thoracic pathologies on CT and chest X-ray: A systematic review. Diagnostics 2021, 11, 2206. [Google Scholar] [CrossRef]
- Kho, A.N.; Pacheco, J.A.; Peissig, P.L.; Rasmussen, L.; Newton, K.M.; Weston, N.; Crane, P.K.; Pathak, J.; Chute, C.G.; Bielinski, S.J.; et al. Electronic Medical Records for Genetic Research: Results of the eMERGE Consortium. Sci. Transl. Med. 2011, 3, 79re1. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, H.; Karimi, S.; Nguyen, A. Matching patients to clinical trials using semantically enriched document representation. J. Biomed. Inform. 2020, 105, 103406. [Google Scholar] [CrossRef] [PubMed]
- Spasic, I.; Krzeminski, D.; Corcoran, P.; Balinsky, A. Cohort Selection for Clinical Trials from Longitudinal Patient Records: Text Mining Approach. JMIR Med. Inform. 2019, 7, e15980. [Google Scholar] [CrossRef]
- Mathias, J.S.; Gossett, D.; Baker, D.W. Use of electronic health record data to evaluate overuse of cervical cancer screening. J. Am. Med. Inform. Assoc. 2012, 19, e96. [Google Scholar] [CrossRef]
- Strom, B.L.; Schinnar, R.; Jones, J.; Bilker, W.B.; Weiner, M.G.; Hennessy, S.; Leonard, C.E.; Cronholm, P.F.; Pifer, E. Detecting pregnancy use of non-hormonal category X medications in electronic medical records. J. Am. Med. Inform. Assoc. 2011, 18 (Suppl. S1), 81–86. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peissig, P.L.; Santos Costa, V.; Caldwell, M.D.; Rottscheit, C.; Berg, R.L.; Mendonca, E.A.; Page, D. Relational machine learning for electronic health record-driven phenotyping. J. Biomed. Inform. 2014, 52, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Ferrão, J.C.; Oliveira, M.D.; Janela, F.; Martins, H.M.G.; Gartner, D. Can structured EHR data support clinical coding? A data mining approach. Health Syst. 2020, 10, 138–161. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, G.R.; Pineda, A.L.; Bear Don’t Walk, O.J.; Zehnder, A.M.; Ayyar, S.; Page, R.L. FasTag: Automatic text classification of unstructured medical narratives. PLoS ONE 2020, 15, e0234647. [Google Scholar] [CrossRef] [PubMed]
- Schuemie, M.J.; Sen, E.; ’t Jong, G.W.; Van Soest, E.M.; Sturkenboom, M.C.; Kors, J.A. Automating classification of free-text electronic health records for epidemiological studies. Pharmacoepidemiol. Drug Saf. 2012, 21, 651–658. [Google Scholar] [CrossRef]
- Tam, C.S.; Gullick, J.; Saavedra, A.; Vernon, S.T.; Figtree, G.A.; Chow, C.K.; Cretikos, M.; Morris, R.W.; William, M.; Morris, J.; et al. Combining structured and unstructured data in EMRs to create clinically-defined EMR-derived cohorts. BMC Med. Inform. Decis. Mak. 2021, 21, 91. [Google Scholar] [CrossRef] [PubMed]
- Shivade, C.; Raghavan, P.; Fosler-Lussier, E.; Embi, P.J.; Elhadad, N.; Johnson, S.B.; Lai, A.M. A review of approaches to identifying patient phenotype cohorts using electronic health records. J. Am. Med. Inform. Assoc. 2014, 21, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.L.; Hsu, T.J.; Hsieh, C.H.; Singaravelan, A. Applying Convolutional Neural Networks to Predict the ICD-9 Codes of Medical Records. Sensors 2020, 20, 7116. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.A.; Holmgren, A.R.; Noorbaloochi, S. Accuracy of Veterans Administration databases for a diagnosis of rheumatoid arthritis. Arthritis Rheum. 2004, 51, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Kandula, S.; Zeng-Treitler, Q.; Chen, L.; Salomon, W.L.; Bray, B.E. A bootstrapping algorithm to improve cohort identification using structured data. J. Biomed. Inform. 2011, 44 (Suppl. S1), S63–S68. [Google Scholar] [CrossRef]
- Perry, T.; Zha, H.; Oster, M.E.; Frias, P.A.; Braunstein, M. Utility of a Clinical Support Tool for Outpatient Evaluation of Pediatric Chest Pain. AMIA Annu. Symp. Proc. 2012, 2012, 726. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3540476/ (accessed on 3 March 2022). [PubMed]
- Callahan, A.; Shah, N.H.; Chen, J.H. Research and Reporting Considerations for Observational Studies Using Electronic Health Record Data. Ann. Intern. Med. 2020, 172 (Suppl. S11), S79. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.Q.; Teixeira, P.L.; Mo, H.; Cronin, R.M.; Warner, J.L.; Denny, J.C. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. 2016, 23, e20. [Google Scholar] [CrossRef] [PubMed]
- Fisher, E.S.; Whaley, F.S.; Krushat, W.M.; Malenka, D.J.; Fleming, C.; Baron, J.A.; Hsia, D.C. The accuracy of Medicare’s hospital claims data: Progress has been made, but problems remain. Am. J. Public Health. 1992, 82, 243–248. [Google Scholar] [CrossRef]
- Reker, D.M.; Hamilton, B.B.; Duncan, P.W.; Yeh, S.C.J.; Rosen, A. Stroke: Who’s counting what? J. Rehabil. Res. Dev. 2001, 38, 281–289. Available online: https://pubmed.ncbi.nlm.nih.gov/11392661/ (accessed on 3 March 2022). [PubMed]
- Chescheir, N.; Meints, L. Prospective study of coding practices for cesarean deliveries. Obstet. Gynecol. 2009, 114 Pt 1, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Al Achkar, M.; Kengeri-Srikantiah, S.; Yamane, B.M.; Villasmil, J.; Busha, M.E.; Gebke, K.B. Billing by residents and attending physicians in family medicine: The effects of the provider, patient, and visit factors. BMC Med. Educ. 2018, 18, 136. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Fu, Z.; Shah, A.; Chen, Y.; Peterson, N.B.; Chen, Q.; Mani, S.; Levy, M.A.; Dai, Q.; Denny, J.C. Extracting and Integrating Data from Entire Electronic Health Records for Detecting Colorectal Cancer Cases. AMIA Annu. Symp. Proc. 2011, 2011, 1564. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3243156/ (accessed on 4 March 2022).
- Fernández-Breis, J.T.; Maldonado, J.A.; Marcos, M.; Legaz-García, M.D.C.; Moner, D.; Torres-Sospedra, J.; Esteban-Gil, A.; Martínez-Salvador, B.; Robles, M. Leveraging electronic healthcare record standards and semantic web technologies for the identification of patient cohorts. J. Am. Med. Inform. Assoc. 2013, 20, e288–e296. [Google Scholar] [CrossRef]
- Virani, S.S.; Akeroyd, J.M.; Ahmed, S.T.; Krittanawong, C.; Martin, L.A.; Slagle, S.; Gobbel, G.T.; Matheny, M.E.; Ballantyne, C.M.; Petersen, L.A.; et al. The Use of Structured Data Elements to Identify ASCVD Patients with Statin-Associated Side Effects: Insights from the Department of Veterans Affairs. J. Clin. Lipidol. 2019, 13, 797. [Google Scholar] [CrossRef]
- Ford, E.; Carroll, J.A.; Smith, H.E.; Scott, D.; Cassell, J.A. Extracting information from the text of electronic medical records to improve case detection: A systematic review. J. Am. Med. Inform. Assoc. 2016, 23, 1007–1015. [Google Scholar] [CrossRef]
- Li, L.; Chase, H.S.; Patel, C.O.; Friedman, C.; Weng, C. Comparing ICD9-Encoded Diagnoses and NLP-Processed Discharge Summaries for Clinical Trials Pre-Screening: A Case Study. AMIA Annu. Symp. Proc. 2008, 2008, 404. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2656007/ (accessed on 4 March 2022).
- Friedman, C.; Shagina, L.; Lussier, Y.; Hripcsak, G. Automated encoding of clinical documents based on natural language processing. J. Am. Med. Inform. Assoc. 2004, 11, 392–402. [Google Scholar] [CrossRef]
- Chiaramello, E.; Pinciroli, F.; Bonalumi, A.; Caroli, A.; Tognola, G. Use of “off-the-shelf” information extraction algorithms in clinical informatics: A feasibility study of MetaMap annotation of Italian medical notes. J. Biomed. Inform. 2016, 63, 22–32. [Google Scholar] [CrossRef]
- Faes, L.; Wagner, S.K.; Fu, D.J.; Jack, D.; Liu, X.; Korot, E.; Ledsam, J.R.; Back, T.; Chopra, R.; Pontikos, N.; et al. Automated deep learning design for medical image classification by health-care professionals with no coding experience: A feasibility study. Lancet Digit. Health 2019, 1, e232–e242. [Google Scholar] [CrossRef]
| Group | Number of Patients (%) |
|---|---|
| Thyroid and parathyroid pathology | 2509 (38.92) |
| Salivary gland pathology | 283 (4.39) |
| Head and neck skin pathology | 841 (13.04) |
| Oral cavity | 618 (9.58) |
| Hypopharynx/larynx | 659 (10.22) |
| Oropharynx | 431 (6.68) |
| Nasopharynx | 38 (0.58) |
| Isolated cervical lymph nodes | 363 (5.63) |
| Nasal cavity and sinuses | 66 (1.02) |
| Other | 638 (9.89) |
| Total | 6446 (100) |
| Groups N (%) | UADT | Thyroid and Parathyroid Pathology | Head and Neck Skin Pathology | Other | Salivary Gland Pathology | Macro-Average |
|---|---|---|---|---|---|---|
| 438 (33.4) | 538 (41.0) | 174 (13.3) | 111 (8.5) | 51 (3.9) | 1312 (100) | |
| Accuracy | 0.91 | 0.96 | 0.98 | 0.93 | 0.98 | 0.95 |
| Recall | 0.89 | 0.96 | 0.92 | 0.58 | 0.78 | 0.83 |
| Precision | 0.84 | 0.95 | 0.93 | 0.70 | 0.84 | 0.85 |
| Specificity | 0.92 | 0.97 | 0.99 | 0.97 | 0.99 | 0.97 |
| F1-score | 0.87 | 0.96 | 0.93 | 0.63 | 0.81 | 0.84 |
| Groups N (%) | Isolated Cervical Lymph Nodes | Oral Cavity | Oro-Pharynx | Hypopharynx/Larynx | Nasal Cavity and Sinuses | Naso-Pharynx | Macro-Average |
|---|---|---|---|---|---|---|---|
| 71 (16.3) | 120 (27.6) | 88 (22.2) | 142 (32.6) | 8 (1.8) | 6 (1.4) | 435 (100) | |
| Accuracy | 0.93 | 0.90 | 0.88 | 0.91 | 0.99 | 0.99 | 0.93 |
| Recall | 0.77 | 0.81 | 0.70 | 0.91 | 0.73 | 0.63 | 0.76 |
| Precision | 0.77 | 0.84 | 0.73 | 0.82 | 1 | 0.83 | 0.83 |
| Specificity | 0.96 | 0.94 | 0.93 | 0.92 | 1 | 1 | 0.96 |
| F1-score | 0.77 | 0.82 | 0.72 | 0.86 | 0.84 | 0.71 | 0.79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Culié, D.; Schiappa, R.; Contu, S.; Scheller, B.; Villarme, A.; Dassonville, O.; Poissonnet, G.; Bozec, A.; Chamorey, E. Validation and Improvement of a Convolutional Neural Network to Predict the Involved Pathology in a Head and Neck Surgery Cohort. Int. J. Environ. Res. Public Health 2022, 19, 12200. https://doi.org/10.3390/ijerph191912200
Culié D, Schiappa R, Contu S, Scheller B, Villarme A, Dassonville O, Poissonnet G, Bozec A, Chamorey E. Validation and Improvement of a Convolutional Neural Network to Predict the Involved Pathology in a Head and Neck Surgery Cohort. International Journal of Environmental Research and Public Health. 2022; 19(19):12200. https://doi.org/10.3390/ijerph191912200
Chicago/Turabian StyleCulié, Dorian, Renaud Schiappa, Sara Contu, Boris Scheller, Agathe Villarme, Olivier Dassonville, Gilles Poissonnet, Alexandre Bozec, and Emmanuel Chamorey. 2022. "Validation and Improvement of a Convolutional Neural Network to Predict the Involved Pathology in a Head and Neck Surgery Cohort" International Journal of Environmental Research and Public Health 19, no. 19: 12200. https://doi.org/10.3390/ijerph191912200
APA StyleCulié, D., Schiappa, R., Contu, S., Scheller, B., Villarme, A., Dassonville, O., Poissonnet, G., Bozec, A., & Chamorey, E. (2022). Validation and Improvement of a Convolutional Neural Network to Predict the Involved Pathology in a Head and Neck Surgery Cohort. International Journal of Environmental Research and Public Health, 19(19), 12200. https://doi.org/10.3390/ijerph191912200







