Effects of Oxytetracycline/Lead Pollution Alone and in the Combined Form on Antibiotic Resistance Genes, Mobile Genetic Elements, and Microbial Communities in the Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Experimental Design
2.3. Sample Collection
2.4. Real-Time Quantitative PCR (qPCR)
2.5. 16 S rRNA Gene High-Throughput Sequencing
2.6. Data Processing
3. Results and Discussion
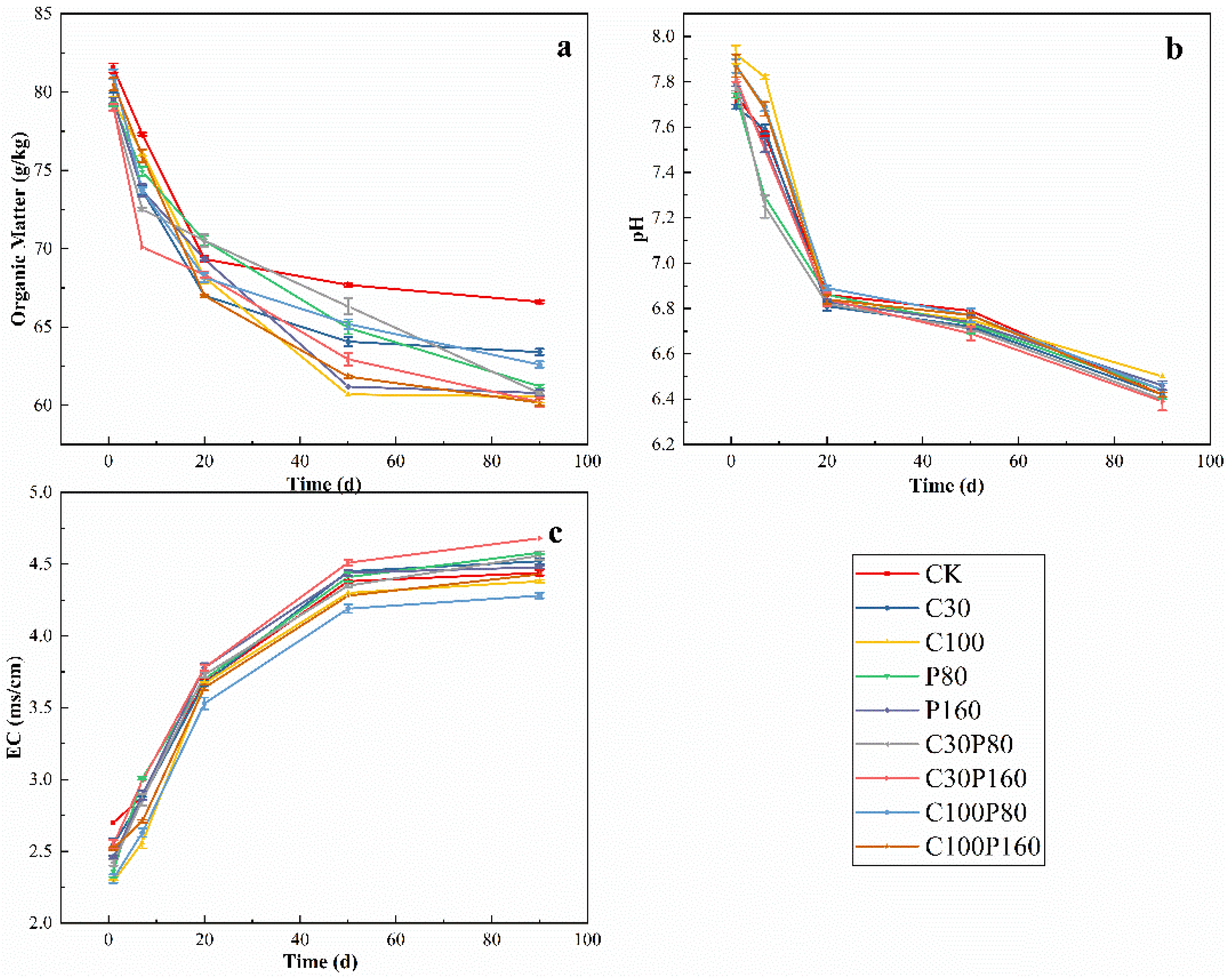
3.1. Soil’s Physical and Chemical Properties
3.2. ARGs and MGEs
3.3. The Soil Microbial Community Evolution
3.4. Relationship between Physical and Chemical Properties of Soil, Microbial Community, ARGs and MGEs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donald, K. Time to Deal with Antibiotics. Science 2013, 342, 777. [Google Scholar] [CrossRef]
- Barrete, A.P.; Ringwald, P. Global Report on Antimalarial Efficacy and Drug Resistance: 2000–2010; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Kalashnikov, M.; Mueller, M.; Mcbeth, C.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. Rapid phenotypic stress-based microfluidic antibiotic susceptibility testing of Gram-negative clinical isolates. Sci. Rep. 2017, 7, 8031. [Google Scholar] [CrossRef]
- Burki, T.K. Superbugs: An Arms Race Against Bacteria. Lancet Respir. Med. 2018, 6, 668. [Google Scholar] [CrossRef] [PubMed]
- Jechalke, S.; Heuer, H.; Siemens, J.; Amelung, W.; Smalla, K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014, 22, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, E.A.; Seyfried, E.; Mcmahon, K.D. Tetracycline Resistance Genes in Activated Sludge Wastewater Treatment Plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef]
- Schmitt, H.; Stoob, K.; Hamscher, G.; Smit, E.; Seinen, W. Tetracyclines and Tetracycline Resistance in Agricultural Soils: Microcosm and Field Studies. Microb. Ecol. 2006, 51, 267–276. [Google Scholar] [CrossRef]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A global perspective on the use, sales, exposure pathways, occurrence, fate, and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 7, 725–759. [Google Scholar] [CrossRef]
- Caban, J.R.; Kuppusamy, S.; Kim, J.H.; Yoon, Y.-E.; Kim, S.Y.; Lee, Y.B. Green manure amendment enhances microbial activity and diversity in antibiotic-contaminated soil. Appl. Soil Ecol. 2018, 129, 72–76. [Google Scholar] [CrossRef]
- Abdu, N.; Abdullahi, A.A.; Abdulkadir, A. Heavy metals and soil microbes. Environ. Chem. Lett. 2016, 15, 65–84. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, F.; Li, H.; Wu, H.; Zhu, J. Contribution of environmental factors on the distribution of antibiotic resistance genes in agricultural soil. Eur. J. Soil Biol. 2020, 102, 103269. [Google Scholar] [CrossRef]
- Mazhar, S.H.; Li, X.; Rashid, A.; Su, J.; Rensing, C. Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 2021, 755, 142702. [Google Scholar] [CrossRef] [PubMed]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial Resistance in Agriculture. mBio 2016, 7, e02227-15. [Google Scholar] [CrossRef] [PubMed]
- Hua, F.; Wang, H.; Lin, C.; Yu, Y. Prevalence of Antibiotic Resistance Genes and Bacterial Pathogens in Long-Term Manured Greenhouse Soils as Revealed by Metagenomic Survey. Environ. Sci. Technol. 2015, 49, 1095–1104. [Google Scholar] [CrossRef]
- Leclercq, S.O.; Chao, W.; Sui, Z.; Hai, W.; Jie, F. A multiplayer game: Species of Clostridium, Acinetobacter, and Pseudomonas are responsible for the persistence of antibiotic resistance genes in manure-treated soils. Environ. Microbiol. 2016, 18, 3494–3508. [Google Scholar] [CrossRef]
- Hu, H.-W.; Han, X.-M.; Shi, X.-Z.; Wang, J.-T.; Han, L.-L.; Chen, D.; He, J.-Z. Temporal changes of antibiotic-resistance genes and bacterial communities in two contrasting soils treated with cattle manure. FEMS Microbiol. Ecol. 2016, 92, 2. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, J.; Mao, D.; Luo, Y. Effect of the selective pressure of sub-lethal level of heavy metals on the fate and distribution of ARGs in the catchment scale. Environ. Pollut. 2017, 220, 900–908. [Google Scholar] [CrossRef]
- Smalla, K.; Heuer, H.; Götz, A.; Niemeyer, D.; Krögerrecklenfort, E.; Tietze, E. Exogenous Isolation of Antibiotic Resistance Plasmids from Piggery Manure Slurries Reveals a High Prevalence and Diversity of IncQ-Like Plasmids. Appl. Environ. Microb. 2000, 66, 4854–4862. [Google Scholar] [CrossRef]
- Shea, K.M. Antibiotic resistance: What is the impact of agricultural uses of antibiotics on children’s health? Pediatrics 2003, 112, 253–258. [Google Scholar] [CrossRef]
- Lu, H.; Xu, Y.; Xu, J.; Ling, J.; Xie, G. Dissemination of antibiotic resistance genes (ARGs) by rainfall on a cyclic economic breeding livestock farm. Int. Biodeterior. Biodegrad. 2019, 138, 114–121. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene richnesss associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235–236, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Qi, Y.; Le, Z.; Han, F.; Li, F.; Yang, H.; Zhang, T.; Feng, Y.; Liu, R.; Sun, Y. The Interactions between Antibiotic Resistance Genes and Heavy Metals Pollution under the Co-selective Pressure Influenced the Bioenzyme Activity. Front. Chem. 2021, 9, 1565. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lan, B.; Fei, H.; Wang, S.; Zhu, G. Heavy metal could drive co-selection of antibiotic resistance in terrestrial subsurface soils. J. Hazard. Mater. 2020, 411, 124848. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-W.; Wang, J.-T.; Li, J.; Li, J.-J.; Ma, Y.-B.; Chen, D.; He, J.-Z. Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ. Microbiol. 2016, 18, 3896–3909. [Google Scholar] [CrossRef] [PubMed]
- Boxall, A.B.; Kolpin, D.W.; Halling-Sørensen, B.; Tolls, J. Peer Reviewed: Are Veterinary Medicines Causing Environmental Risks? Environ. Sci. Technol. 2003, 37, 286A–294A. [Google Scholar] [CrossRef]
- Li, Y.-W.; Wu, X.-L.; Mo, C.-H.; Tai, Y.-P.; Huang, X.-P.; Xiang, L. Investigation of Sulfonamide, Tetracycline, and Quinolone Antibiotics in Vegetable Farmland Soil in the Pearl River Delta Area, Southern China. J. Agric. Food Chem. 2011, 59, 7268–7276. [Google Scholar] [CrossRef]
- Luo, Y.; Feng, L.; Jia, R.; Yang, G.; Yang, Q.; Mu, J. Variation in microbial populations and antibiotic resistance genes in mariculture sediments in the present of the seaweed Ulva fasciata and under the selective pressure of oxytetracycline. Ecotox. Environ. Saf. 2020, 204, 111114. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Lou, C.; Zhai, W.; Tang, X.; Hashmi, M.Z.; Murtaza, R.; Li, Y.; Liu, X.; Xu, J. Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci. Total Environ. 2018, 635, 995–1003. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Zhu, L.; Ge, W.; Wang, J. Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci. Total Environ. 2017, 593–594, 10–17. [Google Scholar] [CrossRef]
- Guo, H.; Xue, S.; Nasir, M.; Lv, J.; Gu, J. Role of Bentonite on the Mobility of Antibiotic Resistance Genes, and Microbial Community in Oxytetracycline and Cadmium Contaminated Soil. Front. Microbiol. 2018, 9, 2722. [Google Scholar] [CrossRef]
- Li, N.; Chen, J.; Liu, C.; Yang, J.; Zhu, C.; Li, H. Cu and Zn exert a greater influence on antibiotic resistance and its transfer than doxycycline in agricultural soils. J. Hazard. Mater. 2021, 423, 127042. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, X.; Wang, J.; Wang, J.; Zhu, L.; Ge, W. Macrolide- and quinolone-resistant bacteria and resistance genes as indicators of antibiotic resistance gene contamination in farmland soil with manure application. Ecol. Indic. 2019, 106, 105456. [Google Scholar] [CrossRef]
- Fernández-Calviño, D.; Bååth, E. Co-selection for antibiotic tolerance in Cu-polluted soil is detected at higher Cu-concentrations than increased Cu-tolerance. Soil Biol. Biochem. 2013, 57, 953–956. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, Y.; Zeng, C.; Li, Y.; Wei, Z. Assessment contributions of physicochemical properties and bacterial community to mitigate the bioavailability of heavy metals during composting based on structural equation models. Bioresource Technol. 2019, 289, 121657. [Google Scholar] [CrossRef]
- Álvarez-Rogel, J.; Peñalver-Alcalá, A.; González-Alcaraz, M.N. Spontaneous vegetation colonizing abandoned metal(loid) mine tailings consistently modulates climatic, chemical and biological soil conditions throughout seasons. Sci. Total Environ. 2022, 838, 1. [Google Scholar] [CrossRef]
- Yanli, F.; Ying, Z.; Hao, D.; Jing, L.; Weiyi, Z.; Yingying, S.; Yanqiu, S. Mechanisms of the effects of humic acid on antibiotic resistance genes and microbial communities in Cd-contaminated soils. Process Saf. Environ. Prot. 2022, 160, 62–69. [Google Scholar] [CrossRef]
- Luo, Y.; Mao, D.; Rysz, M.; Zhou, Q.; Zhang, H.; Xu, L.; Alvarez, P.J.J. Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ. Sci. Technol. 2010, 44, 19, 7220–7225. [Google Scholar] [CrossRef]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular ecology of tetracycline resistance: Development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microb. 2001, 67, 22–32. [Google Scholar] [CrossRef]
- Pei, R.; Kim, S.-C.; Carlson, K.H.; Pruden, A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Frank, T.; Gautier, V.; Talarmin, A.; Bercion, R.; Arlet, G. Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). J. Antimicrob. Chemother. 2007, 59, 742–745. [Google Scholar] [CrossRef]
- Shi, R.-Y.; Liu, Z.-D.; Li, Y.; Jiang, T.; Xu, M.; Li, J.-Y.; Xu, R.-K. Mechanisms for increasing soil resistance to acidification by long-term manure application. Soil Tillage Res. 2018, 185, 77–84. [Google Scholar] [CrossRef]
- Zhou, D.-M.; Hao, X.-Z.; Wang, Y.-J.; Dong, Y.-H.; Cang, L. Copper and Zn uptake by radish and pakchoi as affected by application of livestock and poultry manures. Chemosphere 2005, 59, 167–175. [Google Scholar] [CrossRef]
- Antil, R.S.; Singh, M. Effects of organic manures and fertilizers on organic matter and nutrients status of the soil. Agron. Soil. Sci. 2007, 53, 519–528. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.-G. Does organically produced lettuce harbor higher richness of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Qiao, M.; Zhang, B.; Cheng, W.-D.; Zhu, Y.-G. Richness and Diversity of Tetracycline Resistance Genes in Soils Adjacent to Representative Swine Feedlots in China. Sci. Technol. 2010, 44, 6933–6939. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Sun, W.; Gu, J.; Wang, X.-J.; Sun, J.-J.; Yin, Y.-N.; Duan, M.-L. Variable effects of oxytetracycline on antibiotic resistance gene richness and the bacterial community during aerobic composting of cow manure. J. Hazard. Mater. 2016, 315, 61–69. [Google Scholar] [CrossRef]
- Kong, W.D.; Zhu, Y.G.; Fu, B.J.; Marschner, P.; He, J.Z. The veterinary antibiotic oxytetracycline and Cu influence functional diversity of the soil microbial community. Environ. Pollut. 2006, 143, 129–137. [Google Scholar] [CrossRef]
- Tongyi, Y.; Yanpeng, L.; Xingang, W.; Fen, Y.; Jun, L.; Yubin, T. Co-selection for antibiotic resistance genes is induced in a soil amended with zinc. Soil Use Manag. 2020, 36, 328–337. [Google Scholar] [CrossRef]
- Pei, R.; Cha, J.; Carlson, K.H.; Pruden, A. Response of Antibiotic Resistance Genes (ARG) to Biological Treatment in Dairy Lagoon Water. Environ. Sci. Technol. 2007, 41, 5108–5113. [Google Scholar] [CrossRef] [PubMed]
- Sardar, M.F.; Zhu, C.; Geng, B.; Ahmad, H.R.; Song, T.; Li, H. The fate of antibiotic resistance genes in cow manure composting: Shaped by temperature-controlled composting stages. Bioresource Technol. 2020, 320, 124403. [Google Scholar] [CrossRef] [PubMed]
- Huerta, B.; Marti, E.; Gros, M.; López, P.; Pompêo, M.; Armengol, J.; Barceló, D.; Balcázar, J.L.; Rodríguez-Mozaz, S.; Marcé, R. Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci. Total Environ. 2013, 456–457, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Z.; Xiang, Y.; Xu, R.; Zheng, Y.; Lu, Y.; Jia, M.; Sun, S.; Cao, J.; Xiong, W. Evolutions of antibiotic resistance genes (ARGs), class 1 integron-integrase (intl 1) and potential hosts of ARGs during sludge anaerobic digestion with the iron nanoparticles addition. Sci. Total Environ. 2020, 724, 138248. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fu, Q.; Wen, Q.; Wu, Y.; Bao, H.; Guo, J. Microbial community competition rather than high-temperature predominates ARGs elimination in swine manure composting. J. Hazard. Mater. 2021, 423, 127149. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Huang, X.-H.; Sun, J.-Z.; Graham, D.W.; Xie, B. Antibiotic Resistance Genes and Associated Microbial Community Conditions in Aging Landfill Systems. Environ. Sci. Technol. 2017, 51, 12859–12867. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, G.; Hou, S.; Zhang, T.; Li, Z.; Liang, F. Distribution of ARGs and MGEs among glacial soil, permafrost, and sediment using metagenomic analysis. Environ. Pollut. 2017, 234, 339–346. [Google Scholar] [CrossRef]
- Amachawadi, R.G.; Scott, H.M.; Vinasco, J.; Tokach, M.D.; Dritz, S.S.; Nelssen, J.L.; Nagaraja, T.G. Effects of In-Feed Copper, Chlortetracycline, and Tylosin on the Prevalence of Transferable Copper Resistance Gene, tcrB, Among Fecal Enterococci of Weaned Piglets. Foodborne Pathog. Dis. 2015, 12, 670–678. [Google Scholar] [CrossRef]
- Chapman, J.S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 2003, 51, 271–276. [Google Scholar] [CrossRef]





| Samples | Moisture Content (%) | pH | Organic Carbon (g/kg) | Electric Conductivity (ms/cm) |
|---|---|---|---|---|
| Topsoil | 10.34 | 8.38 | 17.92 | 1.214 |
| Chicken manure | 8.63 | 7.12 | 50.24 | 2.845 |
| Groups | CK | C30 | C100 | P80 | P160 | C30P80 | C30P160 | C100P80 | C100P160 |
|---|---|---|---|---|---|---|---|---|---|
| OTC (mg/kg) | 0 | 30 | 100 | 0 | 0 | 30 | 30 | 100 | 100 |
| Pb (mg/kg) | 0 | 0 | 0 | 80 | 160 | 80 | 160 | 80 | 160 |
| Gene Type | Genes | Primer | Fragment Length/bp |
|---|---|---|---|
| Tetracycline genes | tetB [38] | F: CGAAGTAGGGGTTGAGACGC R: AGACCAAGACCCGCTAATGAA | 192 |
| tetQ [39] | F: AGAATCTGCTGTTTGCCAGTG R: CGGAGTGTCAATGATATTGCA | 169 | |
| Sulfonamides genes | sul1 [37] | F: CGCACCGGAAACATCGCTGCAC R: TGAAGTTCCGCCGCAAGGCTCG | 163 |
| sul2 [37] | F: TCCGGTGGAGGCCGGTATCTGG R: CGGGAATGCCATCTGCCTTGAG | 191 | |
| sul3 [40] | F: TCCGTTCAGCGAATTGGTGCAG R: TTCGTTCACGCCTTACACCAGC | 143 | |
| EMGs | intl1 [41] | F: CTGGATTTCGATCACGGCACG R: ACATGCGTGTAAATCATCGTCG | 473 |
| intl2 [37] | F: GTTATTTTATTGCTGGGATTAGGC R: TTTTACGCTGCTGTATGGTGC | 166 | |
| 16 S rRNA | 16 S rRNA [37] | F: CCTACGGGAGGCAGCAG R: ATTACCGCGGCTGCTGG | 193 |
| Treatments | CK | C30 | C100 | P80 | P160 | C30P80 | C30P160 | C100P80 | C100P160 |
|---|---|---|---|---|---|---|---|---|---|
| tetB | 0.0030 ± 3 × 10−4 ab | 0.0028 ± 1.5 × 10−4 b | 0.0022 ± 1.5 × 10−4 c | 0.0014 ± 1.5 × 10−4 × 10 | 0.0021 ± 1.3 × 10−4 d | 0.0033 ± 3 × 10−4 a | 0.0022 ± 3 × 10−4 c | 0.0025 ± 9 × 10−5 c | 0.0022 ± 1.11 × 10−4 c |
| tetQ | 2.6150 × 10−4 ± 5 × 10−5 c | 2.2212 × 10−4 ± 5 × 10−5 c | 3.0329 × 10−4 ± 5 × 10−5 a | 2.8212 × 10−4 ± 5 × 10−5 bc | 3.0022 × 10−4 ± 5 × 10−5 ab | 2.4392 × 10−4 ± 5 × 10−5 c | 2.6967 × 10−4 ± 5 × 10−5 b | 2.0902 × 10−4 ± 5 × 10−5 d | 2.1732 × 10−4 ± 5 × 10−5 d |
| sul1 | 0.0404 ± 3 × 10−4 c | 0.0329 ± 6 × 10−4 c | 0.0422 ± 3 × 10−4 b | 0.0245 ± 6 × 10−4 d | 0.0361 ± 3 × 10−4 bc | 0.0277 ± 6 × 10−4 d | 0.033 ± 3 × 10−4 c | 0.0414 ± 3 × 10−4 bc | 0.0568 ± 0.0015 a |
| sul2 | 0.0139 ± 2 × 10−4 a | 0.0114 ± 2 × 10−4 ab | 0.0036 ± 2 × 10−4 c | 0.0140 ± 2 × 10−4 b | 0.0016 ± 2 × 10−4 a | 0.0156 ± 2 × 10−4 c | 0.0107 ± 6.6 × 10−4 c | 0.0041 ± 2 × 10−4 bc | 0.0157 ± 6 × 10−4 a |
| sul3 | 4.6702 × 10−6 ± 0 × 10 | 6.3394 × 10−6 ± 3 × 10−7 c | 4.8222 × 10−6 ± 5.4 × 10−7 d × 10 | 7.4022 × 10−6 ± 0 bc | 5.7073 × 10−6 ± 2.6 × 10−7 d | 5.6950 × 10−6 ± 1.5 × 10−7 cd | 7.1370 × 10−6 ± 2 × 10−7 bc | 1.2162 × 10−5 ± 1.2 × 10−6 a | 7.3663 × 10−6 ± 7 × 10−7 b |
| intl1 | 0.0025 ± 2 × 10−4 c | 2.6225 × 10-4 ± 2.6 × 10−5 f | 5.3958 × 10−4 ± 5 × 10−5 d | 2.5314 × 10−4 ± 2.5 × 10−5 f | 2.5633 × 10−4 ± 2.5 × 10−6 f | 0.0071 ± 7.1 × 10−5 a | 3.3561 × 10−4 ± 3.3 × 10−5 × 10f | 1.2884 × 10−4 ± 1.2 × 10−5 g | 0.0056 ± 2.8 × 10−4 b |
| intl2 | 3.4871 × 10−4 ± 0 bc | 2.7539 × 10−4 ± 2.7 × 10−5 c | 6.3766 × 10−4 ± 3.3 × 10−5 a | 2.8952 × 10−4 ± 2.8 × 10−5 c | 4.0077 × 10−4 ± 2.8 × 10−5 b | 2.7865 × 10−4 ± 2.7 × 10−5 c | 3.3495 × 10−4 ± 3.3 × 10−5 c | 3.2769 × 10−4 ± 3.2 × 10−5 c | 2.9727 × 10−4 ± 2.9 × 10−5 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Li, Z.; Shao, Y.; Fu, Y.; Zhang, W.; Shao, Y.; Zhu, Y. Effects of Oxytetracycline/Lead Pollution Alone and in the Combined Form on Antibiotic Resistance Genes, Mobile Genetic Elements, and Microbial Communities in the Soil. Int. J. Environ. Res. Public Health 2022, 19, 15619. https://doi.org/10.3390/ijerph192315619
Guo T, Li Z, Shao Y, Fu Y, Zhang W, Shao Y, Zhu Y. Effects of Oxytetracycline/Lead Pollution Alone and in the Combined Form on Antibiotic Resistance Genes, Mobile Genetic Elements, and Microbial Communities in the Soil. International Journal of Environmental Research and Public Health. 2022; 19(23):15619. https://doi.org/10.3390/ijerph192315619
Chicago/Turabian StyleGuo, Tengfei, Zhaoyi Li, Yanqiu Shao, Yanli Fu, Weiyi Zhang, Yingying Shao, and Ying Zhu. 2022. "Effects of Oxytetracycline/Lead Pollution Alone and in the Combined Form on Antibiotic Resistance Genes, Mobile Genetic Elements, and Microbial Communities in the Soil" International Journal of Environmental Research and Public Health 19, no. 23: 15619. https://doi.org/10.3390/ijerph192315619
APA StyleGuo, T., Li, Z., Shao, Y., Fu, Y., Zhang, W., Shao, Y., & Zhu, Y. (2022). Effects of Oxytetracycline/Lead Pollution Alone and in the Combined Form on Antibiotic Resistance Genes, Mobile Genetic Elements, and Microbial Communities in the Soil. International Journal of Environmental Research and Public Health, 19(23), 15619. https://doi.org/10.3390/ijerph192315619







