Forecasting Weekly Dengue Cases by Integrating Google Earth Engine-Based Risk Predictor Generation and Google Colab-Based Deep Learning Modeling in Fortaleza and the Federal District, Brazil
Abstract
:1. Introduction
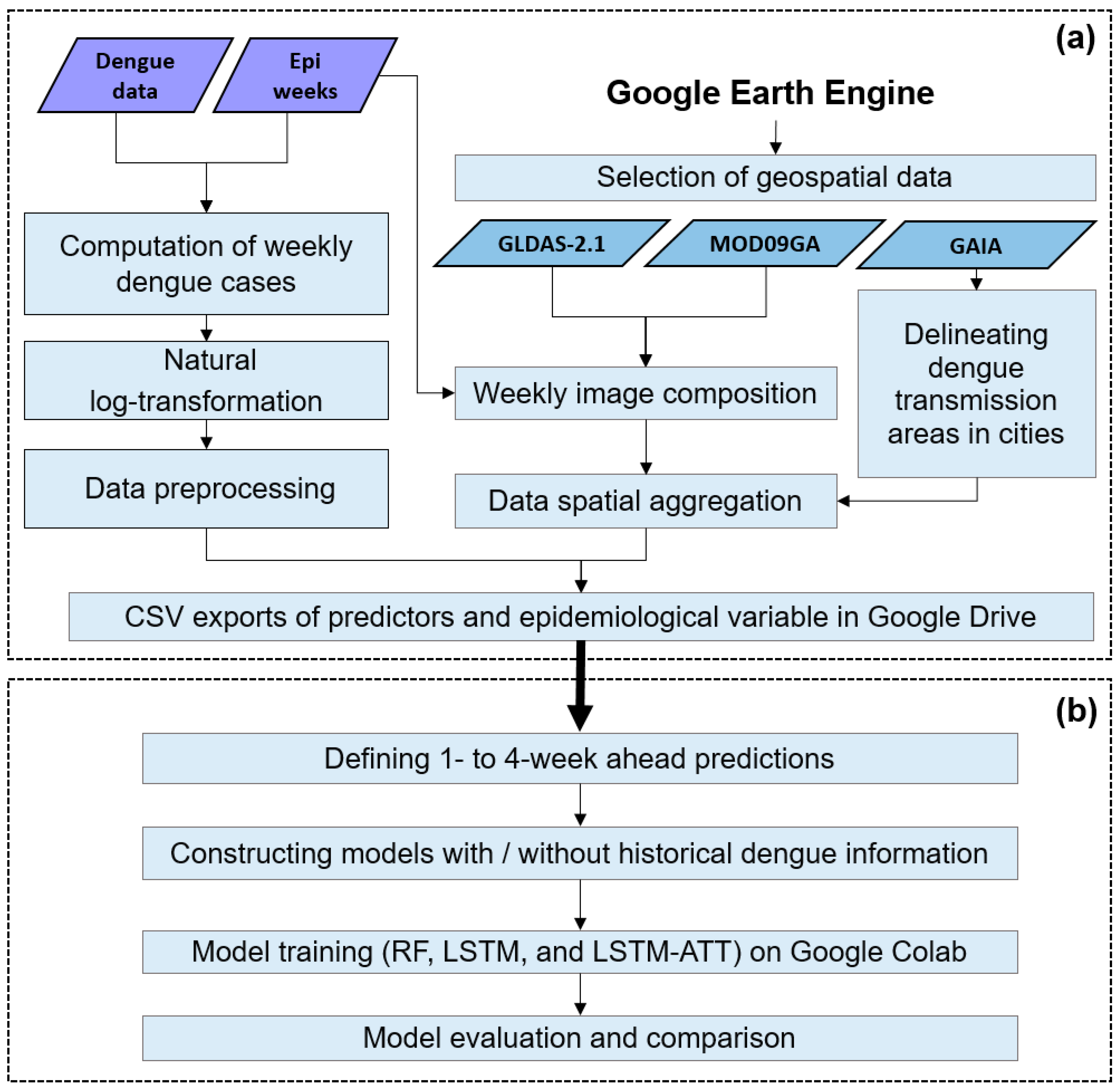
2. The Framework for Dengue Risk Prediction Based on GEE and Google Colab
2.1. Models
2.1.1. LSTM
2.1.2. LSTM with Attention Mechanism
2.1.3. RF
2.2. Model Evaluation
2.3. Multi-Date Ahead Prediction Scenarios
3. Experiments
3.1. Generating the Time Series of Weekly Dengue Cases
3.2. Delineating the Dengue Transmission Areas and Generating the Time Series of Risk Predictors Based on the GEE Platform
3.3. Model Construction, Training, and Evaluation Using Google Colab
4. Results
4.1. Time Series of Climate and Environmental Factors and Weekly Dengue Cases
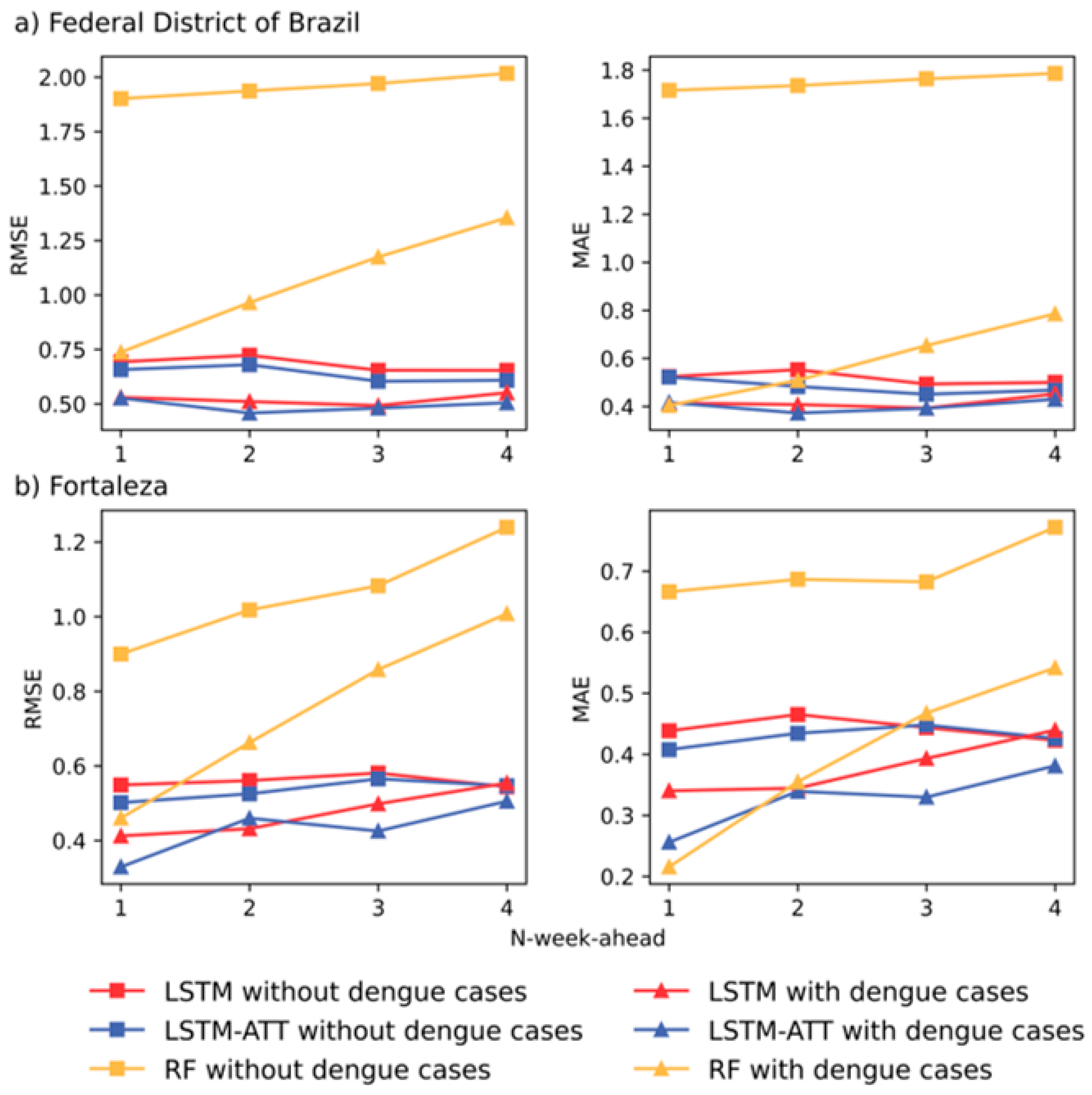
4.2. Outcomes of RF, LSTM and LSTM-ATT Modeling
- RF models frequently have higher prediction errors than LSTM and LSTM-ATT models, and introducing historical dengue data as one of the input features can improve the performance of RF models for 1- to 4-week ahead predictions (see the yellow lines in Figure 4). Most of the predicted curves of RF models differed greatly from those of the observed cases, especially on the dataset of the Federal District of Brazil (Figure 5c).
- All the blue and red lines are relatively stable in Figure 4, which suggests a slight difference in the accuracies of the 1- to 4-week ahead predictions for both LSTM and LSTM-ATT models.
- The red curves with squares are frequently above the blue curves with squares in Figure 4, which suggests that LSTM modeling can benefit from an attention mechanism, which can further achieve performance lift in most cases. Similarly, the red curves with triangles are frequently above the blue curve with triangles in Figure 4, which suggests that LSTM-ATT modeling can also benefit from an attention mechanism.
5. Discussion
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A
Appendix B
| Models | Federal District of Brazil | Fortaleza | ||||
| RMSE | MAE | RMSE | MAE | |||
| LSTM | LSTM without dengue cases | 1-week | 0.6929 | 0.5238 | 0.5488 | 0.4385 |
| 2-week | 0.7231 | 0.5527 | 0.5606 | 0.4652 | ||
| 3-week | 0.6540 | 0.4930 | 0.5808 | 0.4437 | ||
| 4-week | 0.6535 | 0.4999 | 0.5439 | 0.4229 | ||
| LSTM with dengue cases | 1-week | 0.5299 | 0.4137 | 0.4123 | 0.3403 | |
| 2-week | 0.5105 | 0.4074 | 0.4317 | 0.3446 | ||
| 3-week | 0.4920 | 0.3931 | 0.4983 | 0.3932 | ||
| 4-week | 0.5509 | 0.4519 | 0.5539 | 0.4397 | ||
| LSTM-ATT without dengue cases | 1-week | 0.6570 | 0.5222 | 0.5017 | 0.4077 | |
| 2-week | 0.6798 | 0.4827 | 0.5251 | 0.4345 | ||
| 3-week | 0.6037 | 0.4505 | 0.5653 | 0.4481 | ||
| 4-week | 0.6083 | 0.4677 | 0.5467 | 0.4260 | ||
| LSTM-ATT with dengue cases | 1-week | 0.5265 | 0.4162 | 0.3292 | 0.2560 | |
| 2-week | 0.4579 | 0.3723 | 0.4598 | 0.3394 | ||
| 3-week | 0.4805 | 0.3920 | 0.4254 | 0.3298 | ||
| 4-week | 0.5049 | 0.4295 | 0.5053 | 0.3811 | ||
| RF | RF without dengue cases | 1-week | 1.9010 | 1.7157 | 0.8998 | 0.6661 |
| 2-week | 1.9362 | 1.7358 | 1.0179 | 0.6866 | ||
| 3-week | 1.9702 | 1.7637 | 1.0827 | 0.6824 | ||
| 4-week | 2.0166 | 1.7864 | 1.2393 | 0.7717 | ||
| RF with dengue cases | 1-week | 0.7371 | 0.4041 | 0.4601 | 0.2156 | |
| 2-week | 0.9646 | 0.5087 | 0.6626 | 0.3550 | ||
| 3-week | 1.1738 | 0.6531 | 0.8581 | 0.4675 | ||
| 4-week | 1.3540 | 0.7855 | 1.0079 | 0.5417 | ||
References
- Horstick, O.; Tozan, Y.; Wilder-Smith, A. Reviewing dengue: Still a neglected tropical disease? PLoS Negl. Trop. Dis. 2015, 9, e0003632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Ryan, S.J.; Carlson, C.J.; Mordecai, E.A.; Johnson, L.R. Global expansion and redistribution of Aedes-borne virus transmission risk with climate change. PLoS Negl. Trop. Dis. 2019, 13, e0007213. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Kou, S.C.; Lu, F.; Brownstein, J.S.; Brooke, N.; Santillana, M. Advances in using Internet searches to track dengue. PLoS Comput. Biol. 2017, 13, e1005607. [Google Scholar] [CrossRef] [Green Version]
- Withanage, G.P.; Viswakula, S.D.; Nilmini Silva Gunawardena, Y.I.; Hapugoda, M.D. A forecasting model for dengue incidence in the District of Gampaha, Sri Lanka. Parasites Vectors 2018, 11, 262. [Google Scholar] [CrossRef] [PubMed]
- Polwiang, S. The time series seasonal patterns of dengue fever and associated weather variables in Bangkok (2003–2017). BMC Infect. Dis. 2020, 20, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estallo, E.L.; Benitez, E.M.; Lanfri, M.A.; Scavuzzo, C.M.; Almirón, W.R. MODIS Environmental Data to Assess Chikungunya, Dengue, and Zika Diseases Through Aedes (Stegomia) aegypti Oviposition Activity Estimation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 5461–5466. [Google Scholar] [CrossRef]
- Jain, R.; Sontisirikit, S.; Iamsirithaworn, S.; Prendinger, H. Prediction of dengue outbreaks based on disease surveillance, meteorological and socio-economic data. BMC Infect. Dis. 2019, 19, 272. [Google Scholar] [CrossRef]
- Li, Z.; Gurgel, H.; Xu, L.; Yang, L.; Dong, J. Improving Dengue Forecasts by Using Geospatial Big Data Analysis in Google Earth Engine and the Historical Dengue Information-Aided Long Short Term Memory Modeling. Biology 2022, 11, 169. [Google Scholar] [CrossRef]
- Zhao, N.; Charland, K.; Carabali, M.; Nsoesie, E.O.; Maheu-Giroux, M.; Rees, E.; Yuan, M.; Garcia Balaguera, C.; Jaramillo Ramirez, G.; Zinszer, K. Machine learning and dengue forecasting: Comparing random forests and artificial neural networks for predicting dengue burden at national and sub-national scales in Colombia. PLoS Negl. Trop. Dis. 2020, 14, e0008056. [Google Scholar] [CrossRef]
- Buczak, A.L.; Baugher, B.; Moniz, L.J.; Bagley, T.; Babin, S.M.; Guven, E. Ensemble method for dengue prediction. PLoS ONE 2018, 13, e0189988. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ong, J.H.Y.; Rajarethinam, J.; Yap, G.; Ng, L.C.; Cook, A.R. Neighbourhood level real-time forecasting of dengue cases in tropical urban Singapore. BMC Med. 2018, 16, 129. [Google Scholar] [CrossRef] [Green Version]
- Marti, R.; Li, Z.; Catry, T.; Roux, E.; Mangeas, M.; Handschumacher, P.; Gaudart, J.; Tran, A.; Demagistri, L.; Faure, J.-F.; et al. A Mapping Review on Urban Landscape Factors of Dengue Retrieved from Earth Observation Data, GIS Techniques, and Survey Questionnaires. Remote Sens. 2020, 12, 932. [Google Scholar] [CrossRef] [Green Version]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Quackenbush, L.; Adeli, S.; Brisco, B. Google Earth Engine for geo-big data applications: A meta-analysis and systematic review. ISPRS J. Photogramm. Remote Sens. 2020, 164, 152–170. [Google Scholar] [CrossRef]
- Amani, M.; Ghorbanian, A.; Ahmadi, S.A.; Kakooei, M.; Moghimi, A.; Mirmazloumi, S.M.; Moghaddam, S.H.A.; Mahdavi, S.; Ghahremanloo, M.; Parsian, S.; et al. Google Earth Engine Cloud Computing Platform for Remote Sensing Big Data Applications: A Comprehensive Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 5326–5350. [Google Scholar] [CrossRef]
- Wimberly, M.C.; Nekorchuk, D.M.; Kankanala, R.R. Cloud-based applications for accessing satellite Earth observations to support malaria early warning. Sci. Data 2022, 9, 208. [Google Scholar] [CrossRef]
- Frake, A.N.; Peter, B.G.; Walker, E.D.; Messina, J.P. Leveraging big data for public health: Mapping malaria vector suitability in Malawi with Google Earth Engine. PLoS ONE 2020, 15, e0235697. [Google Scholar] [CrossRef]
- Carvajal, T.M.; Viacrusis, K.M.; Hernandez, L.F.T.; Ho, H.T.; Amalin, D.M.; Watanabe, K. Machine learning methods reveal the temporal pattern of dengue incidence using meteorological factors in metropolitan Manila, Philippines. BMC Infect. Dis. 2018, 18, 183. [Google Scholar] [CrossRef]
- Baquero, O.S.; Santana, L.M.R.; Chiaravalloti-Neto, F. Dengue forecasting in São Paulo city with generalized additive models, artificial neural networks and seasonal autoregressive integrated moving average models. PLoS ONE 2018, 13, e0195065. [Google Scholar] [CrossRef]
- Liu, D.; Guo, S.; Zou, M.; Chen, C.; Deng, F.; Xie, Z.; Hu, S.; Wu, L. A dengue fever predicting model based on Baidu search index data and climate data in South China. PLoS ONE 2019, 14, e0226841. [Google Scholar] [CrossRef]
- Mussumeci, E.; Codeco Coelho, F. Large-scale multivariate forecasting models for Dengue—LSTM versus random forest regression. Spat Spatiotemporal Epidemiol. 2020, 35, 100372. [Google Scholar] [CrossRef]
- Benedum, C.M.; Shea, K.M.; Jenkins, H.E.; Kim, L.Y.; Markuzon, N. Weekly dengue forecasts in Iquitos, Peru; San Juan, Puerto Rico; and Singapore. PLoS Negl. Trop. Dis. 2020, 14, e0008710. [Google Scholar] [CrossRef]
- Liu, K.; Yin, L.; Zhang, M.; Kang, M.; Deng, A.-P.; Li, Q.-L.; Song, T. Facilitating fine-grained intra-urban dengue forecasting by integrating urban environments measured from street-view images. Infect. Dis. Poverty 2021, 10, 40. [Google Scholar] [CrossRef]
- Xu, J.; Xu, K.; Li, Z.; Meng, F.; Tu, T.; Xu, L.; Liu, Q. Forecast of Dengue Cases in 20 Chinese Cities Based on the Deep Learning Method. Int. J. Env. Res. Public Health 2020, 17, 453. [Google Scholar] [CrossRef] [Green Version]
- Bomfim, R.; Pei, S.; Shaman, J.; Yamana, T.; Makse, H.A.; Andrade, J.S., Jr.; Lima Neto, A.S.; Furtado, V. Predicting dengue outbreaks at neighbourhood level using human mobility in urban areas. J. R. Soc. Interface 2020, 17, 20200691. [Google Scholar] [CrossRef]
- Nguyen, V.-H.; Tuyet-Hanh, T.T.; Mulhall, J.; Minh, H.V.; Duong, T.Q.; Chien, N.V.; Nhung, N.T.T.; Lan, V.H.; Minh, H.B.; Cuong, D.; et al. Deep learning models for forecasting dengue fever based on climate data in Vietnam. PLoS Negl. Trop. Dis. 2022, 16, e0010509. [Google Scholar] [CrossRef]
- Akhtar, M.; Kraemer, M.U.G.; Gardner, L.M. A dynamic neural network model for predicting risk of Zika in real time. BMC Med. 2019, 17, 171. [Google Scholar] [CrossRef] [Green Version]
- Aiken, E.L.; Nguyen, A.T.; Viboud, C.; Santillana, M. Toward the use of neural networks for influenza prediction at multiple spatial resolutions. Sci. Adv. 2021, 7, eabb1237. [Google Scholar] [CrossRef]
- Carneiro, T.; NóBrega, R.V.M.D.; Nepomuceno, T.; Bian, G.B.; Albuquerque, V.H.C.D.; Filho, P.P.R. Performance Analysis of Google Colaboratory as a Tool for Accelerating Deep Learning Applications. IEEE Access 2018, 6, 61677–61685. [Google Scholar] [CrossRef]
- Bisong, E. Google colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Bisong, E., Ed.; Apress: Berkeley, CA, USA, 2019; pp. 59–64. [Google Scholar]
- Tsunoda, T.; Cuong, T.C.; Dong, T.D.; Yen, N.T.; Le, N.H.; Phong, T.V.; Minakawa, N. Winter refuge for Aedes aegypti and Ae. albopictus mosquitoes in Hanoi during Winter. PLoS ONE 2014, 9, e95606. [Google Scholar] [CrossRef]
- Maciel-de-Freitas, R.; Neto, R.B.; Gonçalves, J.M.; Codeço, C.T.; Lourenço-de-Oliveira, R. Movement of dengue vectors between the human modified environment and an urban forest in Rio de Janeiro. J. Med. Entomol. 2006, 43, 1112–1120. [Google Scholar] [CrossRef]
- Lacroix, R.; Delatte, H.; Hue, T.; Reiter, P. Dispersal and survival of male and female Aedes albopictus (Diptera: Culicidae) on Réunion Island. J. Med. Entomol. 2009, 46, 1117–1124. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1–15. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Z.; Kong, D.; Han, H.; Zhao, Y. EA-LSTM: Evolutionary attention-based LSTM for time series prediction. Knowl. -Based Syst. 2019, 181, 104785. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Huang, M.; Zhu, X.; Zhao, L. Attention-based LSTM for aspect-level sentiment classification. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 606–615. [Google Scholar]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Routledge: London, UK, 2017. [Google Scholar]
- Hyndman, R.J.; Koehler, A.B. Another look at measures of forecast accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Gurgel, H.; Li, M.; Dessay, N.; Gong, P. Urban Land Expansion from Scratch to Urban Agglomeration in the Federal District of Brazil in the Past 60 Years. Int. J. Environ. Res. Public Health 2022, 19, 1032. [Google Scholar] [CrossRef]
- da Silva Neto, S.R.; Tabosa de Oliveira, T.; Teixiera, I.V.; Medeiros Neto, L.; Souza Sampaio, V.; Lynn, T.; Endo, P.T. Arboviral disease record data—Dengue and Chikungunya, Brazil, 2013–2020. Sci. Data 2022, 9, 198. [Google Scholar] [CrossRef]
- Drumond, B.; Angelo, J.; Xavier, D.R.; Catao, R.; Gurgel, H.; Barcellos, C. Dengue spatiotemporal dynamics in the Federal District, Brazil: Occurrence and permanence of epidemics. Cien Saude Colet 2020, 25, 1641–1652. [Google Scholar] [CrossRef]
- MacCormack-Gelles, B.; Lima Neto, A.S.; Sousa, G.S.; Nascimento, O.J.; Machado, M.M.T.; Wilson, M.E.; Castro, M.C. Epidemiological characteristics and determinants of dengue transmission during epidemic and non-epidemic years in Fortaleza, Brazil: 2011–2015. PLoS Negl. Trop. Dis. 2018, 12, e0006990. [Google Scholar] [CrossRef]
- Charlesworth, S.M.; Kligerman, D.C.; Blackett, M.; Warwick, F. The Potential to Address Disease Vectors in Favelas in Brazil Using Sustainable Drainage Systems: Zika, Drainage and Greywater Management. Int. J. Environ. Res. Public Health 2022, 19, 2860. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Li, X.; Wang, J.; Bai, Y.; Chen, B.; Hu, T.; Liu, X.; Xu, B.; Yang, J.; Zhang, W.; et al. Annual maps of global artificial impervious area (GAIA) between 1985 and 2018. Remote Sens. Environ. 2020, 236, 111510. [Google Scholar] [CrossRef]
- Rodell, M.; Houser, P.R.; Jambor, U.; Gottschalck, J.; Mitchell, K.; Meng, C.-J.; Arsenault, K.; Cosgrove, B.; Radakovich, J.; Bosilovich, M.; et al. The Global Land Data Assimilation System. Bull. Am. Meteorol. Soc. 2004, 85, 381. [Google Scholar] [CrossRef] [Green Version]
- Vermote, E.; Wolfe, R. MOD09GA MODIS/Terra Surface Reflectance Daily L2G Global 1kmand 500m SIN Grid V006. NASA EOSDIS Land Processes DAAC. Available online: https://doi.org/10.5067/MODIS/MOD09GA.006 (accessed on 15 July 2022).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Li, Z.; Dong, J. Big Geospatial Data and Data-Driven Methods for Urban Dengue Risk Forecasting: A Review. Remote Sens. 2022, 14, 5052. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, T.; Liu, K.; Xia, Y.; Lu, Y.; Jing, Q.; Yang, Z.; Hu, W.; Lu, J. Developing a Time Series Predictive Model for Dengue in Zhongshan, China Based on Weather and Guangzhou Dengue Surveillance Data. PLoS Negl. Trop. Dis. 2016, 10, e0004473. [Google Scholar] [CrossRef] [Green Version]
- McGough, S.F.; Clemente, L.; Kutz, J.N.; Santillana, M. A dynamic, ensemble learning approach to forecast dengue fever epidemic years in Brazil using weather and population susceptibility cycles. J. R. Soc. Interface 2021, 18, 20201006. [Google Scholar] [CrossRef]
- Kiang, M.V.; Santillana, M.; Chen, J.T.; Onnela, J.P.; Krieger, N.; Engo-Monsen, K.; Ekapirat, N.; Areechokchai, D.; Prempree, P.; Maude, R.J.; et al. Incorporating human mobility data improves forecasts of Dengue fever in Thailand. Sci. Rep. 2021, 11, 923. [Google Scholar] [CrossRef]
- Sanchez, L.; Vanlerberghe, V.; Alfonso, L.; Marquetti, M.d.C.; Guzman, M.G.; Bisset, J.; van der Stuyft, P. Aedes aegypti larval indices and risk for dengue epidemics. Emerg. Infect. Dis. 2006, 12, 800–806. [Google Scholar] [CrossRef] [Green Version]
- Ong, J.; Aik, J.; Ng, L.C. Short Report: Adult Aedes abundance and risk of dengue transmission. PLoS Negl. Trop. Dis. 2021, 15, e0009475. [Google Scholar] [CrossRef]
- Shahid, F.; Zameer, A.; Muneeb, M. Predictions for COVID-19 with deep learning models of LSTM, GRU and Bi-LSTM. Chaos Solitons Fractals 2020, 140, 110212. [Google Scholar] [CrossRef] [PubMed]
- Bracher, J.; Ray, E.L.; Gneiting, T.; Reich, N.G. Evaluating epidemic forecasts in an interval format. PLoS Comput. Biol. 2021, 17, e1008618. [Google Scholar] [CrossRef] [PubMed]




| Dengue Risk Predictors and Epidemiological Variables | Data Sources | Spatial Resolution | Temporal Resolution | Period | |
|---|---|---|---|---|---|
| Climate | Precipitation per week (Rsum) | GLDAS-2.1 | 27,000 m | 3-hourly | 2000 to present |
| Mean temperature per week (Tmean) | |||||
| Mean relative humidity per week (RHmean) | |||||
| Environment | Mean NDVI per week (NDVImean) | MOD09GA | 500 m | Daily | 2000 to present |
| Epidemiology | Number of dengue cases per week | Brazilian arboviral disease by [41] | City-level | Weekly | 2013–2020 |
| Dengue transmission areas | 1 km buffer around the imperious surface in cities | GAIA | 30 m | Annual | 2017 |
| Parameters | LSTM without Dengue Cases | LSTM with Dengue Cases | LSTM-ATT without Dengue Cases | LSTM-ATT with Dengue Cases |
|---|---|---|---|---|
| Time step | 12 | 12 | 12 | 12 |
| Loss function | MSE | MSE | MSE | MSE |
| Number of units | 64 | 64 | 64 | 64 |
| Epoch | 1000 | 1500 | 1500 | 2000 |
| Batch size | 12 | 12 | 12 | 12 |
| Learning rate | 0.005 | 0.003 | 0.005 | 0.003 |
| Optimizer | Adam | Adam | Adam | Adam |
| Attention Size | - | - | 64 | 64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z. Forecasting Weekly Dengue Cases by Integrating Google Earth Engine-Based Risk Predictor Generation and Google Colab-Based Deep Learning Modeling in Fortaleza and the Federal District, Brazil. Int. J. Environ. Res. Public Health 2022, 19, 13555. https://doi.org/10.3390/ijerph192013555
Li Z. Forecasting Weekly Dengue Cases by Integrating Google Earth Engine-Based Risk Predictor Generation and Google Colab-Based Deep Learning Modeling in Fortaleza and the Federal District, Brazil. International Journal of Environmental Research and Public Health. 2022; 19(20):13555. https://doi.org/10.3390/ijerph192013555
Chicago/Turabian StyleLi, Zhichao. 2022. "Forecasting Weekly Dengue Cases by Integrating Google Earth Engine-Based Risk Predictor Generation and Google Colab-Based Deep Learning Modeling in Fortaleza and the Federal District, Brazil" International Journal of Environmental Research and Public Health 19, no. 20: 13555. https://doi.org/10.3390/ijerph192013555




