Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Consideration
2.2. Real Time Case Notification and MBS-Lab Deployment
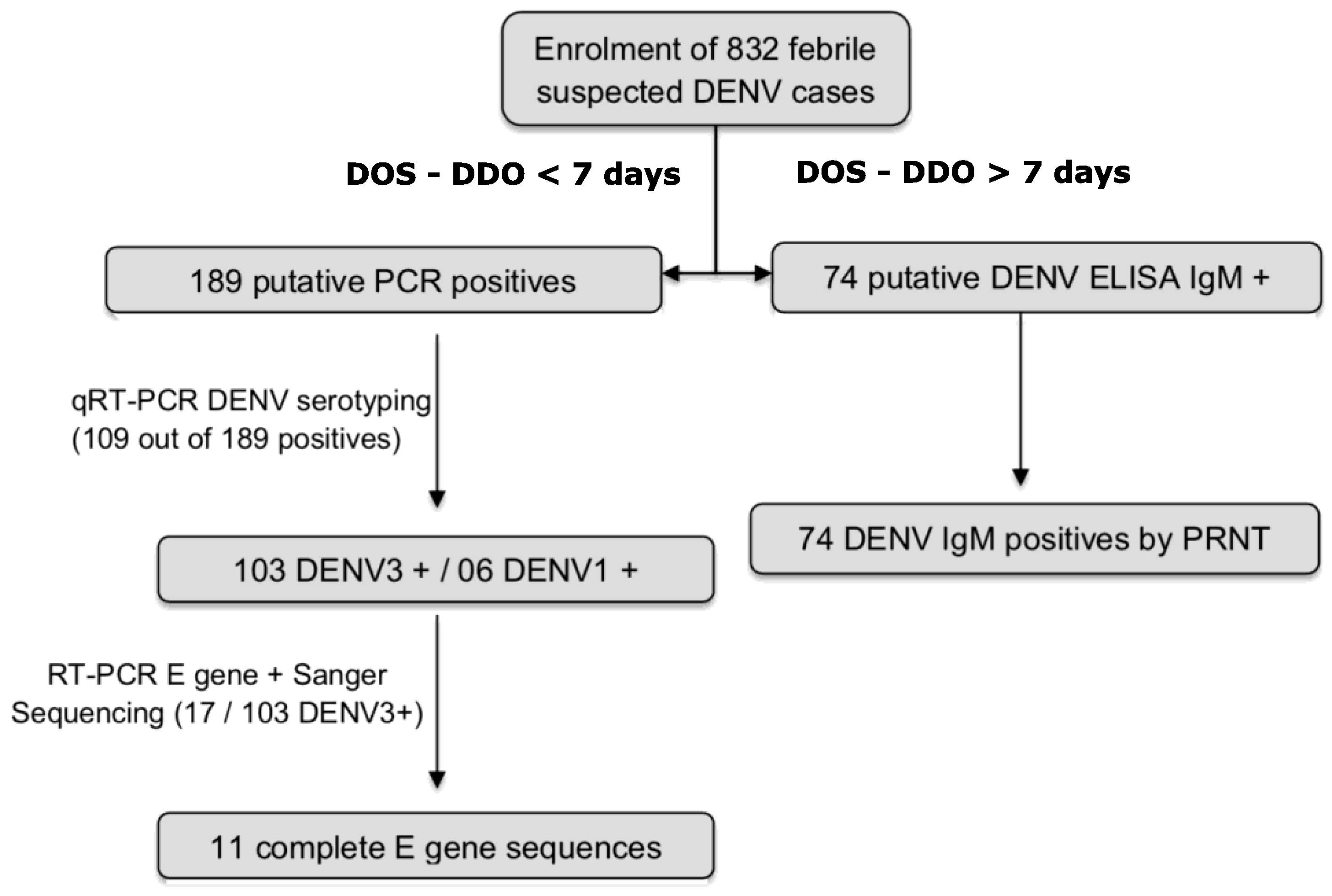
2.3. Patient Enrolment and Sample Collection
2.4. RNA Extraction
2.5. Dengue Virus Detection by qRT-PCR
2.6. Dengue Virus Antibody Detection by Serological Assays
2.7. Data Management and Real Time Case Reporting
2.8. Complete E Gene Sequencing
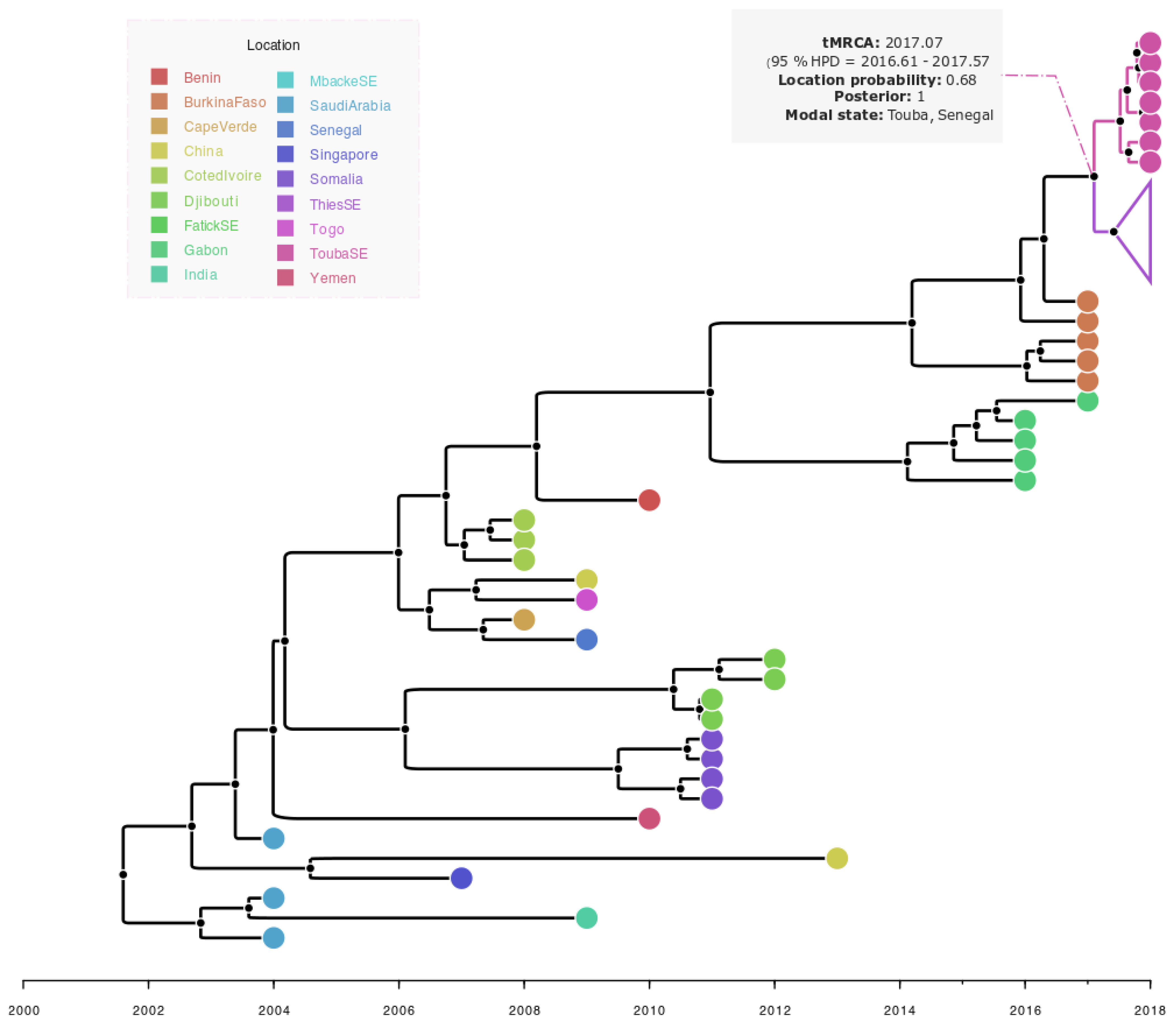
2.9. Dataset Phylogenetic and Phylogeographic Analysis
3. Results
3.1. Epidemiology, Demographic and Clinical Characteristics
3.2. Detection of DENV RNA and Serotyping
3.3. Detection of DENV IgM
3.4. Phylogenetic and Bayesian Analysis of Detected DENV-3 Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Murray, N.E.A.; Quam, M.B.; Wilder-Smith, A. Epidemiology of dengue: Past, present and future prospects. Clin. Epidemiol. 2013, 5, 299–309. [Google Scholar] [PubMed]
- Mairuhu, A.T.A.; Wagenaar, J.; Brandjes, D.P.M.; van Gorp, E.C.M. Dengue: An arthropod-borne disease of global importance. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-de-Lima, V.H.; Lima-Camara, T.N. Natural vertical transmission of dengue virus in Aedes aegypti and Aedes albopictus: A systematic review. Parasites Vectors 2018, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Kashif Zahoor, M.; Rasul, A.; Asif Zahoor, M.; Sarfraz, I.; Zulhussnain, M.; Rasool, R.; Majeed, H.N.; Jabeen, F.; Ranian, K. Dengue Fever: A General Perspective. In Dengue Fever—A Resilient Threat in the Face of Innovation [Internet]; Abelardo Falcón-Lezama, J., Betancourt-Cravioto, M., Tapia-Conyer, R., Eds.; IntechOpen: London, UK, 2019; Available online: https://www.intechopen.com/books/dengue-fever-a-resilient-threat-in-the-face-of-innovation/dengue-fever-a-general-perspective (accessed on 21 November 2022).
- Chen, R.; Vasilakis, N. Dengue—Quo tu et quo vadis? Viruses 2011, 3, 1562–1608. [Google Scholar] [CrossRef] [PubMed]
- Hotta, S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J. Infect. Dis. 1952, 90, 1–9. [Google Scholar] [CrossRef]
- Whitehead, S.S.; Blaney, J.E.; Durbin, A.P.; Murphy, B.R. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 2007, 5, 518–528. [Google Scholar] [CrossRef]
- WHO; TDR (Eds.) Dengue: Guidelines for Diagnosis, Treatment, Prevention, and Control; TDR; World Health Organization: Geneva, Switzerland, 2009; 147p. [Google Scholar]
- Lim, J.K.; Carabali, M.; Lee, J.S.; Lee, K.S.; Namkung, S.; Lim, S.K.; Ridde, V.; Fernandes, J.; Lell, B.; Matendechero, S.H.; et al. Evaluating dengue burden in Africa in passive fever surveillance and seroprevalence studies: Protocol of field studies of the Dengue Vaccine Initiative. BMJ Open 2018, 8, e017673. [Google Scholar] [CrossRef]
- Kokernot, R.H.; Smithburn, K.C.; Weinbren, M.P. Neutralizing antibodies to arthropod-borne viruses in human beings and animals in the Union of South Africa. J. Immunol. 1956, 77, 313–323. [Google Scholar]
- Carey, D. Dengue viruses from febrile patients in nigeria, 1964–1968. Lancet 1971, 297, 105–106. [Google Scholar] [CrossRef]
- Faye, O.; Ba, Y.; Faye, O.; Talla, C.; Diallo, D.; Chen, R.; Mondo, M.; Ba, R.; Macondo, E.; Siby, T.; et al. Urban Epidemic of Dengue Virus Serotype 3 Infection, Senegal, 2009. Emerg. Infect. Dis. 2014, 20, 456–459. [Google Scholar] [CrossRef]
- Fall, C.; Cappuyns, A.; Faye, O.; Pauwels, S.; Fall, G.; Dia, N.; Diagne, M.M.; Diagne, C.T.; Niang, M.; Mbengue, A.; et al. Field evaluation of a mobile biosafety laboratory in Senegal to strengthen rapid disease outbreak response and monitoring. Afr. J. Lab. Med. 2020, 20, 456–459. Available online: http://www.ajlmonline.org/index.php/AJLM/article/view/1041 (accessed on 12 September 2022). [CrossRef] [PubMed]
- Dieng, I.; Hedible, B.G.; Diagne, M.M.; El Wahed, A.A.; Diagne, C.T.; Fall, C.; Richard, V.; Vray, M.; Weidmann, M.; Faye, O.; et al. Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal. Diagnostics 2020, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Dieng, I.; Diarra, M.; Diagne, M.M.; Faye, M.; Dior Ndione, M.H.; Ba, Y.; Diop, M.; Ndiaye, E.H.; Zanotto, P.M.D.A.; Diop, B.; et al. Field Deployment of a Mobile Biosafety Laboratory Reveals the Co-Circulation of Dengue Viruses Serotype 1 and Serotype 2 in Louga City, Senegal, 2017. J. Trop. Med. 2021, 2021, 8817987. [Google Scholar] [CrossRef]
- Wagner, D.; de With, K.; Huzly, D.; Hufert, F.; Weidmann, M.; Breisinger, S.; Eppinger, S.; Kern, W.V.; Bauer, T.M. Nosocomial Acquisition of Dengue. Emerg. Infect. Dis. 2004, 10, 1872–1873. [Google Scholar] [CrossRef] [PubMed]
- Ba, F.; Loucoubar, C.; Faye, O.; Fall, G.; Noelle Mbaye, R.N.P.; Sembene, M.; Diallo, M.; Balde, A.T.; Sall, A.A.; Faye, O. Retrospective analysis of febrile patients reveals unnoticed epidemic of zika fever in Dielmo, Senegal, 2000. Clin. Microbiol. Infect. Dis. 2018, 3, 1–9. [Google Scholar] [CrossRef]
- Luo, L.; Liang H ying Jing Q long He, P.; Yuan, J.; Di, B.; Bai, Z.-J.; Wang, Y.-L.; Zheng, X.-L.; Yang, Z.-C. Molecular characterization of the envelope gene of dengue virus type 3 newly isolated in Guangzhou, China, during 2009–2010. Int. J. Infect. Dis. 2013, 17, e498–e504. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. Available online: https://academic.oup.com/ve/article/doi/10.1093/ve/vey016/5035211 (accessed on 14 September 2020). [CrossRef]
- Diallo, M.; Ba, Y.; Sall, A.A.; Diop, O.M.; Ndione, J.A.; Mondo, M.; Girault, L.; Mathiot, C. Amplification of the Sylvatic Cycle of Dengue Virus Type 2, Senegal, 1999–2000: Entomologic Findings and Epidemiologic Considerations. Emerg. Infect. Dis. 2003, 9, 362–367. [Google Scholar] [CrossRef]
- Diagne, C.T.; Barry, M.A.; Ba, Y.; Faye, O.; Sall, A.A. Dengue epidemic in Touba, Senegal: Implications for the Grand Magal Pilgrimage for travellers. J. Travel Med. 2019, 26, tay123. [Google Scholar] [CrossRef]
- Dieng, I.; Ndione, M.H.D.; Fall, C.; Diagne, M.M.; Diop, M.; Gaye, A.; Barry, M.A.; Diop, B.; Ndiaye, M.; Bousso, A.; et al. Multifoci and multiserotypes circulation of dengue virus in Senegal between 2017 and 2018. BMC Infect. Dis. 2021, 21, 867. [Google Scholar] [CrossRef]
- Hu, H.; Nigmatulina, K.; Eckhoff, P. The scaling of contact rates with population density for the infectious disease models. Math. Biosci. 2013, 244, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Kuo, L.L.; Yang, K.D.; Lin, P.S.; Lu, P.L.; Lin, C.C.; Chang, K.; Chen, T.C.; Lin, W.R.; Lin, C.Y.; et al. Laboratory diagnostics of dengue fever: An emphasis on the role of commercial dengue virus nonstructural protein 1 antigen rapid test. J. Microbiol. Immunol. Infect. 2013, 46, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Mwanyika, G.O.; Mboera, L.E.G.; Rugarabamu, S.; Ngingo, B.; Sindato, C.; Lutwama, J.J.; Paweska, J.T.; Misinzo, G. Dengue Virus Infection and Associated Risk Factors in Africa: A Systematic Review and Meta-Analysis. Viruses 2021, 13, 536. [Google Scholar] [CrossRef]
- Dieng, I.; Cunha, M.d.P.; Diagne, M.M.; Sembène, P.M.; Zanotto, P.M.d.A.; Faye, O.; Faye, O.; Sall, A.A. Origin and Spread of the Dengue Virus Type 1, Genotype V in Senegal, 2015–2019. Viruses 2021, 13, 57. [Google Scholar] [CrossRef]
- Tan, K.K.; Zulkifle, N.I.; Sulaiman, S.; Pang, S.P.; NorAmdan, N.; MatRahim, N.; Abd-Jamil, J.; Shu, M.-H.; Mahadi, N.M.; AbuBakar, S. Emergence of the Asian lineage dengue virus type 3 genotype III in Malaysia. BMC Evol. Biol. 2018, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Ushijima, Y.; Loembe, M.M.; Bikangui, R.; Nguema-Ondo, G.; Mpingabo, P.I.; Zadeh, V.R.; Pemba, C.M.; Kurosaki, Y.; Igasaki, Y.; et al. Re-emergence of dengue virus serotype 3 infections in Gabon in 2016–2017, and evidence for the risk of repeated dengue virus infections. Int. J. Infect. Dis. 2020, 91, 129–136. [Google Scholar] [CrossRef]
- Tarnagda, Z.; Cissé, A.; Bicaba, B.W.; Diagbouga, S.; Sagna, T.; Ilboudo, A.K.; Al, Z.T.E.; Lingani, M.; Sondo, K.A.; Yougbaré, I.; et al. Dengue Fever in Burkina Faso, 2016. Emerg. Infect. Dis. 2018, 24, 170–172. [Google Scholar] [CrossRef]
- Gubler, D.J. Dengue and Dengue Hemorrhagic Fever. Clin. Microbiol. Rev. 1998, 11, 17. [Google Scholar] [CrossRef]
- Ayolabi, C.I.; Olusola, B.A.; Ibemgbo, S.A.; Okonkwo, G.O. Detection of Dengue viruses among febrile patients in Lagos, Nigeria and phylogenetics of circulating Dengue serotypes in Africa. Infect. Genet. Evol. 2019, 75, 103947. [Google Scholar] [CrossRef]
- Franco, L.; Di Caro, A.; Carletti, F.; Vapalahti, O.; Renaudat, C.; Zeller, H.; Tenorio, A. Recent expansion of dengue virus serotype 3 in West Africa. Eurosurveillance 2010, 15, 19490. [Google Scholar] [CrossRef]
- Soo, K.M.; Khalid, B.; Ching, S.M.; Chee, H.Y. Meta-Analysis of Dengue Severity during Infection by Different Dengue Virus Serotypes in Primary and Secondary Infections. Huy NT, éditeur. PLoS ONE 2016, 11, e0154760. [Google Scholar] [CrossRef] [PubMed]
- Gaye, A.; Ndiaye, T.; Sy, M.; Deme, A.B.; Thiaw, A.B.; Sene, A.; Ndiaye, C.; Diedhiou, Y.; Mbaye, A.M.; Ndiaye, I.; et al. Genomic investigation of a dengue virus outbreak in Thiès, Senegal, in 2018. Sci. Rep. 2021, 11, 10321. [Google Scholar] [CrossRef] [PubMed]




| Confirmed Cases (N = 263) | Negative (N = 569) | Total (N = 832) | p Value | |
|---|---|---|---|---|
| Age | <0.001 1 | |||
| Median | 18 | 21 | 20 | |
| Q1, Q3 | 9.5, 26.0 | 12.0, 32.0 | 12.0, 30.0 | |
| Group age, in year | <0.001 2 | |||
| Unknown | 12 | 27 | 39 | |
| <15 | 91 (36.3%) | 165 (30.4%) | 256 (32.3%) | |
| (15–30) | 115 (45.8%) | 204 (37.6%) | 319 (40.2%) | |
| (30–45) | 32 (12.7%) | 114 (21.0%) | 146 (18.4%) | |
| (45–80) | 13 (5.2%) | 59 (10.9%) | 72 (9.1%) | |
| Sex | 0.111 2 | |||
| F | 130 (49.4%) | 315 (55.4%) | 445 (53.5%) | |
| M | 133 (50.6%) | 254 (44.6%) | 387 (46.5%) | |
| Clinical signs and Symptoms | ||||
| Fever | 213 (81.0%) | 369 (64.9%) | 582 (70.0%) | <0.001 2 |
| Join pain | 154 (58.6%) | 326 (57.3%) | 480 (57.7%) | 0.732 2 |
| Asthenia | 149 (56.7%) | 347 (61.0%) | 496 (59.6%) | 0.237 2 |
| Headache | 243 (92.4%) | 495 (87.0%) | 738 (88.7%) | 0.022 2 |
| Retrorbital pain | 60 (22.8%) | 123 (21.6%) | 183 (22.0%) | 0.698 2 |
| Bleeding | 15 (5.7%) | 19 (3.3%) | 34 (4.1%) | 0.109 2 |
| Muscle pain | 133 (50.6%) | 311 (54.7%) | 444 (53.4%) | 0.272 2 |
| Abdominal pain | 73 (27.8%) | 168 (29.5%) | 241 (29.0%) | 0.601 2 |
| Meningo-encephalitis | 8 (3.0%) | 11 (1.9%) | 19 (2.3%) | 0.320 2 |
| Shock syndrome | 2 (0.8%) | 0 (0.0%) | 2 (0.2%) | 0.037 2 |
| Rash | 10 (3.8%) | 16 (2.8%) | 26 (3.1%) | 0.4452 |
| Profession | 0.256 2 | |||
| Bureaucrat | 21 (8.0%) | 52 (9.1%) | 73 (8.8%) | |
| Trade-market | 17 (6.5%) | 54 (9.5%) | 71 (8.5%) | |
| Farmer | 6 (2.3%) | 21 (3.7%) | 27 (3.2%) | |
| Student | 68 (25.9%) | 131 (23.0%) | 199 (23.9%) | |
| Homemaker | 55 (20.9%) | 142 (25.0%) | 197 (23.7%) | |
| Unspecific activities | 73 (27.8%) | 128 (22.5%) | 201 (24.2%) | |
| Workers | 23 (8.7%) | 41 (7.2%) | 64 (7.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dieng, I.; Fall, C.; Barry, M.A.; Gaye, A.; Dia, N.; Ndione, M.H.D.; Fall, A.; Diop, M.; Sarr, F.D.; Ndiaye, O.; et al. Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal. Int. J. Environ. Res. Public Health 2022, 19, 16912. https://doi.org/10.3390/ijerph192416912
Dieng I, Fall C, Barry MA, Gaye A, Dia N, Ndione MHD, Fall A, Diop M, Sarr FD, Ndiaye O, et al. Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal. International Journal of Environmental Research and Public Health. 2022; 19(24):16912. https://doi.org/10.3390/ijerph192416912
Chicago/Turabian StyleDieng, Idrissa, Cheikh Fall, Mamadou Aliou Barry, Aboubacry Gaye, Ndongo Dia, Marie Henriette Dior Ndione, Amary Fall, Mamadou Diop, Fatoumata Diene Sarr, Oumar Ndiaye, and et al. 2022. "Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal" International Journal of Environmental Research and Public Health 19, no. 24: 16912. https://doi.org/10.3390/ijerph192416912
APA StyleDieng, I., Fall, C., Barry, M. A., Gaye, A., Dia, N., Ndione, M. H. D., Fall, A., Diop, M., Sarr, F. D., Ndiaye, O., Dieng, M., Diop, B., Diagne, C. T., Ndiaye, M., Fall, G., Sylla, M., Faye, O., Loucoubar, C., Faye, O., & Sall, A. A. (2022). Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal. International Journal of Environmental Research and Public Health, 19(24), 16912. https://doi.org/10.3390/ijerph192416912







