Effects of Land Reclamation on Soil Bacterial Community and Potential Functions in Bauxite Mining Area
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of the Study Area
2.2. Soil Sampling and Pre-Treatment
2.3. DNA Extraction, Sequencing, and Sequencing Data Processing
2.4. Soil Physicochemical Properties Analysis
2.5. Data Analysis
3. Results
3.1. Characteristics of Soil Physicochemical Properties in Different Plots
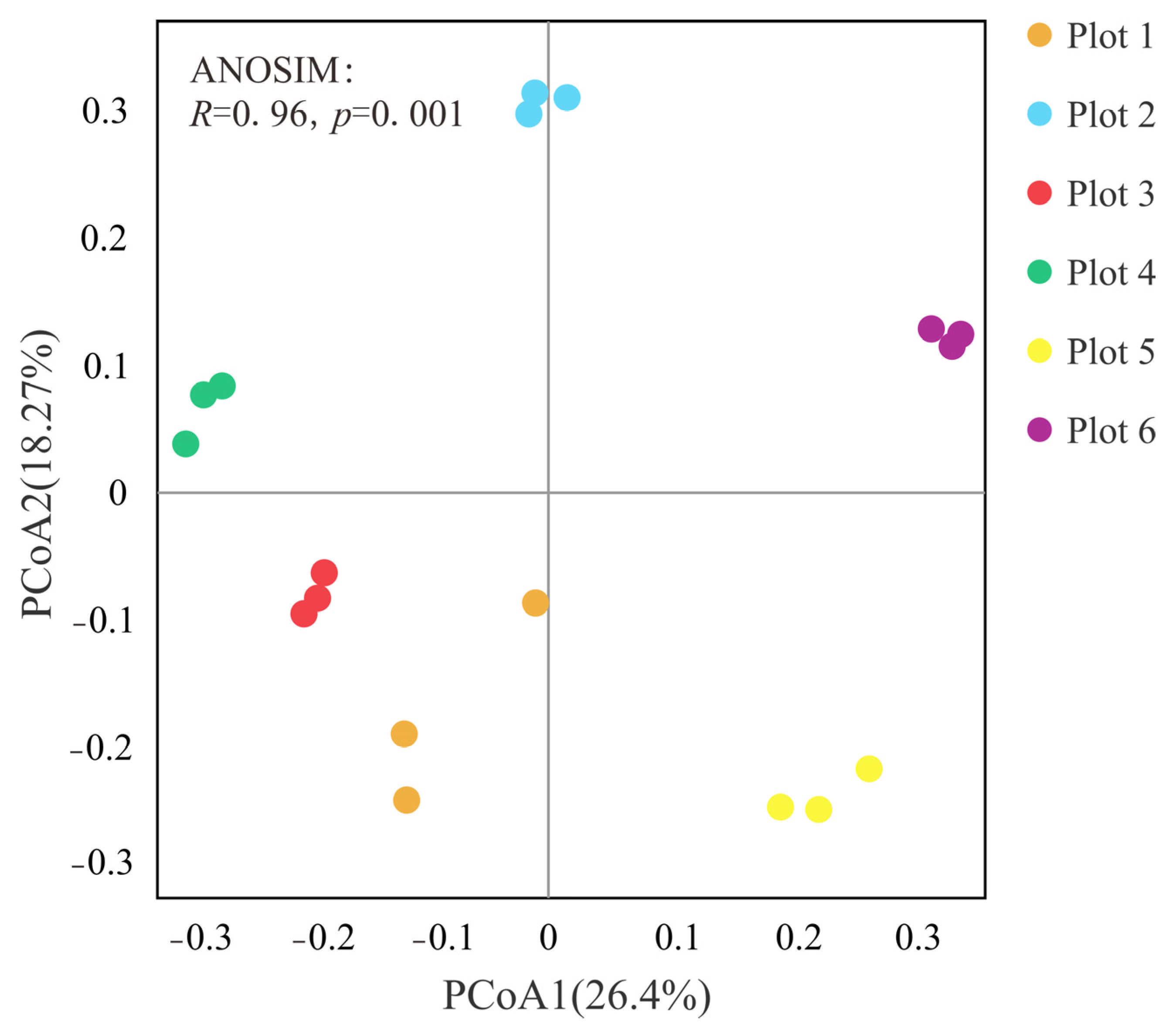
3.2. Bacterial Diversity and Community Composition of Soils in Different Plots
3.2.1. Bacterial Diversity in Different Plots
3.2.2. Bacterial Community Composition of Reclaimed Plots
3.3. Analysis of Soil Bacterial Co-Occurrence Networks in Different Plots
3.4. Correlations between Soil Bacterial Community Structure and Soil Physicochemical Properties
3.5. Functional Prediction of Soil Bacteria in Different Plots
4. Discussion
4.1. The Effects of Reclamation on Soil Nutrient Enhancement
4.2. Driving Effects of Reclamation on Changes in Soil Bacterial Community Structure
4.3. Interactions between Soil C and N Levels and Bacterial Community Structure in Reclaimed Plots
4.4. Effects of Reclamation on Microbial Network Interactions and Keystone Taxa
4.5. The Driving Effects of Reclamation on Potential Functional Changes in Soil Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Bogatyrev, B.A.; Zhukov, V.V.; Tsekhovsky, Y.G. Formation conditions and regularities of the distribution of large and superlarge bauxite deposits. Lithol. Miner. Resour. 2009, 44, 135–151. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Q.; Zhang, Q.; Carranza, E.J.M.; Liu, H.; Liu, X.; Deng, J. Interaction between karst terrain and bauxites: Evidence from quaternary orebody distribution in Guangxi, SW China. Sci. Rep. 2017, 7, 11842. [Google Scholar] [CrossRef] [Green Version]
- Kusin, F.M.; Rahman, M.S.A.; Madzin, Z.; Jusop, S.; Mohamat-Yusuff, F.; Ariffin, M.; Md, Z.M.S. The occurrence and potential ecological risk assessment of bauxite mine-impacted water and sediments in Kuantan, Pahang, Malaysia. Environ. Sci. Pollut. Res. 2017, 24, 1306–1321. [Google Scholar] [CrossRef]
- Adeli, A.; Mclaughlin, M.; Brooks, J.; Read, J.; Willers, J.; Lang, D.; McGrew, R. Age chronosequence effects on restoration quality of reclaimed coal mine soils in Mississippi agroecosystems. Soil Sci. 2013, 178, 335–343. [Google Scholar] [CrossRef]
- Harris, M.A.; Omoregie, S.N. Post-mining deterioration of bauxite overburdens in Jamaica: Storage methods or subsoil dilution? Environ. Geol. 2008, 54, 111–115. [Google Scholar] [CrossRef]
- Li, L.; Li, T.; Meng, H.; Xie, Y.; Zhang, J.; Hong, J. Effects of seven-year fertilization reclamation on bacterial community in a coal mining subsidence area in Shanxi, China. Int. J. Environ. Res. Public Health 2021, 18, 12504. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, Z.; Yang, D.; Li, H.; Liu, B.; Gao, H.; Cao, C.; Zhou, Y.; Li, J.; Li, S. Land use dynamic evolution and driving factors of typical open-pit coal mines in Inner Mongolia. Int. J. Environ. Res. Public Health 2022, 19, 9723. [Google Scholar] [CrossRef]
- Sheoran, V.; Sheoran, A.S.; Poonia, P. Soil reclamation of abandoned mine land by revegetation: A review. Int. J. Soil Sediment Water 2010, 3, 2. [Google Scholar]
- Kumar, S.; Maiti, S.K.; Chaudhuri, S. Soil development in 2–21 years old coalmine reclaimed spoil with trees: A case study from Sonepur-Bazari opencast project, Raniganj coalfield, India. Ecol. Eng. 2015, 84, 311–324. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wen, H.Y.; Chen, L.Q.; Yin, T. Succession of bacterial community structure and diversity in soil along a chronosequence of reclamation and re-vegetation on coal mine spoils in China. PLoS ONE 2014, 9, 12. [Google Scholar] [CrossRef]
- Wang, J.M.; Wang, H.D.; Cao, Y.G.; Bai, Z.K.; Qin, Q. Effects of soil and topographic factors on vegetation restoration in opencast coal mine dumps located in a loess area. Sci. Rep. 2016, 6, 22058. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Zhang, F.H.; Yang, L.; Wang, D.; Wang, W.C. Response of soil bacterial community structure to different reclamation years of abandoned salinized farmland in arid China. Arch. Microbiol. 2019, 201, 1219–1232. [Google Scholar] [CrossRef]
- Li, J.; Xin, Z.; Yan, J.; Li, H.; Chen, J.; Ding, G. Physicochemical and microbiological assessment of soil quality on a chronosequence of a mine reclamation site. Eur. J. Soil. Sci. 2018, 69, 1056–1067. [Google Scholar] [CrossRef]
- Mummey, D.L.; Stahl, P.D.; Buyer, J.S. Soil microbiological properties 20 years after surface mine reclamation: Spatial analysis of reclaimed and undisturbed sites. Soil Biol. Biochem. 2002, 34, 1717–1725. [Google Scholar] [CrossRef]
- Sun, L.; Xun, W.B.; Huang, T.; Zhang, G.S.; Gao, J.S.; Ran, W.; Li, D.C.; Shen, Q.R.; Zhang, R.F. Alteration of the soil bacterial community during parent material maturation driven by different fertilization treatments. Soil Biol. Biochem. 2016, 96, 207–215. [Google Scholar] [CrossRef]
- Vogel, C.; Babin, D.; Pronk, G.J.; Heister, K.; Smalla, K.; Kögel-Knabner, I. Establishment of macro-aggregates and organic matter turnover by microbial communities in long-term incubated artificial soils. Soil Boil. Biochem. 2014, 79, 57–67. [Google Scholar] [CrossRef]
- Uroz, S.; Calvaruso, C.; Turpault, M.; Frey-Klett, P. Mineral weathering by bacteria: Ecology, actors and mechanisms. Trends Microbiol. 2009, 17, 378–387. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, J.M.; Bai, Z.K.; Reading, L. Effects of surface coal mining and land reclamation on soil properties: A review. Earth Sci. Rev. 2019, 191, 12–25. [Google Scholar] [CrossRef]
- Narayanan, M.; Thangabalu, R.; Natarajan, D.; Kumarasamy, S.; Kandasamy, S.; Elfasakhany, A.; Pugazhendhi, A. Reclamation competence of Crotalaria juncea with the amalgamation and influence of indigenous bacteria on a waste dump of bauxite mine. Chemosphere 2021, 279, 130632. [Google Scholar] [CrossRef]
- Vilas, B.; Hiran, F.; Almeida, L.F.J.; Teixeira, R.S.; Souza, I.F.; Silva, I.R. Soil organic carbon recovery and coffee bean yield following bauxite mining. Land Degrad. Dev. 2018, 29, 1565–1573. [Google Scholar] [CrossRef]
- Borges, S.R.; Santos, R.S.; Oliveira, D.M.S.; Souza, I.F.; Verburg, E.E.J.; Pimentel, L.G.; Cruz, R.S.; Silva, I.R. Practices for rehabilitating bauxite-mined areas and an integrative approach to monitor soil quality. Land Degrad. Dev. 2019, 30, 866–877. [Google Scholar] [CrossRef]
- Lewis, D.E.; Chauhan, A.; White, J.R.; Overholt, W.; Green, S.J.; Jasrotia, P.; Wafula, D.; Jagoe, C. Microbial and geochemical assessment of bauxitic un-mined and post-mined chronosequence soils from Mocho mountains, Jamaica. Microb. Ecol. 2012, 64, 738–749. [Google Scholar] [CrossRef]
- Ward, S.C. Soil development on rehabilitated bauxite mines in south-west Australia. Soil Res. 2000, 38, 453–464. [Google Scholar] [CrossRef]
- Greenberg, W.A.; Wilding, L.P. Pre- and post-mined bauxite soils of Jamaica: Physical and chemical properties. Soil Sci. Soc. Am. J. 2007, 71, 181–188. [Google Scholar] [CrossRef]
- Banning, N.C.; Gleeson, D.B.; Grigg, A.H.; Grant, C.D.; Andersen, G.L.; Brodie, E.L.; Murphy, D.V. Soil microbial community successional patterns during forest ecosystem restoration. Appl. Environ. Microb. 2011, 77, 6158–6164. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.H.; Wei, Y.J.; Liao, S.F. Resear on landforms characteristics of karst accumulative bauxite deposit in Pingguo and its metallogenic regularity. Guangxi Geol. 2000, 4, 23–28. [Google Scholar]
- Chen, W. Practice of engineering reclamation of mine goaf in Pingguo bauxite mine. Miner. Resour. Geol. 2013, 27, 78–80. [Google Scholar]
- Qin, C.S.; Zhong, B.G.; Huang, S.J. Application of integrated mining-reclamation technology in Pingguo bauxite mining area. South. Land Resour. 2007, 11, 41–43. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with qiime 2 ‘ s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Lu, R.K. Methods of Soil Agricultural CHEMISTRY Analysis; Chinese Agricultural Science And Technology Press: Beijiing, China, 1999. [Google Scholar]
- Shen, L.D.; Liu, X.; Wu, H.S.; Tian, M.H.; Ran, P.; Liu, J.Q.; Yang, Y.L.; Yang, W.T.; Wang, H.Y. Effect of different fertilization regimes on the vertical distribution of anaerobic ammonium oxidation in paddy soils. Eur. J. Soil Biol. 2020, 99, 103206. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Yuan, J.H.; Zhu, W.B.; Chen, G.L.; Wang, S.Q. Effect of P fertilizer reduction regime on soil olsen-P, root fe-plaque P, and rice P uptake in rice-wheat rotation paddy fields. Pedosphere 2021, 31, 94–102. [Google Scholar] [CrossRef]
- Zhou, J.B.; Jin, Z.J.; Leng, M.; Xiao, X.Y.; Wang, X.T.; Pan, F.J.; Li, Q. Investigation of soil bacterial communities and functionalities within typical karst paddy field soils in southern China. Fresenius Environ. Bull. 2021, 30, 3537–3548. [Google Scholar]
- Brown, M.B. 400: A method for combining non-independent, one-sided tests of significance. Biometrics 1975, 31, 987–992. [Google Scholar] [CrossRef]
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272. [Google Scholar] [CrossRef]
- Karimi, B.; Maron, P.A.; Chemidlin-Prevost Boure, N.; Bernard, N.; Gilbert, D.; Ranjard, L. Microbial diversity and ecological networks as indicators of environmental quality. Environ. Chem. Lett. 2017, 15, 265–281. [Google Scholar] [CrossRef]
- Newman, M. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.N.; Li, Q.; Yan, J.H.; Liu, C.; Zhong, J.X. Vegetation restoration facilitates belowground microbial network complexity and recalcitrant soil organic carbon storage in southwest China karst region. Sci. Total Environ. 2022, 820, 153137. [Google Scholar] [CrossRef]
- Wen, L.; Li, D.J.; Yang, L.Q.; Luo, P.; Chen, H.; Xiao, K.C.; Song, T.Q.; Zhang, W.; He, X.Y.; Chen, H.S.; et al. Rapid recuperation of soil nitrogen following agricultural abandonment in a karst area, southwest China. Biogeochemistry 2016, 129, 341–354. [Google Scholar] [CrossRef]
- Wu, Q.B.; Wang, X.K.; Guo, R. Soil organic carbon stability and influencing factors. Chin. J. Soil Sci. 2005, 36, 105–109. [Google Scholar] [CrossRef]
- Liao, X.; Niu, Y.H.; Liu, D.Y.; Chen, Z.M.; He, T.H.; Luo, J.F.; Lindsey, S.; Ding, W.X. Four-year continuous residual effects of biochar application to a sandy loam soil on crop yield and N2O and NO emissions under maize-wheat rotation. Agric. Ecosyst. Environ. 2020, 302, 107109. [Google Scholar] [CrossRef]
- Shan, L.N.; He, Y.F.; Chen, J.; Huang, Q.; Lian, X.; Wang, H.C.; Liu, Y.L. Nitrogen surface runoff losses from a Chinese cabbage field under different nitrogen treatments in the Taihu lake basin, China. Agric. Water Manag. 2015, 159, 255–263. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef]
- Bai, Z.; Xie, H.T.; Kao-Kniffin, J.; Chen, B.D.; Shao, P.S.; Liang, C. Shifts in microbial trophic strategy explain different temperature sensitivity of CO2 flux under constant and diurnally varying temperature regimes. FEMS Microbiol. Ecol. 2017, 93, fix063. [Google Scholar] [CrossRef]
- Ciancio, A.; Rosso, L.C.; Lopez-Cepero, J.; Colagiero, M. Rhizosphere 16S-its metabarcoding profiles in banana crops are affected by nematodes, cultivation, and local climatic variations. Front. Microbiol. 2022, 13, 1602. [Google Scholar] [CrossRef]
- Zheng, X.Z.; Lin, C.; Guo, B.L.; Yu, J.H.; Ding, H.; Peng, S.Y.; Sveen, T.R.; Zhang, Y.S. Effects of re-vegetation restoration on soil bacterial community structure in degraded land in subtropical China. Eur. J. Soil Biol. 2020, 98, 103184. [Google Scholar] [CrossRef]
- Kari, A.; Nagymáté, Z.; Romsics, C.; Vajna, B.; Tóth, E.; Lazanyi-Kovács, R.; Rizó, B.; Kutasi, J.; Bernhardt, B.; Farkas, É.; et al. Evaluating the combined effect of biochar and pgpr inoculants on the bacterial community in acidic sandy soil. Appl. Soil Ecol. 2021, 160, 103856. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Joseph, C.M.; Yang, G.P.; Phillips, D.A.; Nelson, L.M. Evaluation of bacteria isolated from rice for plant growth promotion and biological control of seedling disease of rice. Can. J. Microbiol. 2001, 47, 916–924. [Google Scholar] [CrossRef]
- Harris, J. Soil microbial communities and restoration ecology: Facilitators or followers? Science 2009, 325, 573–574. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Ahn, J.; Song, J.; Kim, B.; Kim, M.; Joa, J.; Weon, H. Characterization of the bacterial and archaeal communities in rice field soils subjected to long-term fertilization practices. J. Microbiol. 2012, 50, 754–765. [Google Scholar] [CrossRef]
- Solly, E.F.; Weber, V.; Zimmermann, S.; Walthert, L.; Hagedorn, F.; Schmidt, M.W.I. A critical evaluation of the relationship between the effective cation exchange capacity and soil organic carbon content in swiss forest soils. Front. For. Glob. Change 2020, 3, 98. [Google Scholar] [CrossRef]
- Clark, C.M.; Cleland, E.E.; Collins, S.L.; Fargione, J.E.; Gough, L.; Gross, K.L.; Pennings, S.C.; Suding, K.N.; Grace, J.B. Environmental and plant community determinants of species loss following nitrogen enrichment. Ecol. Lett. 2007, 10, 596–607. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.S.; Jin, Z.J.; Jia, Y.H.; Li, Q. Comparison of soil bacteria community structures from three soil land-use between karst and non-karst areas under three kinds of land use. Carsologica Sin. 2019, 38, 164–172. [Google Scholar]
- Jia, Y.H.; Jin, Z.J.; Yuan, W.; Cheng, Y.Y.; Qiu, J.M.; Liang, J.T.; Pan, F.J.; Liu, D.S. Comparison of soil bacterial community structure between paddy fields and dry land in the Huixian karst wetland, China. Environ. Sci. 2019, 40, 3313–3323. [Google Scholar] [CrossRef]
- Shimomura, T.; Yonekawa, Y.; Nagura, H.; Tateyama, M.; Fujiyoshi, Y.; Irie, K. A native prokaryotic voltage-dependent calcium channel with a novel selectivity filter sequence. eLife 2020, 9, e52828. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Freilich, M.A.; Wieters, E.; Broitman, B.R.; Marquet, P.A.; Navarrete, S.A. Species co-occurrence networks: Can they reveal trophic and non-trophic interactions in ecological communities? Ecology 2018, 99, 690–699. [Google Scholar] [CrossRef]
- Luo, F.; Zhong, J.X.; Yang, Y.F.; Zhou, J.Z. Application of random matrix theory to microarray data for discovering functional gene modules. Phys. Rev. E 2006, 73, 031924. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.W.; Yang, Y.F.; Chen, S.; Zhao, M.X.; Zhu, Z.W.; Yang, S.H.; Qu, Y.Y.; Ma, Q.; He, Z.L.; Zhou, J.Z.; et al. Long-term successional dynamics of microbial association networks in anaerobic digestion processes. Water Res. 2016, 104, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.Z.; Deng, Y.; Luo, F.; He, Z.L.; Tu, Q.C.; Zhi, X.Y. Functional molecular ecological networks. mBio 2010, 1, e00169-e00110. [Google Scholar] [CrossRef] [Green Version]
- Herren, C.M.; McMahon, K.D. Keystone taxa predict compositional change in microbial communities. Environ. Microbiol. 2018, 20, 2207–2217. [Google Scholar] [CrossRef] [Green Version]
- Rivett, D.W.; Bell, T. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat. Microbiol. 2018, 3, 767–772. [Google Scholar] [CrossRef]
- Ma, J.; Dong, W.X.; Zhu, Y.F.; Yu, H.C.; Xiao, D.; Chen, F. Impact of land reclamation on the carbon sequestration potential of soil microorganisms in the disturbed mining area of eastern plain. J. China Coal Soc. 2022, 47, 1306–1317. [Google Scholar] [CrossRef]
- Martins, M.L.L.; Tempest, D.W. Metabolic response of bacillus stearothermophilus chemostat cultures to a secondary oxygen limitation. Microbiology 1991, 137, 1391–1396. [Google Scholar] [CrossRef] [Green Version]
- Subbarao, G.V.; Sahrawat, K.L.; Nakahara, K.; Rao, I.M.; Ishitani, M.; Hash, C.T.; Kishii, M.; Bonnett, D.G.; Berry, W.L.; Lata, J.C. A paradigm shift towards low-nitrifying production systems: The role of biological nitrification inhibition (BNI). Ann. Bot. 2013, 112, 297–316. [Google Scholar] [CrossRef] [Green Version]
- Hayatsu, M.; Tago, K.; Saito, M. Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 2008, 54, 33–45. [Google Scholar] [CrossRef]
- Liang, S.C.; Deng, J.J.; Jiang, Y.; Wu, S.J.; Zhou, Y.B.; Zhu, W.X. Functional distribution of bacterial community under different land use patterns based on faprotax function prediction. Pol. J. Environ. Stud. 2020, 29, 1245–1261. [Google Scholar] [CrossRef]
- Subbarao, G.V.; Yoshihashi, T.; Worthington, M.; Nakahara, K.; Ando, Y.; Sahrawat, K.L.; Rao, I.M.; Lata, J.; Kishii, M.; Braun, H. Suppression of soil nitrification by plants. Plant Sci. 2015, 233, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.S.; Xu, B.K.; Han, J.P.; Shi, L.S. Effects of drying-rewetting cycles on ferrous iron-involved denitrification in paddy soils. Water 2021, 13, 3212. [Google Scholar] [CrossRef]
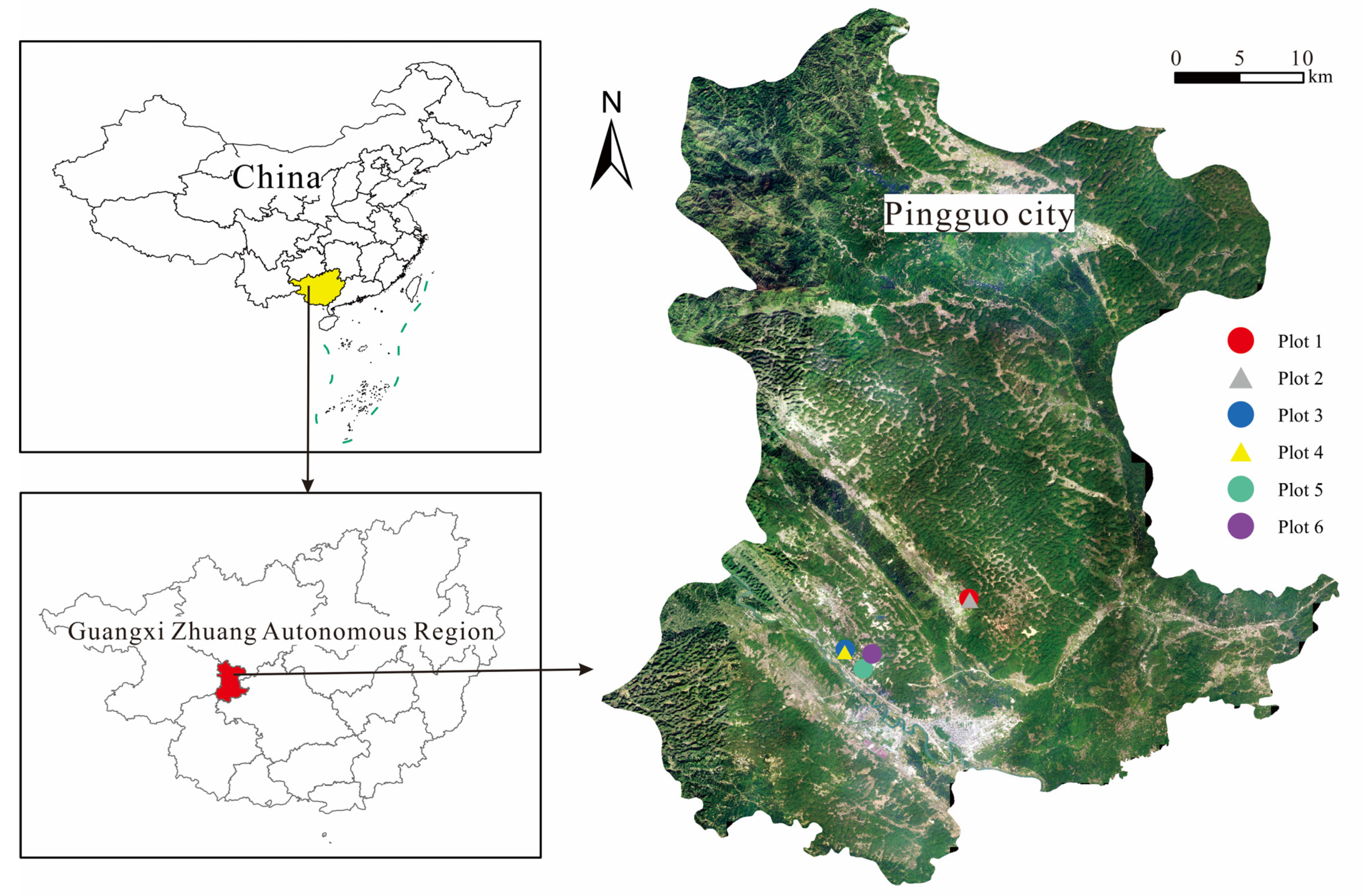




| Plot Types | Plots | Reclamation Year | Vegetation | Management Method | Longitude and Latitude |
|---|---|---|---|---|---|
| Control | Plot 1 | 0 | Leveled plot, no planting | None | 23°25′46″ N, 107°35′21″ E |
| Reclaimed | Plot 2 | 1 | Glycine max | No artificial intervention after planting | 23°25′42″ N, 107°35′23″ E |
| Plot 3 | 4 | Leucaena leucocephala | No artificial intervention after planting | 23°23′21″ N, 107°29′30″ E | |
| Plot 4 | 7 | Neyraudia reynaudiana | No artificial intervention after planting | 23°23′13″ N, 107°29′48″ E | |
| Plot 5 | 10 | Zea mays | Inorganic compound fertilizers mainly, better human management | 23°22′26″ N, 107°30′21″ E | |
| Plot 6 | 12 | Brassica pekinensis | Organic–inorganic fertilizer dispensing, fine human management | 23°23′9″ N, 107°30′46″ E |
| Properties | Plot 1 | Plot 2 | Plot 3 | Plot 4 | Plot 5 | Plot 6 |
|---|---|---|---|---|---|---|
| pH (H2O) | 7.86 ± 0.04 b | 5.95 ± 0.03 f | 8.06 ± 0.03 a | 6.53 ± 0.02 e | 7.27 ± 0.02 c | 6.74 ± 0.04 d |
| SOC (g·kg−1) | 4.22 ± 0.03 e | 6.85 ± 0.11 cd | 6.50 ± 0.03 d | 7.87 ± 0.58 c | 19.31 ± 0.68 b | 33.59 ± 1.61 a |
| DOC (mg·kg−1) | 11.58 ± 0.70 e | 14.42 ± 1.14 d | 16.88 ± 1.16 cd | 18.90 ± 1.66 c | 95.96 ± 4.99 b | 119.77 ± 4.76 a |
| TN (g·kg−1) | 1.37 ± 0.21 e | 2.77 ± 0.15 c | 2.30 ± 0.10 d | 2.43 ± 0.21 cd | 3.47 ± 0.25 b | 4.57 ± 0.23 a |
| AN (mg·kg−1) | 13.92 ± 0.74 e | 32.02 ± 0.75 c | 32.71 ± 0.44 c | 20.20 ± 1.26 d | 52.86 ± 1.07 b | 78.10 ± 1.98 a |
| NH4+-N (mg·kg−1) | 2.48 ± 0.37 d | 6.79 ± 0.84 b | 4.32 ± 0.49 c | 4.23 ± 0.34 c | 6.76 ± 0.41 b | 19.18 ± 1.18 a |
| NO3−-N (mg·kg−1) | 1.33 ± 0.11 d | 1.34 ± 0.08 d | 3.04 ± 0.30 c | 0.47 ± 0.14 e | 4.94 ± 0.25 b | 6.00 ± 0.37 a |
| TP (g·kg−1) | 0.09 ± 0.01 d | 0.12 ± 0.01 d | 0.12 ± 0.01 d | 0.20 ± 0.04 c | 0.58 ± 0.01 b | 3.65 ± 0.09 a |
| AP (mg·kg−1) | 3.73 ± 0.54 e | 5.11 ± 0.73 d | 3.10 ± 0.44 e | 9.58 ± 0.90 c | 27.99 ± 0.62 b | 85.70 ± 0.49 a |
| CEC (cmol+·kg−1) | 3.52 ± 0.75 c | 2.65 ± 0.09 cd | 4.60 ± 0.13 b | 1.92 ± 0.36 d | 4.80 ± 0.44 ab | 5.55 ± 0.76 a |
| Ca2+ (cmol·kg−1) | 2.55 ± 0.09 d | 1.91 ± 0.03 e | 7.68 ± 0.10 a | 1.75 ± 0.03 e | 3.17 ± 0.05 c | 6.18 ± 0.19 b |
| Mg2+ (cmol·kg−1) | 0.06 ± 0.00 e | 0.18 ± 0.00 c | 0.15 ± 0.00 d | 0.15 ± 0.00 d | 0.50 ± 0.01 a | 0.23 ± 0.01 b |
| Sample Plots | Chao1 | ACE | Shannon | Simpson |
|---|---|---|---|---|
| Plot 1 | 1786.03 ± 274.10 ab | 1786.23 ± 274.26 ab | 6.54 ± 0.43 b | 0.996 ± 0.003 b |
| Plot 2 | 1658.67 ± 136.21 b | 1658.73 ± 136.09 b | 6.56 ± 0.10 b | 0.996 ± 0.000 b |
| Plot 3 | 1907.00 ± 40.71 ab | 1907.13 ± 40.64 ab | 6.88 ± 0.06 ab | 0.998 ± 0.000 a |
| Plot 4 | 1959.00 ± 129.55 a | 1959.00 ± 129.55 a | 6.84 ± 0.07 ab | 0.997 ± 0.000 ab |
| Plot 5 | 1865.34 ± 41.01 ab | 1865.60 ± 40.97 ab | 6.98 ± 0.05 a | 0.998 ± 0.000 a |
| Plot 6 | 1920.67 ± 68.96 ab | 1920.82 ± 68.82 ab | 6.94 ± 0.04 a | 0.998 ± 0.000 a |
| Properties | Plot 1 | Plot 2 | Plot 3 | Plot 4 | Plot 5 | Plot 6 |
|---|---|---|---|---|---|---|
| Nodes | 184 | 214 | 228 | 195 | 218 | 208 |
| Edges 1 | 2259 (58.39%) | 1911 (51.70%) | 1968 (51.68%) | 1851 (51.92%) | 1842 (50.81%) | 1773 (52.00%) |
| Modularity | 0.50 | 0.77 | 0.74 | 0.72 | 0.77 | 0.76 |
| Characteristic Path Length | 3.29 | 6.95 | 6.49 | 6.81 | 8.66 | 6.67 |
| Average Number of Neighbors | 24.81 | 17.86 | 17.26 | 18.99 | 16.90 | 17.05 |
| Clustering Coefficient | 0.67 | 0.75 | 0.75 | 0.76 | 0.76 | 0.76 |
| Network Density | 0.14 | 0.08 | 0.08 | 0.10 | 0.08 | 0.08 |
| Diameter of Network | 13 | 22 | 15 | 21 | 26 | 14 |
| Plots | ASV Id | Closeness Centrality | Betweenness Centrality | Degree | Categories |
|---|---|---|---|---|---|
| Plot 1 | ASV 49 | 0.45 | 0.07 | 65 | Blautia |
| ASV 152 | 0.44 | 0.07 | 65 | Bacteroides | |
| ASV 224 | 0.44 | 0.08 | 64 | Ruminococcaceae | |
| ASV 215 | 0.44 | 0.08 | 60 | Clostridiaceae | |
| ASV 248 | 0.39 | 0.02 | 60 | Coprococcus | |
| Plot 2 | ASV 149 | 0.19 | 0.01 | 29 | Rhodospirillaceae |
| ASV 609 | 0.18 | 0.00 | 28 | Syntrophobacteraceae | |
| ASV 431 | 0.19 | 0.01 | 27 | Myxococcaceae | |
| ASV 1065 | 0.18 | 0.00 | 27 | Solibacterales | |
| ASV 127 | 0.18 | 0.00 | 27 | Acidimicrobiales EB1017 | |
| Plot 3 | ASV 806 | 0.19 | 0.02 | 30 | Bacteria WS3 |
| ASV 21 | 0.18 | 0.01 | 30 | Clostridium | |
| ASV 161 | 0.18 | 0.01 | 30 | Acidimicrobiales | |
| ASV 23 | 0.18 | 0.01 | 29 | Peptostreptococcaceae | |
| ASV 463 | 0.20 | 0.06 | 28 | Bacteria WS3 | |
| Plot 4 | ASV 40 | 0.20 | 0.09 | 38 | Peptostreptococcaceae |
| ASV 1802 | 0.20 | 0.03 | 38 | Acidobacteria | |
| ASV 124 | 0.20 | 0.03 | 38 | β-proteobacteria | |
| ASV 183 | 0.20 | 0.04 | 37 | Hyphomicrobium | |
| ASV 529 | 0.20 | 0.03 | 37 | Kibdelosporangium | |
| Plot 5 | ASV 211 | 0.14 | 0.03 | 30 | Proteobacteria |
| ASV 164 | 0.14 | 0.02 | 28 | Gemmatimonadetes | |
| ASV 72 | 0.14 | 0.02 | 27 | Gemmatimonadetes | |
| ASV 469 | 0.14 | 0.02 | 26 | Chloracidobacteria PK29 | |
| ASV 616 | 0.14 | 0.00 | 25 | Acidobacteria Gp6 iii1-15 | |
| Plot 6 | ASV 149 | 0.17 | 0.06 | 30 | Rhodospirillaceae |
| ASV 546 | 0.18 | 0.03 | 29 | Hyphomonadaceae | |
| ASV 1021 | 0.18 | 0.05 | 28 | β-proteobacteria | |
| ASV 174 | 0.17 | 0.03 | 27 | Acidobacteria Gp6 iii1-15 | |
| ASV 514 | 0.17 | 0.01 | 27 | Gemmatimonadetes Gp5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Jin, Z.; Xiong, L.; Tong, L.; Zhu, H.; Zhang, X.; Qin, G. Effects of Land Reclamation on Soil Bacterial Community and Potential Functions in Bauxite Mining Area. Int. J. Environ. Res. Public Health 2022, 19, 16921. https://doi.org/10.3390/ijerph192416921
Li X, Jin Z, Xiong L, Tong L, Zhu H, Zhang X, Qin G. Effects of Land Reclamation on Soil Bacterial Community and Potential Functions in Bauxite Mining Area. International Journal of Environmental Research and Public Health. 2022; 19(24):16921. https://doi.org/10.3390/ijerph192416921
Chicago/Turabian StyleLi, Xuesong, Zhenjiang Jin, Liyuan Xiong, Lingchen Tong, Hongying Zhu, Xiaowen Zhang, and Guangfa Qin. 2022. "Effects of Land Reclamation on Soil Bacterial Community and Potential Functions in Bauxite Mining Area" International Journal of Environmental Research and Public Health 19, no. 24: 16921. https://doi.org/10.3390/ijerph192416921




