1. Introduction
Environmental exposures profoundly affect human health, but community health professionals, such as physicians, nurses, and nursing assistants, lack basic training in environmental health. Compounding this, there is a lack of awareness of genetic susceptibility and its interaction with environmental health hazards [
1,
2,
3]. Such susceptibilities can be specific to different world populations and particular ethnicities. A lack of knowledge is evident among clinical providers serving Tribal community members in the United States. This is especially true in the Western and Midwestern states where more than 600,000 Native community members are exposed to mountain-top mining waste [
4]. Moreover, other Tribal communities have their Tribal lands in close proximity to heavily contaminated U.S. EPA-designated waste sites. Indeed, 13% of the 1344 Superfund sites in the U.S. have a reservation within 10 miles and as of 2013, about 7.5% of Native American homes did not have safe drinking water or basic sanitation [
5,
6,
7,
8]. These same communities also struggle with many gene-associated, environmentally-induced chronic diseases, such as high rates of type II diabetes, cardiovascular diseases, stroke, and several types of cancers [
9,
10,
11,
12,
13,
14]. Therefore, education of health professionals is essential as the first step in improving community health with the ultimate goal of translating this knowledge to the entire community. Thus, a multidisciplinary collaboration among three academic research centers joined with nursing professionals working with Tribal nations for the purpose of piloting a new educational approach to meet community needs. This environmental justice work is significant because it fills a much-needed gap in health delivery and disease prevention. By elucidating complex genetic interactions in health and extending the reach of these understandings, preventive strategies are more likely to be implemented to improve public health for marginalized communities, including indigenous peoples throughout the world.
The National Institutes of Health (NIH) distinguishes between the terms of genetics and genomics in the following way. Genetics is “the study of all of a person’s genes and their inheritance of disease.” Genomics, however, involves “the study of all of a person’s genes including the study of the interaction of genes with each other and with the environment” [
15]. Since genomics is inclusive of gene-environment interactions, this term will be used hereafter in this context. The Precision Medicine Initiative is a future-looking goal in the U.S. that seeks to perfect prevention and treatment strategies by taking individual genomic variability into account. This movement emphasizes the relevance of genomics in nurses’ clinical practice, and points out that nurses need more training and greater confidence in genomics to fully participate in this effort. The rapid pace of scientific discoveries makes educational progress difficult, but nursing education needs to evolve and integrate genomics into the regulatory standards [
16]. The U.S. is not alone in documenting the need for integration of genomics teaching into nursing education, as this is a worldwide issue, including in Hong Kong, Taiwan, and Mainland China [
17]. One major milestone that has been met in the U.S. is the development of the required essential nursing educational competencies in genetics and genomics [
18].
Our novel educational method introduced here includes short modules with modeling kits, providing hands-on learning experiences that faculty at nursing schools can insert into their existing coursework. In the future, these modules could be tailored for self-study and used by professional clinicians and nurses, as well as academic faculty members, to fill continuing educational needs. Here, because our focus was on nursing students’ learning gains after working with unique tactile molecular models, we reviewed learning style preferences of students in nursing schools. Dr. Santhamma James et al. [
19], reporting on learning style preferences of their first-year nursing students, found that tactile learning was ranked the highest, above visual, aural, and read–write methods by students (
p < 0.001). They also noted that rural students had significantly higher preference for visual and tactile learning compared to their metropolitan counterparts. Based on these and additional study results, the authors called for teaching in nursing schools to be both more hands-on and more interactive to improve the effectiveness of the learning process. The overall effects of learning style on the performance of university students have also been well documented by numerous researchers [
20,
21,
22].
Kathleen Calzone and collaborators in 2018 acknowledged that a concerted global effort was needed to address well-known deficits in nurses’ genomic literacy. They defined three primary educational challenges: insufficient curriculum time, insufficient educators capable of teaching genomics (which includes both genetics and environmental health connections), and absence of the competency assessments required for nursing. Only one country (Israel) required a demonstration of genetic/genomic knowledge for general nursing practice [
23]. A survey of 23 nurse leaders from 18 countries revealed that genomic knowledge was basically confined to specialists associated with newborn screening programs, and that worldwide, genomics was often viewed as a niche specialty [
24]. In contrast, Calzone and colleagues in the International Society of Nurses in Genetics (ISONG) view genomics as relevant to all healthcare providers, as it “provides an underpinning for understanding health, risks for manifestations of disease, therapeutic decisions, development of new therapies, and responses to interventions” [
23]. The World Health Organization also affirms nursing’s leadership role, stating that as the largest single healthcare professional group worldwide, nurses have a pivotal role in bringing the benefits of genomics to everyday healthcare [
25].
Some countries have created high-quality, national programs in genomics to fill the gaps in the environmental health and genetics knowledge for the nursing profession. For example, the U.S. National Institute of Nursing Research’s Summer Genetics Institute (SGI) was established about 18 years ago, and annually provides intensive laboratory training in molecular genetics for a group of nurse researchers. However, to bring the benefits of genomics education to everyday health care, which is the goal, broader programs for greater numbers of nurses need to be accommodated [
18].
The main aim of this study was to pilot a novel environmental health and genetics learning framework by combining an interactive, hands-on modeling activity with classical lecture experiences. The purpose was also to demonstrate effective communication of key concepts in environmental health and genetics (e.g., genomics) to health professionals, building the groundwork for translating these concepts to their community members in culturally appropriate language. Our hands-on approach employed the unique MIT DNA and protein models that were designed to provide the users with visual, tactile, and “learning by doing” experiences. With the provided models, participants could independently explore and verify key concepts, generating greater engagement and understanding. In conclusion, this interactive learning experience increased literacy in genomics, and provided insight into how toxic agents are metabolized and how the process is influenced by genetics to produce a wide range of health outcomes. Specifically, the hands-on approach improved environmental health literacy as measured by pre- and post-tests with the participants demonstrating significant learning gains (p < 0.0001). This new approach was successful in building faculty instructors’ and students’ confidence in explaining the molecular mechanisms underlying the actions of environmental chemicals in the body.
2. Materials and Methods
The materials employed in the hands-on modeling study were the MIT Edgerton Center DNA and Protein Sets. They are described here along with the instructional methods used with the nurse study participants. The pre- and post-test materials used to measure nurses’ learning gains with the hands-on educational materials are also included in this section. Since this framework for building environmental health literacy includes lectures on toxicology and exposure biology to accompany the modeling experience, these materials will be briefly described here as well. For nurse audiences, the lectures provided context to the hands-on methods session, focusing on environmental health and impacts on environmental justice communities. For community audiences that included, for example, classroom groups seated in a school auditorium for a lecture, a simple survey form was piloted to assess their learning gains and increased interest in the lecture topic.
2.1. Model Development and Availability
Vandiver invented a flexible double helix model capable of revealing the functional aspects of DNA. The double helix can be used to visualize structural relationships, or easily opened into separate strands and laid flat to simulate cell processes. This functional prototype was first designed from off-the-shelf components. The models used in this research study have now been mass-produced and hold MIT patents. The MIT DNA/RNA kits became available for non-profit purchase in 2016; the protein kits and tRNA kits followed in January 2017. All three modeling kits are currently available and sold online in classroom sets of 14 kits, with each kit typically serving two students. The DNA/RNA models are used in public school districts around the continental U.S. and have been introduced globally. The sets are already serving educational institutions in Singapore, China, Switzerland, Spain, Mexico, Hawaii, and India. MIT’s overall mission is improving science education worldwide and MIT’s non-profit status makes it possible for the MIT Edgerton Center to offer these molecular models at cost with additional support from the NIEHS P30 Environmental Health Research Excellence Centers.
In planning manufacture for global dissemination, the MIT DNA and Protein Sets are well positioned for such distribution, as the plastic components are already mass produced through injection-molded processes where production rates can readily be increased. Instructors in different countries have observed that students with limited English proficiency can follow the written information in the instructional booklets, as the sentences are short and accompanied by photos/diagrams. Additionally, the booklets use scientific vocabulary sparingly, so teachers can add the scientific language that their students are required to learn.
The MIT authors originally created both basic and advanced booklets for teaching how cells make proteins from DNA instructions. More recently, the authors created additional instructional materials that are shown here for teaching hands-on lessons in toxicology using the existing MIT Edgerton Center’s molecular kits. Specifically, new lessons in environmental health were created to demonstrate how genetics play a critical role in a populations’ susceptibility to environmental exposures. In this educational research study, the nursing students initially performed several introductory activities to become familiar with how the models work and to learn the basics about protein form and function. First, they constructed a simplified cell membrane protein using the amino acid subunits. Then, using additional hands-on components, the participants successfully produced the same membrane protein from a gene, using the multiple steps occurring in protein synthesis. After the preparatory steps, students were introduced to a genetic case study in exposure biology. Because the purpose of the study was to evaluate the new instructional capability provided by the functional 3D molecular models, the models are described below in greater detail.
2.2. DNA/RNA and Protein Subunits
The pieces for the models are manufactured by an injection-molded process using ABS plastic with tight quality control and dimensional tolerance. The nucleotide monomers in the DNA/RNA kits can be linked and unlinked to demonstrate cell processes. The DNA nucleotide models are easily formed into long DNA strands by linking the phosphate and sugar components of adjoining nucleotides (
Figure 1). Each base (thymine, adenine, cytosine, and guanine) has a designated color for ease of recognition, and the purine and pyrimidine bases are sized to resemble the differences in nature. The nucleotide sugars have a number 3′ (not visible on
Figure 1) marked on the end that is opposite to the 5′ phosphate group. To further emphasize that two DNA strands should be constructed in opposite directions (antiparallel), the individual bases of both RNA and DNA are marked with prominent arrows pointing toward the 3′ end of the sugar. The large directional arrows in
Figure 1 are marked in white. The 3′ biochemical designations inform advanced students, while novices correctly assemble an antiparallel double helix following the large arrows imprinted on the bases.
Base-pairing of the model nucleotides is accomplished through a ball-and-socket connection, permitting the two DNA strands to twist into a double helix as shown in
Figure 2a. This same connection permits the two sides of the DNA ladder to easily open up when lying flat, demonstrating the weak hydrogen bonds that hold the two DNA strands together. A quick disconnect feature allows for the two DNA strands to open up easily along any section of the DNA ladder to demonstrate the cellular processes of replication, transcription, and DNA repair. A simplified DNA replication fork is shown in
Figure 2b where two small white tags were added to each strand of original DNA. These tags allow participants to observe DNA replication and to discover the semiconservative nature of the process (one strand in the new molecule is always conserved from the original DNA). The process of transcription can also be accomplished using the RNA nucleotides provided in the DNA/RNA kit (
Figure 2c). The strand opposite the gene serves as the template for the mRNA nucleotides. The resulting mRNA sequence matches the sequence of the gene.
Importantly, all the models work together. This includes the DNA, RNA, and tRNA pieces as well as the amino acids in the protein kit. Therefore, protein chains can be produced from mRNA transcripts and the models can also be used to demonstrate the process of translation on the ribosome. These modeling features provide learners with a seamless way to visualize numerous cell processes in molecular biology, and provide the designers with opportunities to publish additional instructional materials related to environmental health.
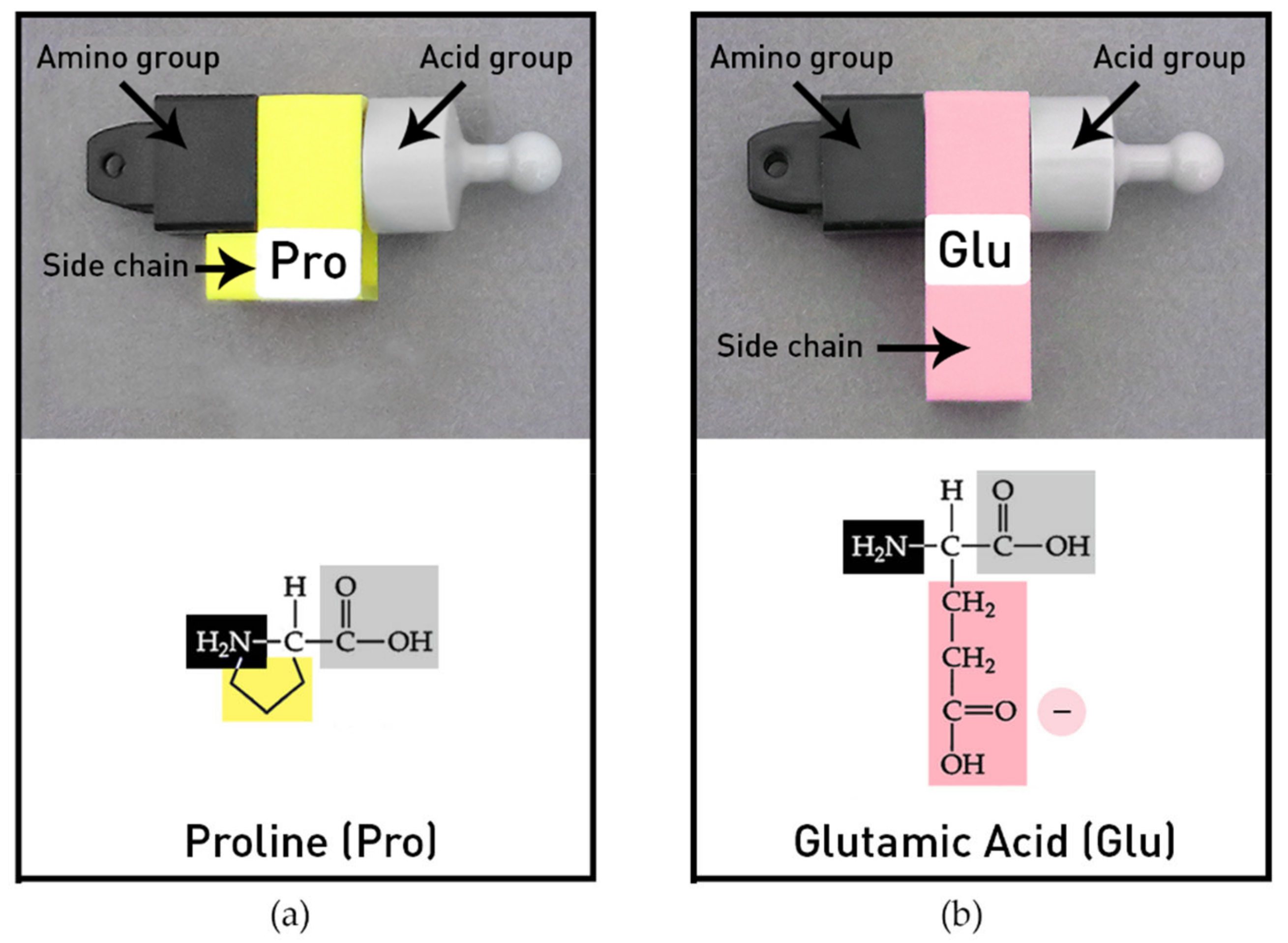
The amino acid models provided in the MIT Protein Set represent the most common twenty amino acids with many distinctive functional features. The individual amino acids have generalized side chains (-R groups) with shapes and colors to represent their different chemical properties. Amino acids can be joined end to end (amino group to acid group) by peptide bonds to create different sequences and chain lengths. The model protein chains are flexible and can be folded into classical biochemical forms, such as helices and beta pleated sheets, owing to the design of the amino acids’ side chains that can rotate freely, with the exception of proline. Proline’s side chain has an extra bond and thus its side chain cannot rotate (
Figure 3a). Two of the twenty common amino acids in the protein kit are shown in
Figure 3. The colors of the side chains indicate the four chemical groups. Hydrophobic amino acids are yellow and the hydrophilic amino acids include three colors: basic amino acid side chains are blue (positive), acid side chains are pink (negative), and polar side chains are green (polar). The chemical diagrams illustrate how the side chains (-R groups) were stylized to indicate relative differences in size and shape in the models.
2.3. Form and Function
To learn how the amino acid sequence can affect function of the channel proteins in membranes, participants assembled individual amino acids into a protein chain with a prescribed sequence and then coiled each chain into a helix; in
Figure 4, the side chains (-R groups) of the amino acids face outwards (towards their surroundings). For the next step, the participants decided how to position all four of these transmembrane proteins into the model cell membrane to create a working pore. A functioning channel protein will have hydrophilic amino acids facing inwards, to create the pore opening. These charged positive and negative side chains on the amino acids facilitate the passage of ions through the cell membrane.
Figure 4b shows a top-down view of a cell membrane in which a channel protein is represented by four short amino acid chains spanning the thickness of lipid bilayer. In reality, a protein chain in a cell membrane has ten helical turns as opposed to two shown in this lesson. This simplified protein permits learners to later assemble the same channel protein starting from a gene and using the molecular processes of transcription and translation. The lesson plan also includes genes for demonstrating how a single nucleotide substitution can produce a non-functional membrane protein, as well as a second example where a single nucleotide substitution codes for the same amino acid and the protein remains functional.
2.4. Protein Synthesis
The tRNA kit, working together with the other models and materials, demonstrates how messenger RNA (mRNA) determines the order of the amino acids creating a new protein on the ribosome. As shown in
Figure 5, each tRNA carries a specific amino acid. The RNA nucleotides on the bottom of the tRNA, labeled as the anticodon, bind to the mRNA on the ribosome. The tRNA kit includes the ribosome mat for demonstrating translation as shown in
Figure 6 and
Figure 7. To assist in visualizing the steps before using the models, instructors showed a five-minute MIT Edgerton Center video where a pair of hands correctly models translation on the ribosome mat [
26]. In
Figure 6, the two green overlapping ovals represent the large and small ribosomal subunits, which unite to form the working ribosome in the cytoplasm. The ribosome mat includes several orange shapes, marking the position for the mRNA and tRNA molecules. Each tRNA molecule moves through three locations on the ribosome, known as the E, P, and A sites, labeled on the ribosome mat.
The messenger RNA molecule, responsible for selecting the order of the amino acids, binds to the ribosome first. Next, the tRNA molecules carrying specific amino acids bind to the mRNA (
Figure 7). In
Figure 7a, the arrow indicates that the protein chain comes off of the tRNA in the P site. That chain will bind to the end of the amino acid held in site A. In
Figure 7b, a peptide bond is formed at the A site. This action shows how elongation of the protein chain occurs. The blue amino acid (Arg) stays attached to its tRNA in site A. In
Figure 7c, both the mRNA and tRNA with the protein chain have moved over to the left by three nucleotide spaces (compared to
Figure 7b). This move causes the empty tRNA to exit via the E site. This movement also opens up the A site for another tRNA to enter. The next tRNA selected will have an anticodon that base-pairs with AGC on the mRNA. This process continues until a release factor binds to the mRNA, releasing it from the ribosome.
2.5. Teacher Training Videos and Classroom Resources
The resources provided with MIT DNA and Protein Sets serve as excellent materials for self-instruction, strengthening faculty knowledge in preparation for teaching genomics. Additionally, training videos posted on the MIT Edgerton Center website demonstrate classroom instructional techniques for each set; also posted online are 12 three-minute videos for the hands-on activities. One example shows how to put together a protein chain from individual amino acids and how to fold the chain into an alpha helix. These practical and quick videos ensure that participants can build the structures, permitting them to focus on the concepts. The videos also show common building errors that help students avoid time-consuming missteps, assisting the instructor in keeping the class together. Summarizing videos are also provided and shown after the hands-on activities. Reviewing concepts with the videos strengthens the experience, helping the students recognize which facts are most important to learn. Lastly, students if absent can make up class time on their own if provided with a modeling kit, access to online videos, and the instructional booklets. All videos are listed on the MIT Edgerton Center DNA and Protein Set YouTube playlist [
26].
2.6. A Case-Based Study with Cytochrome P450 (CYP450) Proteins
Cytochrome P450 (CYP) proteins, found in abundance in liver and kidney, help to break down toxicants and drugs by transforming them into water soluble molecules that can be excreted in the urine. However, CYP proteins can also convert relatively non-toxic molecules into potent DNA-damaging agents that cause cancer. A key concept for health professionals to recognize is that different world populations have variations (polymorphisms) in their CYP genes. These gene variants can have consequences in protein structure, making a population less or more sensitive to a specific toxicant, influencing health outcomes. A new case-based lesson using the MIT protein models was devised that presented three hypothetical examples (Carey, Yeshna, and Paxton) with CYP protein variants and where participants were guided to explore possible genetic susceptibilities. The protein variants illustrated in the examples were based on well-known CYP variants [
27,
28,
29,
30,
31,
32,
33,
34,
35,
36]. Native American populations in the U.S. were selected as relevant examples for the participants in the learning framework and for worldwide indigenous populations, more broadly.
After completing the introductory protein activities, nurse participants explored CYP protein function by constructing a two-dimensional representation of the protein’s active site. As shown in
Figure 8, construction of the active site with the amino acids was guided by individual CYP Protein Mats, and participants worked in teams of two. As substrate binding initiates the metabolic process, each active site was tested for its ability to bind a specific environmental chemical or drug. The toxicant substrates were designed as color-coded paper cards to be positioned inside the active site. Hydrophobic and hydrophilic interactions, as well as substrate size, determined whether binding could occur. Polycyclic aromatic hydrocarbons (PAHs) were provided as a classroom example as this substrate can be converted into a potent carcinogen by the metabolic action of specific CYP proteins [
33,
34,
35,
36].
Case studies with Paxton and Yeshna have two amino acids clearly marked as variants on their CYP Protein Mats (
Figure 9). These variant amino acids and the ethnic heritage for each person match the research for their variant CYP proteins. Multiple CYP protein active sites were combined and simplified into two-dimensional structures to represent the CYP active site of each person. However, the models accurately depict the CYP proteins’ hydrophobic or hydrophilic interaction with the substrates. The models capture the altered interactions of the amino acids in the active site as well as amino acid sequence variations leading to the change in the proteins’ function.
Case-based studies for the study participants included representative scenarios to illustrate genetic susceptibility to environmental toxicants and drugs.
Figure 9b,c, illustrates a case-based study that compares Yeshna’s (
Figure 9b) and Paxton’s (
Figure 9c) experiences with exposure to cigarette smoke. Because of their CYP variants, they will have different health outcomes.
The study participants discovered that Yeshna’s CYP protein binds and metabolizes nicotine easily and it leaves her body quickly. The broad hydrophobic patch of amino acids (in yellow) provides multiple sites for nicotine to bind (
Figure 10a,b). Yeshna’s metabolism makes her have more frequent cravings for nicotine and she has greater difficulty quitting her smoking habit. Yeshna’s CYP protein active site also binds benzo[a]pyrene well as a result of its hydrophobic interactions with this substrate. Thus, a higher percentage of the benzo[a]pyrene molecules are converted into potent cancer-causing agents. This process explains why Yeshna has a higher risk of getting cancer from smoking.
In comparison, Paxton has an easier time quitting smoking and a lower risk of lung cancer (
Figure 10c,d). The two hydrophilic amino acid variants (in blue) prevent the hydrophobic nicotine from binding well and it is metabolized poorly. Because nicotine stays in Paxton’s body longer, his nicotine cravings are decreased. In addition, his CYP protein binds and metabolizes benzo[a]pyrene poorly and produces fewer carcinogens.
These case studies, combined with tactile learning, lead to a conclusion that genetic variations in CYP proteins can affect human behavior, as well as cancer risk. After the students had worked with the models and discovered the potential health effects caused by genetic variations in CYP proteins, research results in figures graphically representing CYP2A6 variant distributions across different world geographic populations (e.g., Africa, Southeast Asia, and the U.S.) were presented for discussion [
32]. The students noted the distribution of genetic variants in CYP protein types in these world populations were different. Genetic variations in populations produce changes in CYP protein amino acid sequences, causing major disparities in drug reactions, addictive behaviors, and cancer risk.
An example of the distribution of one CYP variant found in the U.S. was examined more closely. CYP2A6 is the name of this protein, and the symbol (*) after the name indicates a specific variant. CYP2A6 is the CYP protein that increases smoking desire by decreasing nicotine levels in the blood. More than 60% of the tested North Plains and Southwest Native American populations have the variant CYP2A6*1B compared to ~30% of Caucasian populations as in the case with Yeshna. In the case study presented to the students, Yeshna’s CYP proteins were modeled after typical Native American protein variants and Paxton’s CYP proteins were modeled after typical Caucasian protein variants [
29,
30,
31,
32].
2.7. Study Design and Content for the Models
To ensure familiarity with molecular representations and to teach the molecular content pertinent to genetics and environmental heath literacy, the hands-on component included the following four lessons: (1) protein structure and function, (2) DNA structure and function and mRNA transcription, (3) protein synthesis including translation on the ribosome, and (4) case studies on genetic susceptibility. The study session was three hours in duration, including the pre- and post-test times. Additionally, a ten-minute break was given halfway through the session.
2.8. Survey Method for the Models
The survey method for this study employed the MIT DNA and Protein Set with a pre-test–post-test method in which data were collected from participants at the beginning and end of the session. Data were collected from two tribally-affiliated sites. As one of the sites did not have classroom access to the internet for test taking online, data were collected uniformly at both sites by distributing printed questionnaires. This method of administrating the tests avoided any potential bias. To assure confidentiality, each participant was given instructions for creating a unique alphanumeric identifier code and asked to use the same code on both the pre-test and post-test versions. The questionnaire was primarily multiple choice with some true/false questions. In the pre-test, a brief set of open-ended questions were included concerning demographics and prior education in the topics to be taught.
Data were entered into Excel spreadsheets by the Tufts University CTSI staff. Responses were entered twice to check for accuracy. Using an answer key provided by the principal investigator from MIT, a scoring algorithm was created in which each correct answer was awarded a point. The 18-item questionnaire was scored from 0 (lowest score) to 17 (highest score). The “difference” was the post-test score minus the pre-test score.
2.9. Study Sites and Subjects
Two geographic sites were involved: Site 1 was the Ramapo College Nursing Program, in Ramapo, New Jersey, where advanced undergraduate nursing students in the Bachelor of Science in Nursing (BSN) Program support public health programs for the citizens of the Ramapo Lenape Tribe. Site 2 consisted of a diverse group of health professionals from the Cheyenne River Sioux Reservation. The primary partner at this site was the Missouri Breaks Industries Research, Inc. (MBIRI) in Eagle Butte, South Dakota, which is a research center employing 35 nurses. Our Project Team also partnered with the Oglala Lakota College, Eagle Butte Campus Associate Nursing Program where additional students and instructors participated. The pre-test and post-test data from the New Jersey and South Dakota sites were designated as “Nurse Data Ramapo (Site 1)” and “Missouri Breaks (MBIRI) (Site 2)” respectively, and presented together (n = 116).
2.10. Targeted Lectures in Toxicology
Our educational framework to improve public health knowledge includes targeted lectures on environmental health and toxicology in combination with the hands-on modeling exercises and case-based lessons. These lectures included the authors’ community-based environmental health research among Indigenous peoples of the U.S. At the Ramapo College in NJ, the nursing students received three environmental health/toxicology lectures (each ~40 min in duration with 20 min for questions and answers): (1) Environmental Health and Toxicology Basics, (2) Marginalized Communities and Environmental Contamination and Intervention Strategies, and (3) Metabolism of Environmental Toxicants. Similarly, three lectures were given at the Oglala Lakota College in South Dakota, on the Cheyenne River Sioux Tribal Reservation. Two presentations were also given to community members. One was an open evening lecture sponsored by Oglala Lakota College, and the second was for high school students given during the school day at Wakpala High School on the Standing Rock Tribal Reservation. An audience feedback form was piloted with the high school students to survey self-reported learning about health and the environment.
The feedback form, on a half-page of paper, was a modified SAMI Card, an all-purpose assessment form originally created by the St. Louis Science Center (
Figure 11). Our four-question, five-minute survey for this pilot was brief with a space for optional comments: two were Likert scale questions with four option-check boxes. Emoji faces clarified the ranking system [
37,
38]. The first Likert question measured interest in the lecture topic, and the second question measured self-reported learning about health and the environment. The next two questions were open-ended: “Tell us something that was new to you,” and, “Did you learn what you wanted to know?”
4. Discussion
4.1. Previous Studies
The nursing students’ robust educational gains reported here further substantiate an earlier study published in the
Journal of Nursing Education by the authors and their extended team [
39]. This earlier study achieved learning gains with the same pre- and post-tests significance values of
p < 0.0001, demonstrating the hands-on learning approach is well-suited to teaching the molecular basis of genetic susceptibility. This earlier study also demonstrated the modeling was effective across a broad nursing audience, as nurses were of different ages, educational and cultural backgrounds, and were employed in numerous nursing fields. These earlier workshops, held annually on the MIT campus between the years 2007 and 2012, attracted practicing nurses from major hospitals, research institutions, and also faculty from nursing schools across the U.S. The workshops’ purpose was to teach key molecular concepts in genetics/genomics and to offer participants Continuing Education Units (CEUs) for the workshop; U.S. nurses are required to earn CEUs annually by state rules. These studies served as the basis for an expanded environmental health educational framework presented here [
39].
The previous nurse workshops conducted on the MIT campus also explored the long-term recall of the molecular biology concepts, asking participants for a personal or work-related example of how they had used their newly acquired genetic knowledge. An email request was sent to 50 participants who had attended workshops one or two years prior. In this way, the two cohort groups completing responses were replying three to fifteen months after their workshops. The survey returned 12 volunteered online responses, including the three examples quoted here: (1) “Articles that I read make so much more sense to me now,” (2) “In reviewing new research protocols, I have a better understanding of the mechanism by which the drug’s action is being researched”, and (3) “I have been able to explain the changes occurring in the flu viruses to people who do not understand the importance of vaccination” [
39]. In the latter example, the practicing nurse reported she could now, knowing more about mutations and genetics, explain the need for annual flu vaccinations.
Taken together, the results of previous and current studies support the anticipated outcome that hands-on experiences with the MIT DNA and protein models lead to an increase in nurses’ scientific literacy. From previous studies it has been shown that nurses retained these knowledge gains through specific situations where they applied genetic concepts. Examples given included continuing educational endeavors such as reading research journals, and communicating difficult-to-understand public health information to patients, such as explaining why flu vaccinations are needed on an annual basis.
4.2. Major Findings and Implications from Current Work
Nursing students in the current study experienced the same hands-on genetics lessons as in the prior study, but in addition, we piloted the case-based CYP protein lesson demonstrating genetic susceptibility, as previously detailed in Methods and Results. The authors plan to continue developing curricula utilizing the modeling kits to explain additional genetics and environmental health concepts. In this way, the next generation of nurses will be better prepared to care for marginalized populations who live on lands where environmental justice contamination issues are present. Additionally, because nurses learn in our framework by actively practicing with genetic models, this educational approach is well positioned for international adoption. “Learning by doing” is a universal teaching method and it relies less on language for conveyance of essential concepts [
19,
20,
21,
22].
In both MIT studies, instructors taught protein synthesis in a novel and non-traditional manner. Protein synthesis, also known as the “central dogma of molecular biology” is summarized as: “DNA ➞ RNA ➞ protein.” Thus, biology teachers have traditionally organized their curriculum in this order. They begin by teaching DNA, continue by describing the role of RNA, and complete the lesson with the creation of a protein. However, since students are unfamiliar with protein structure and function, learning this multistep process is confusing. In contrast, the MIT curriculum introduces proteins first, before DNA. This pedagogical innovation results in improving students’ grasp of the process. In our curriculum, students first build, fold, and place a model channel protein into a mock cell membrane. Then students simulate protein synthesis (DNA ➞ RNA ➞ protein), constructing the same channel protein again. By teaching the structure and function of this channel protein first, students understand the process of protein synthesis more easily and can anticipate the amino acid sequence required for a functional channel protein as well.
Proteins are consistently neglected in secondary education. Evaluations and testimony from high school biology teachers teaching protein synthesis using our models and the “proteins first” method report that it took less time to teach protein synthesis, and that their students grasped the concepts better [
40]. One suggestion offered by Vandiver as to why proteins remain unfamiliar to students is that educators have been hampered by lack of tactile protein models. Contrast this absence with the abundance of DNA models commercially available from educational supply houses. Additionally, DNA is visually present in multimedia, and its iconic structure is well recognized.
4.3. Discussion of Nurses’ Learning Gains
Results from the current study revealed nursing students’ weak foundational knowledge about proteins and DNA. Approximately one quarter of the students did not initially recognize proteins as essential cellular components. Constructing two model proteins with important cellular roles produced a significant increase in students’ understanding. The pre-tests also revealed nursing students’ initial confusion with DNA organization, including the terms: genes, chromosomes, and nucleotides. Excellent learning gains were recorded for these key definitions by visualizing the physical nucleotide models and using them to create genes.
A lack of knowledge about the role of proteins in health was also evident, as recognition of cytochrome P450 enzymes’ function in metabolic homeostasis was initially low. In addition, the importance of genetic variation in protein function was not universally recognized. Overall, the post-test responses indicated that the case-based lesson using the protein models effectively demonstrated both CYP protein function and the effects of genetic variations on health outcomes. A deeper understanding of CYP proteins is particularly useful to healthcare professionals, assisting them in recognizing why some people are more sensitive to chemical exposures than others.
A revolutionary advantage of working with the models could be dismantling one of the psychological barriers to biochemistry. Proteins are typically introduced to students as atomic chemical structures. Chemistry’s symbolic language can be both intimidating and difficult to visualize. The models are more cognitively inviting with their colors and shapes that evoke a sense of playfulness, reducing a fear of failure. Overall, the learning process is far more engaging, and the opportunity for hands-on practice provides for greater feedback, which helps students to assemble their own cognitive images for a deeper understanding. Additionally, students working with the kits in pairs can assist and support each other. Anecdotally, instructors noted that even the less interested students asked questions about the biological processes.
4.4. Community Education Outreach and Applicability
Although the major focus of this project was teaching healthcare professionals serving Indigenous populations, in response to requests of our Tribal partner at MBIRI (Eagle Butte, SD, USA) the researchers piloted additional methods to reach Tribal youth and community members. The authors presented lectures and provided hands-on lessons at four different school systems in the region, including Timberlake HS (Timberlake, SD, USA), Windswept School (Eagle Butte, SD, USA), Wakpala School (Mission Township, SD, USA), and Dupree HS (Dupree, SD, USA). Additionally, the Oglala Lakota College in Eagle Butte hosted a presentation provided by one of the authors, speaking on environmental health and toxicology, which drew a large number of community members as well as college students to an evening event. These sessions were encouraging, and the authors plan to do more community-based events with Tribal partners and rural populations in the future.