Metabolic and Proteomic Profiles Reveal the Response of the ASD-Associated Resistant Strain 6-1 of Lactobacillus plantarum to Propionic Acid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Resistance to Propionic Acid
2.2. Protein Extraction of Resistant Strain Exposed to Propionic Acid
2.3. Peptide and Protein Identification
2.4. Analysis of Proteins’ Differential Expression
2.5. Protein Data Analysis
2.6. Lipopeptide Detection by MALDI-TOF-MS Analysis
3. Results and Discussion
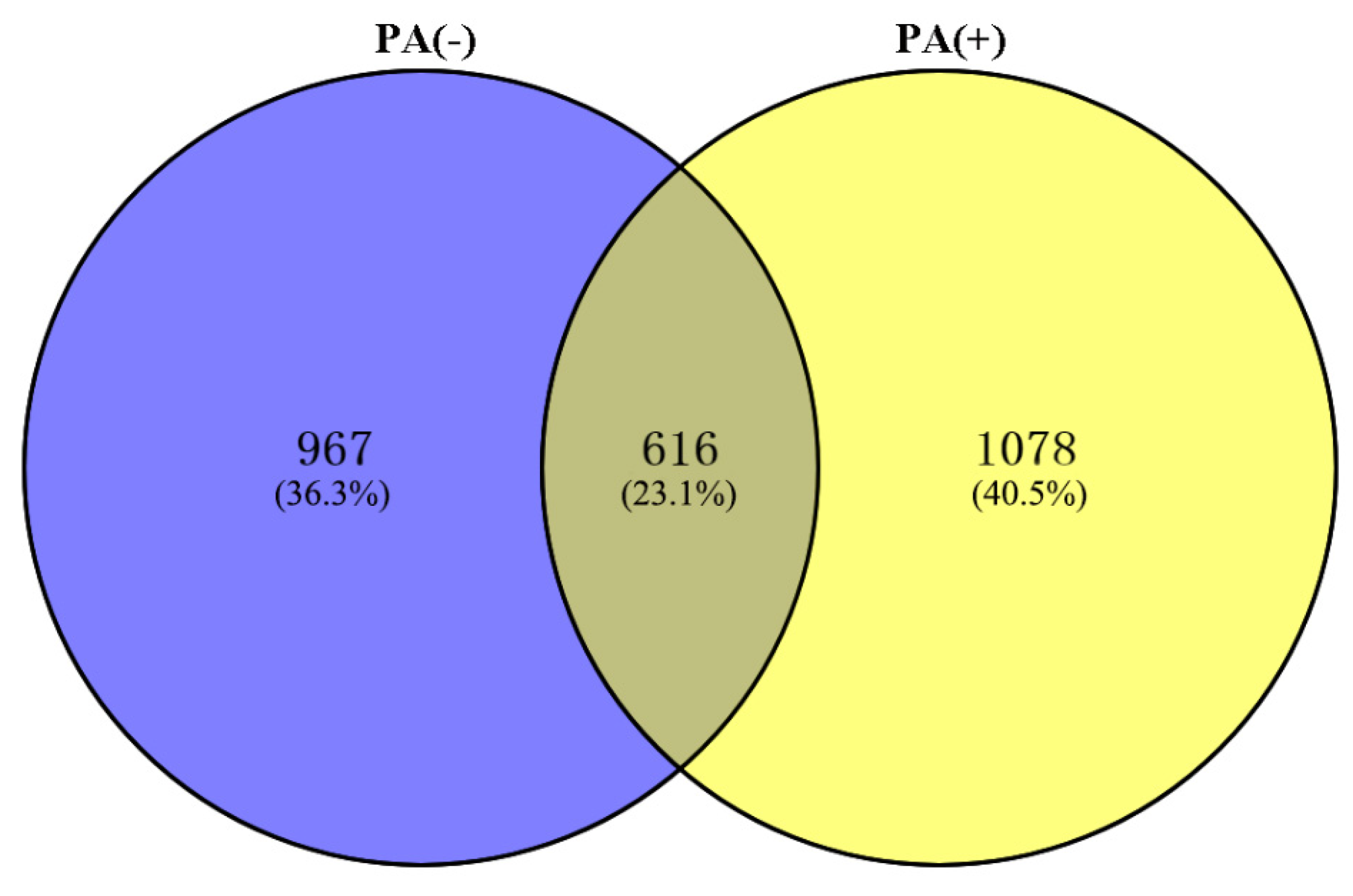
3.1. Number of Identified Total Proteins
3.2. Differentially Expressed Proteins
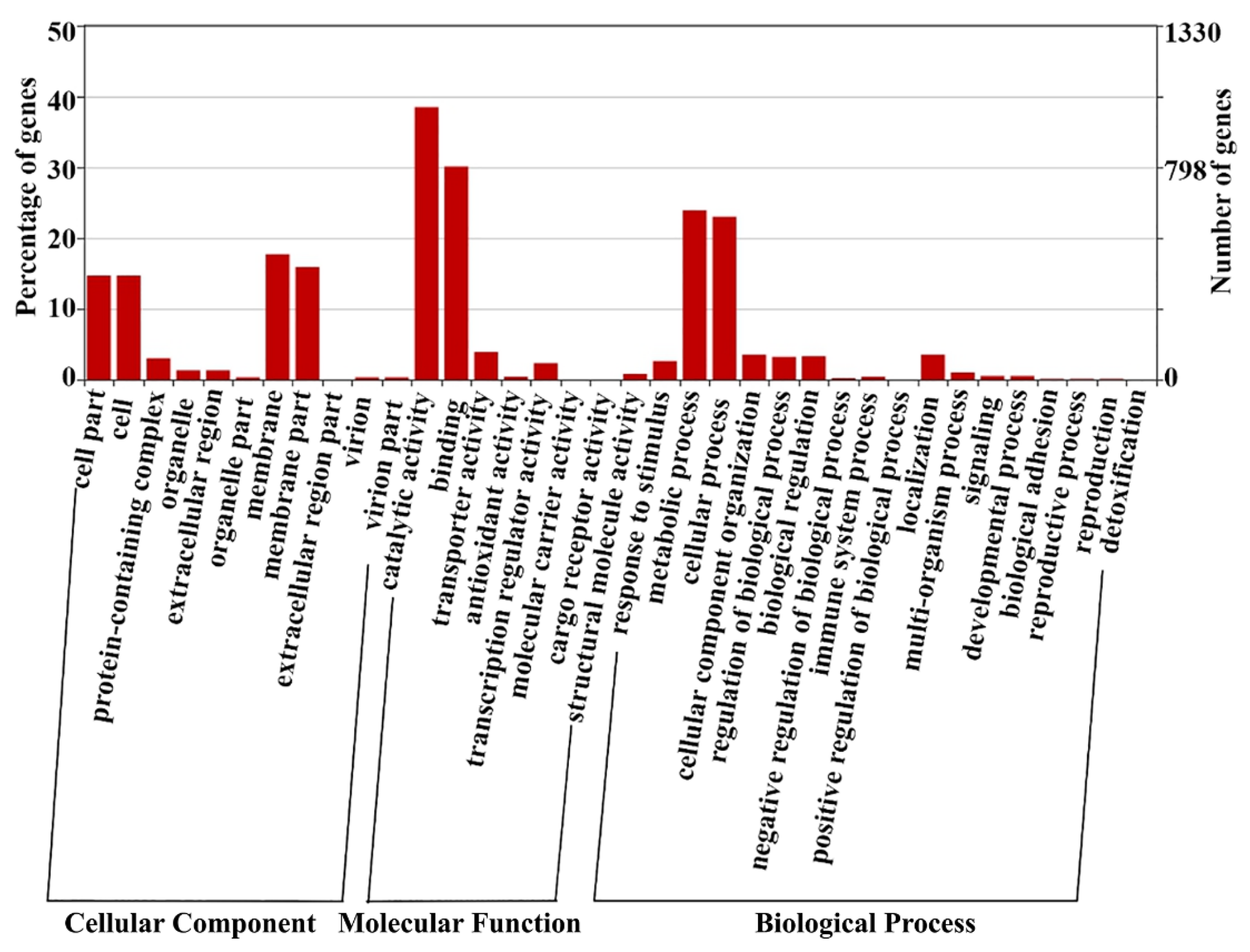
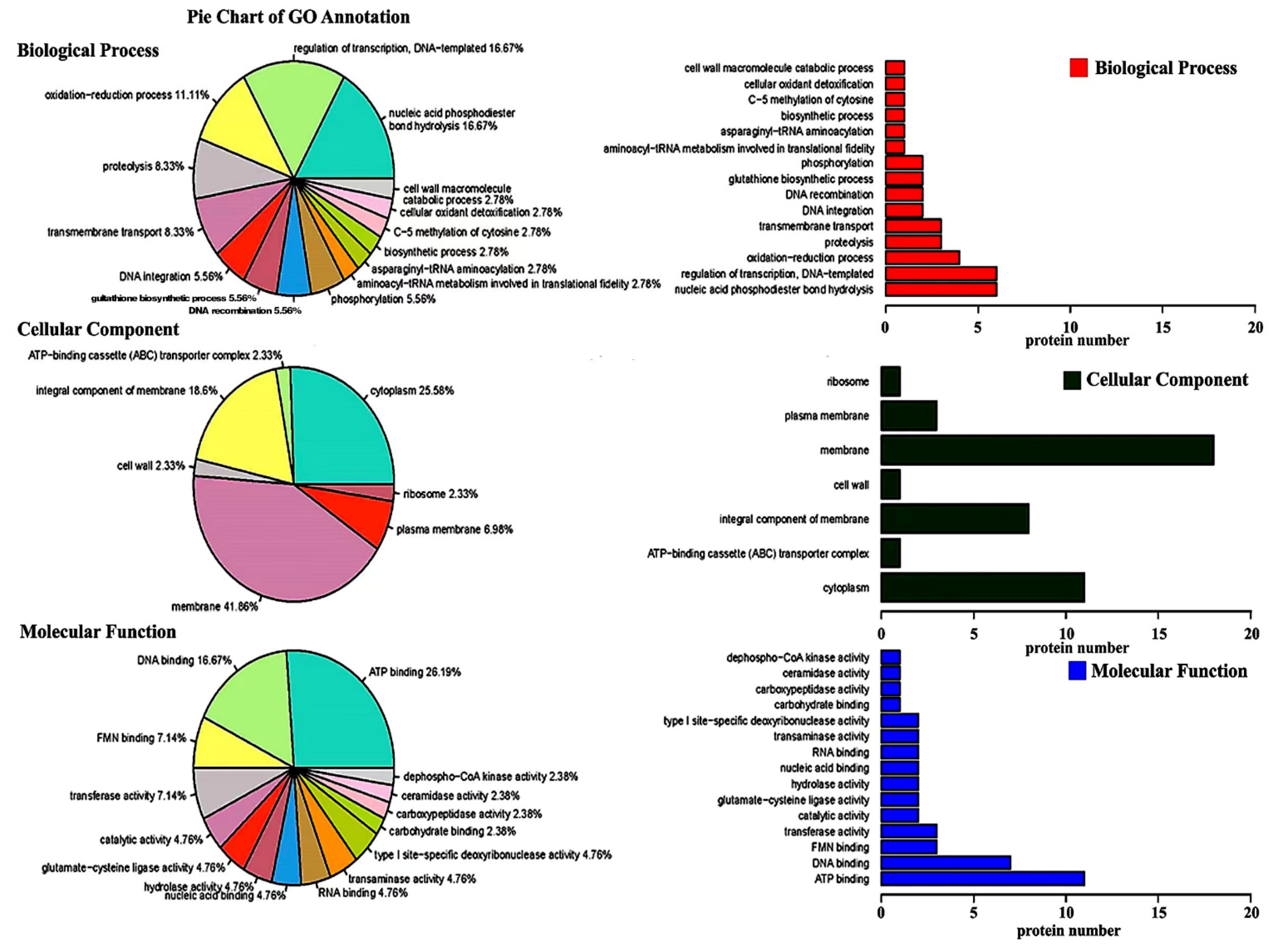
3.3. Gene Ontology Enrichment Analysis
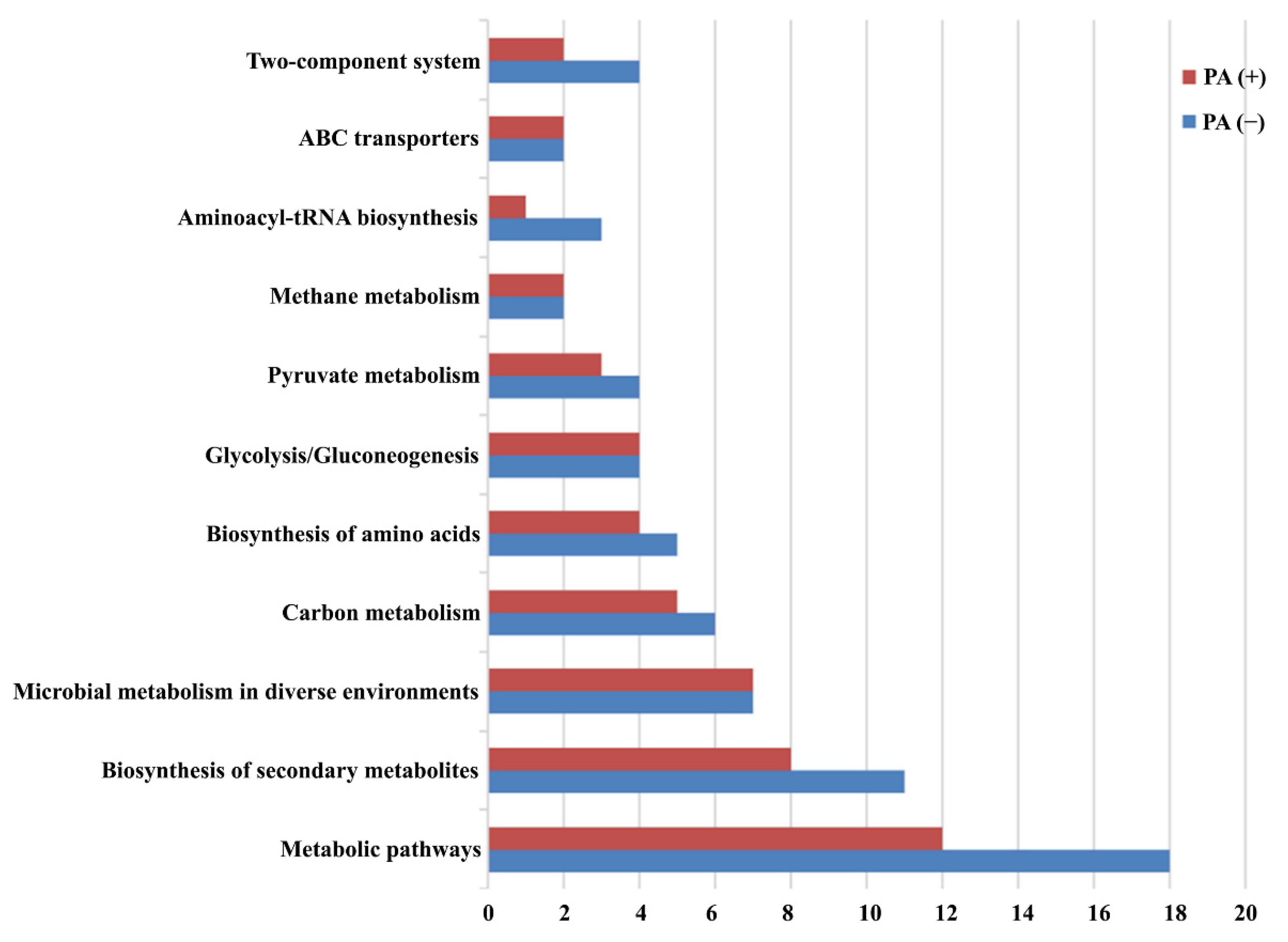
3.4. Pathway Enrichment Analysis
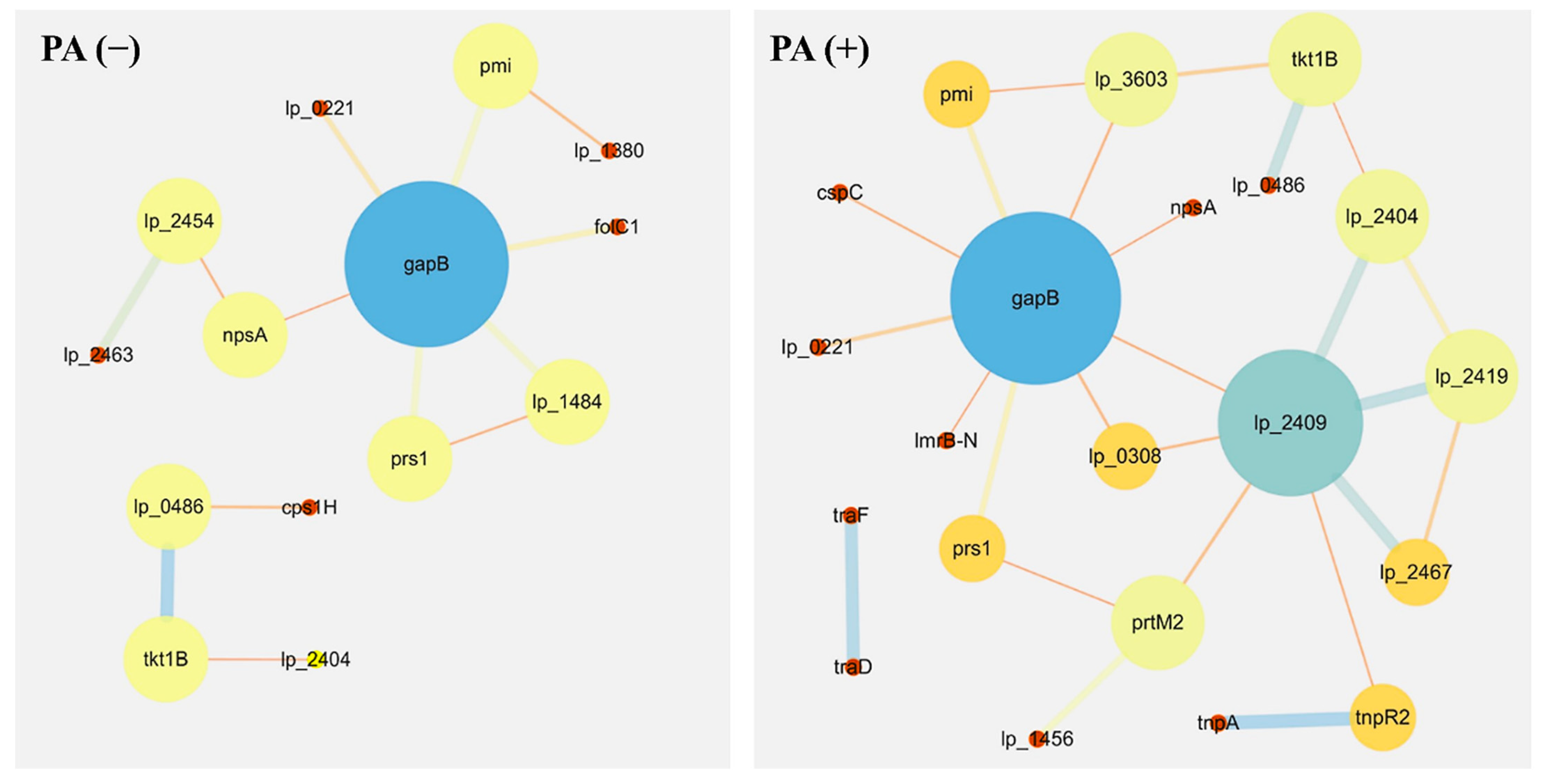
3.5. Protein–Protein Interaction Networks
3.6. MALDI–TOF Identification of Lipopeptides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, R.R.; Wu, Z.F.; Wang, S.; Zhang, M.; Zhou, G.L.; Li, B. Isolation, identification and characterization of propionic acid bacteria associated with autistic spectrum disorder. Microb. Pathog. 2020, 147, 104371. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.R.; Xu, Y.; Wang, Y.G.; Qiu, F.Y. Chemical environment influence the incidence of childhood autism. Asian. J. Chem. 2013, 25, 8835–8837. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Elhefnawy, A.M.; Azouz, H.G.; Roshdy, Y.S.; Ashry, M.H.; Ibrahim, A.E.; Meheissen, M.A. Study of the gut microbiome profile in children with autism spectrum disorder: A single tertiary hospital experience. J. Mol. Neurosci. 2020, 70, 887–896. [Google Scholar] [CrossRef]
- Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Pessah, I.; Van de Water, J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav. Immun. 2011, 25, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Andreo-Martínez, P.; Rubio-Aparicio, M.; Sánchez-Meca, J.; Veas, A.; Martínez-González, A.E. A Meta-analysis of Gut Microbiota in Children with Autism. J. Autism Dev. Disord. 2021, 52, 1374–1387. [Google Scholar] [CrossRef] [PubMed]
- Bojovic, K.; Ignjatovic, D.; Bajic, S.S.; Milutinovic, D.V.; Tomic, M.; Golic, N.; Tolinacki, M. Gut microbiota dysbiosis associated with altered production of short chain fatty acids in Children with neurodevelopmental disorders. Front. Cell Infect. Microbiol. 2020, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.X.; Ma, W.; Zhang, J.; He, Y.; Wang, J. Analysis of gut microbiota profiles and microbe-disease associations in children with autism spectrum disorders in China. Sci. Rep. 2018, 8, 13981. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.L.; Yu, R.R.; Ahmed, T.; Jiang, H.B.; Zhang, M.C.; Lv, L.Q.; Alhumaydhi, F.A.; Allemailem, K.S.; Li, B. Biosynthesis and characterization of zinc oxide nanoparticles and their impact on the composition of gut microbiota in healthy and ADHD children. Front. Microbiol. 2021, 12, 2051. [Google Scholar]
- Zou, R.; Xu, F.F.; Wang, Y.Z.; Duan, M.M.; Guo, M.; Zhang, Q.; Zhao, H.Y.; Zheng, H.J. Changes in the gut microbiota of children with autism spectrum disorder. Autism Res. 2020, 13, 1614–1625. [Google Scholar] [CrossRef]
- Abdellatif, B.; McVeigh, C.; Bendriss, G.; Chaari, A. The promising role of probiotics in managing the altered gut in autism spectrum disorders. Int. J. Mol. Sci. 2020, 21, 4159. [Google Scholar] [CrossRef]
- Panzer, A.R.; Lynch, S.V. Gut Microbial Regulation of Autism Spectrum Disorder Symptoms. Trends Endocrinol. Metab. 2020, 31, 809–811. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Mazmanian, S.K. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 2015, 17, 565–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharon, G.; Cruz, N.J.; Kang, D.W.; Gandal, M.J.; Wang, B.; Kim, Y.M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 2019, 177, 1600–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitschke, A.; Deonandan, R.; Konkle, A.T. The link between autism spectrum disorder and gut microbiota: A scoping review. Autism 2020, 24, 1328–1344. [Google Scholar] [CrossRef] [PubMed]
- Nogay, N.H.; Nahikian-Nelms, M. Can we reduce autism-related gastrointestinal and behavior problems by gut microbiota based dietary modulation? A review. Nutrit. Neurosci. 2021, 24, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Vázquez, L.; Van Ginkel Riba, G.; Arija, V.; Canals, J. Composition of gut microbiota in children with autism spectrum disorder: A systematic review and meta-analysis. Nutrients 2020, 12, 792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, D.; Cheon, K.-A. Alteration of gut microbiota in autism spectrum disorder: An overview. J. Kor. Acad Child Ado. 2020, 31, 131. [Google Scholar] [CrossRef]
- Li, Q.R.; Han, Y.; Dy, A.B.C.; Hagerman, R.J. The gut microbiota and autism spectrum disorders. Front. Cell. Neurosci. 2017, 11, 120. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.F.; Cao, S.Q.; Zhang, X.W. Modulation of gut microbiota–brain axis by probiotics, prebiotics, and diet. J. Agric. Food Chem. 2015, 63, 7885–7895. [Google Scholar] [CrossRef]
- Maigoro, A.Y.; Lee, S. Gut Microbiome-Based Analysis of Lipid A Biosynthesis in Individuals with Autism Spectrum Disorder: An In Silico Evaluation. Nutrients 2021, 13, 688. [Google Scholar] [CrossRef]
- Israelyan, N.; Margolis, K.G. Serotonin as a link between the gut-brain-microbiome axis in autism spectrum. Pharmacol. Res. 2018, 132, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, W.A.; Meguid, N.A.; Bjorklund, G.; Eid, E.M.; Farid, M.; Mohamed, S.K.; Wakeel, K.E.; Chirumbolo, S.; Elsaeid, A.; Hammad, D.Y. Impact of Clostridium Bacteria in Children with Autism Spectrum Disorder and Their Anthropometric Measurements. J. Mol. Neurosci. 2020, 70, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef]
- Taniya, M.A.; Chung, H.J.; Al Mamun, A.; Alam, S.; Aziz, M.A.; Emon, N.U.; Islam, M.M.; Hong, S.T.S.; Podder, B.R.; Mimi, A.A.; et al. Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front. Cell. Infect. Microbiol. 2022, 12, 915701. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezawada, N.; Phang, T.H.; Hold, G.L.; Hansen, R. Autism spectrum disorder and the gut microbiota in children: A systematic review. Nutr. Metab. 2020, 76, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Bundgaard-Nielsen, C.; Knudsen, J.; Leutscher, P.D.; Lauritsen, M.B.; Nyegaard, M.; Hagstrøm, S.; Sørensen, S. Gut microbiota profiles of autism spectrum disorder and attention deficit/hyperactivity disorder: A systematic literature review. Gut Microbes 2020, 11, 1172–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; De Caro, C.; Comegna, M. Gut microbiota features in young children with autism spectrum disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, Z.; Mao, X.H.; Liu, Q.S.; Guo, M.C.; Zhuang, Y.Y.; Liu, Z.; Chen, K.; Chen, J.Y.; Xu, R.; Tang, J.M. Altered gut microbial profile is associated with abnormal metabolism activity of autism spectrum disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef]
- Ding, H.T.; Taur, Y.; Walkup, J.T. Gut microbiota and autism: Key concepts and findings. J. Autism Dev. Disord. 2017, 47, 480–489. [Google Scholar] [CrossRef]
- Ding, X.; Xu, Y.R.; Zhang, X.L.; Zhang, L.L.; Duan, G.Q.; Song, C.L.; Li, Z.H.; Yang, Y.Y.; Wang, Y.Z.; Wang, X.Y.; et al. Gut microbiota changes in patients with autism spectrum disorders. J. Psych. Res. 2020, 129, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.E. Metabolic and mitochondrial disorders associated with epilepsy in children with autism spectrum disorder. Epilepsy Behav. 2015, 47, 147–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.C.; Lee, C.H.; Wang, H. Exploring the association of autism spectrum disorders and constipation through analysis of the gut microbiome. Int. J. Environ. Res. Public Health 2021, 18, 667. [Google Scholar] [CrossRef]
- Olesova, D.; Galba, J.; Piestansky, J.; Celusakova, H.; Repiska, G.; Babinska, K.; Ostatnikova, D.; Katina, S.; Kovac, A. A novel UHPLC-MS method targeting urinary metabolomic markers for autism spectrum disorder. Metabolites 2020, 10, 443. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L.; Shin, H.D.; Chen, R.R.; Zhang, J.; Li, J.H.; Du, G.C.; Shi, Z.P.; Chen, J. Systems-level understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the microenvironment levels: Mechanism and application. J. Biotechnol. 2013, 167, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Meeking, M.M.; MacFabe, D.F.; Mepham, J.R.; Foley, K.A.; Tichenoff, L.J.; Boon, F.H.; Kavaliers, M.; Ossenkopp, K.P. Propionic acid induced behavioural effects of relevance to autism spectrum disorder evaluated in the hole board test with rats. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2020, 97, 109794. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, X.; Chen, R.; Huangfu, W.; Xie, G. Antibacterial activity of chitosan solution against Xanthomonas pathogenic bacteria isolated from Euphorbia pulcherrima. Carbohydr. Polym. 2008, 72, 287–292. [Google Scholar] [CrossRef]
- Shrivastava, N.; Jiang, L.; Li, P.; Sharma, A.K.; Luo, X.Y.; Wu, S.L.; Pandey, R.; Gao, Q.K.; Lou, B.G. Proteomic approach to understand the molecular physiology of symbiotic interaction between Piriformospora indica and Brassica napus. Sci. Rep. 2018, 8, 5773. [Google Scholar] [CrossRef]
- Cao, J.Y.; Xu, Y.P.; Cai, X.Z. TMT-based quantitative proteomics analyses reveal novel defense mechanisms of Brassica napus against the devastating necrotrophic pathogen Sclerotinia sclerotiorum. J. Proteomics 2016, 143, 265–277. [Google Scholar] [CrossRef]
- Abdallah, Y.; Yang, M.; Zhang, M.; Masum, M.M.I.; Ogunyemi, S.O.; Hossain, A.; An, Q.; Yan, C.; Li, B. Plant growth promotion and suppression of bacterial leaf blight in rice by Paenibacillus polymyxa Sx3. Lett. Appl. Microbiol. 2019, 68, 423–429. [Google Scholar] [CrossRef]
- Fanaei, M.; Jurcic, K.; Emtiazi, G. Detection of simultaneous production of kurstakin, fengycin and surfactin lipopeptides in Bacillus mojavensis using a novel gel-based method and MALDI-TOF spectrometry. World J. Microbiol. Biotechnol. 2021, 37, 97. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.; Mazumder, A.; Grace, T. Role of the Gut Microbiome in Autism Spectrum Disorders. Adv. Exp. Med. Biol. 2019, 1118, 253–269. [Google Scholar] [PubMed]
- Srikantha, P.; Mohajeri, M.H. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willyard, C. How gut microbes could drive brain disorders. Nature 2021, 590, 22–25. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heunis, T.; Deane, S.; Smit, S.; Dicks, L.M.T. Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J. Proteome Res. 2014, 13, 4028–4039. [Google Scholar] [CrossRef]
- Ingham, C.J.; Beerthuyzen, M.; Vlieg, J.V. Population Heterogeneity of Lactobacillus plantarum WCFS1 Microcolonies in Response to and Recovery from Acid Stress. Appl. Environ. Microb. 2008, 74, 7750–7758. [Google Scholar] [CrossRef] [Green Version]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [Green Version]
- Călinescu, O.; Paulino, C.; Kühlbrandt, W.; Fendler, K. Keeping it simple, transport mechanism and pH regulation in Na+/H+ exchangers. J. Biol. Chem. 2014, 289, 13168–13176. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Q.; Xiao, Y.; Zhao, J.; Tian, F.W.; Zhang, H.; Narbad, A.; Chen, W. Identification of key proteins and pathways in cadmium tolerance of Lactobacillus plantarum strains by proteomic analysis. Sci. Rep. 2017, 7, 1182. [Google Scholar] [CrossRef] [PubMed]
- de Faria, A.F.; Stéfani, D.; Vaz, B.G.; Silva, Í.S.; Garcia, J.S.; Eberlin, M.N.; Grossman, M.J.; Alves, O.L.; Durrant, L.R. Purification and structural characterization of fengycin homologues produced by Bacillus subtilis LSFM-05 grown on raw glycerol. J. Ind. Microbiol. Biotechnol. 2011, 38, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Wei, Z.; Guan, Z.; Cai, Y.; Liao, X. Antifungal activity of isolated Bacillus amyloliquefaciens SYBC H47 for the biocontrol of peach gummosis. PLoS ONE 2016, 11, e0162125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimkić, I.; Stanković, S.; Nišavić, M.; Petković, M.; Ristivojević, P.; Fira, D.; Berić, T. The profile and antimicrobial activity of Bacillus lipopeptide extracts of five potential biocontrol strains. Front. Microbiol. 2017, 8, 925. [Google Scholar] [CrossRef]
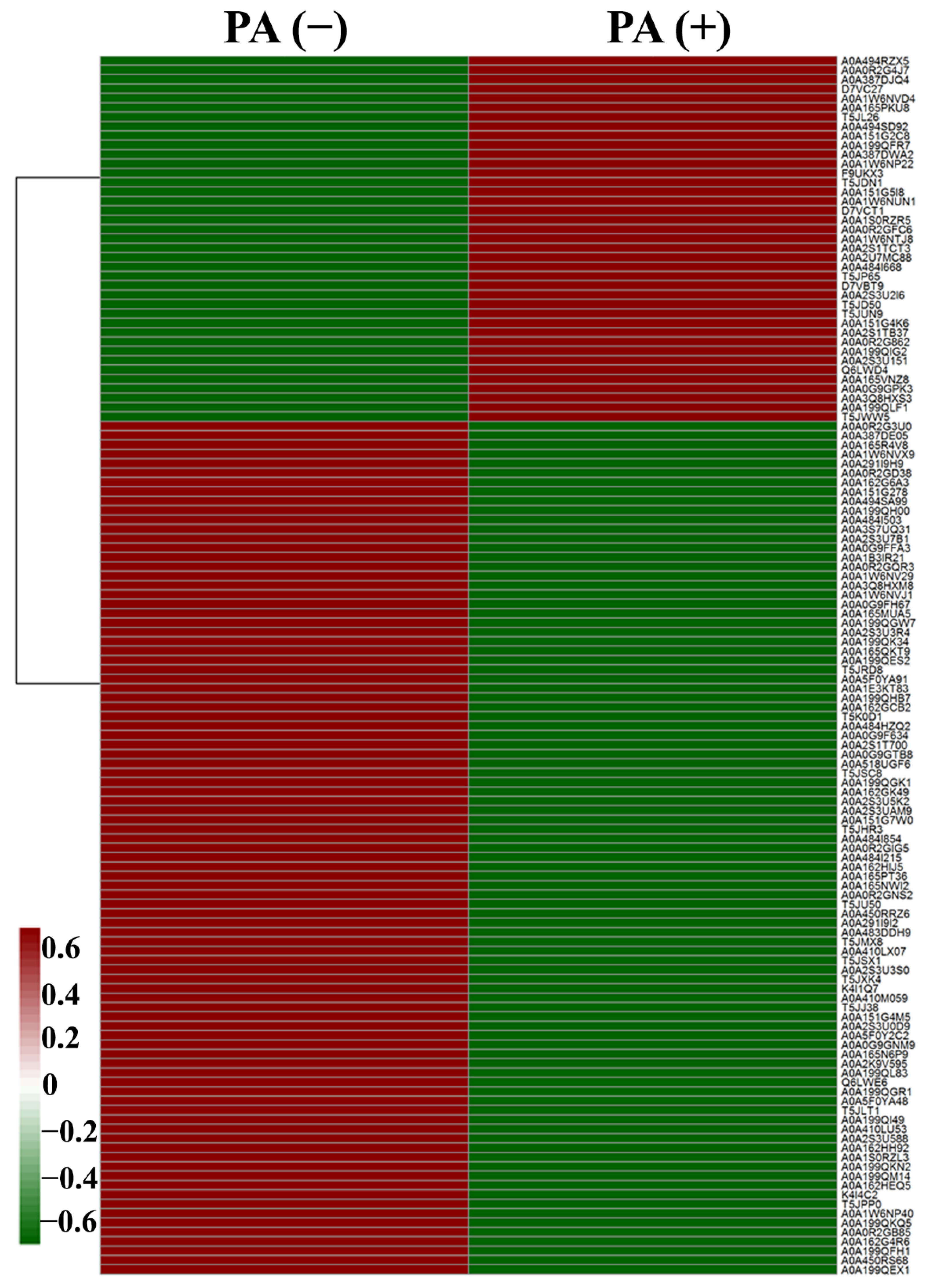
| UniProt Accession | Function Description | Fold Change |
|---|---|---|
| Down-regulation (>10-fold change) | ||
| A0A0G9F634 | Transposase | 0.02 |
| A0A0G9FH67 | Aminotransferase | 0.09 |
| A0A2S1T700 | Uncharacterized protein | 0.03 |
| A0A0R2G3U0 | PTS family mannose fructose sorbose porter component IID | 0.09 |
| A0A2S3U5K2 | ATP-dependent zinc metalloprotease FtsH | 0.04 |
| A0A2S3U7B1 | Glutathione-disulfide reductase | 0.07 |
| A0A0R2GIG5 | Flagellar protein FliS | 0.01 |
| A0A151G278 | Integrase | 0.10 |
| A0A151G4M5 | Uncharacterized protein | 0.06 |
| A0A3Q8HXM8 | Uncharacterized protein | 0.02 |
| A0A162G4R6 | DNA replication protein phage-associated | 0.06 |
| A0A162G6A3 | Putative membrane protein | 0.10 |
| A0A162GCB2 | Uncharacterized protein | 0.06 |
| A0A410LU53 | Uncharacterized protein | 0.08 |
| A0A410LX07 | Tape measure protein | 0.07 |
| A0A162HH92 | Uncharacterized protein | 0.02 |
| A0A165MUA5 | Type I restriction enzyme R protein | 0.09 |
| A0A484I215 | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase | 0.00 |
| A0A165NWI2 | GMP reductase | 0.05 |
| A0A484I503 | Uncharacterized protein | 0.07 |
| A0A484I854 | Usp domain-containing protein | 0.09 |
| A0A165QKT9 | Uncharacterized protein | 0.06 |
| A0A494SA99 | Coenzyme A biosynthesis bifunctional protein CoaBC | 0.01 |
| A0A518UGF6 | Restriction endonuclease subunit S | 0.10 |
| A0A199QEX1 | Aminotransferase | 0.01 |
| A0A5F0Y2C2 | POLAc domain-containing protein | 0.05 |
| A0A5F0YA48 | Bifunctional glutamate-cysteine ligase GshA/glutathione synthetase GshB | 0.05 |
| A0A199QGR1 | Transposase | 0.06 |
| A0A199QH00 | UDP-glucose 6-dehydrogenase | 0.05 |
| K4I4C2 | Tail tape measure | 0.10 |
| T5JJ38 | TetR family transcriptional regulator | 0.01 |
| A0A1B3IR21 | Relaxase superfamily protein | 0.05 |
| A0A1S0RZL3 | Uncharacterized protein | 0.03 |
| T5JSC8 | Oligopeptidase PepB | 0.02 |
| T5JU50 | Serine tRNA ligase | 0.01 |
| A0A1W6NVX9 | Conjugal transfer protein | 0.05 |
| T5K0D1 | Asparagine tRNA ligase | 0.07 |
| Up-regulation (>10 fold change) | ||
| A0A1W6NVD4 | Uncharacterized protein | 11.55 |
| T5JP65 | HTH lacI-type domain-containing protein | 17.40 |
| T5JUN9 | 2,5-diketo-D-gluconic acid reductase | 24.93 |
| T5JWW5 | Chromosome partition protein Smc | 18.05 |
| T5JL26 | 5-formyltetrahydrofolate cyclo-ligase | 24.40 |
| T5JD50 | Glutathione synthetase | 210.94 |
| A0A484I668 | Uncharacterized protein | 11.40 |
| A0A199QFR7 | Protein kinase | 72.45 |
| D7VBT9 | Site-specific recombinase, phage integrase family | 11.89 |
| D7VC27 | Glycosyltransferase, group 1 family protein | 10.29 |
| Q6LWD4 | Putative transposase | 17.71 |
| A0A387DJQ4 | Terminase small subunit | 24.72 |
| A0A151G5I8 | Uncharacterized protein | 119.47 |
| A0A165PKU8 | Functional role page for anaerobic nitric oxidereductase transcription regulator NorR | 10.90 |
| A0A2S3U2I6 | Calcium-transporting ATPase | 18.16 |
| A0A2U7MC88 | RpoD (fragment) | 18.86 |
| A0A2S3U151 | DNA helicase | 106.37 |
| Node | External_id | Function | Role in Metabolic and Synthesis Pathway |
|---|---|---|---|
| tkt1B | 220668.lp_0491 | transketolase, pyrimidine-binding domain | + |
| lp_2463 | 220668.lp_2463 | prophage P2b protein 18, major capsid protein | + |
| lp_1484 | 220668.lp_1484 | hypothetical protein | + |
| gapB | 220668.lp_0789 | glyceraldehyde 3-phosphate dehydrogenase | + |
| pmi | 220668.lp_2384 | mannose-6-phosphate isomerase | + |
| folC1 | 220668.lp_2321 | formylTHF-polyglutamate synthase/folyl-polyglutamate synthase/hydrofolate synthase | + |
| cps1H | 220668.lp_1184 | glycosyltransferase (rhamnosyltransferase), family 2 (GT2) | + |
| lp_2454 | 220668.lp_2454 | prophage P2a protein 3; DNA adenine methylase | + |
| lp_2404 | 220668.lp_2404 | prophage P2a protein 53 | + |
| Node | External_id | Function | Role in Metabolic and Synthesis Pathway |
|---|---|---|---|
| lp_2419 | 220668.lp_2419 | prophage P2a protein 38; minor head protein | + |
| tkt1B | 220668.lp_0491 | transketolase, pyrimidine-binding domain | + |
| lp_2467 | 220668.lp_2467 | prophage P2b protein 14, terminase small subunit | + |
| lp_2409 | 220668.lp_2409 | prophage P2a protein 48; tape measure protein | + |
| prtM2 | 220668.lp_3193 | peptidyl-prolyl isomerase | + |
| gapB | 220668.lp_0789 | glyceraldehyde 3-phosphate dehydrogenase | + |
| pmi | 220668.lp_2384 | mannose-6-phosphate isomerase | + |
| lp_3603 | 220668.lp_3603 | sugar-phosphate aldolase | + |
| cspC | 220668.lp_0997 | cold shock protein CspC | + |
| lp_2404 | 220668.lp_2404 | prophage P2a protein 53 | + |
| m/z | Peak Area | m/z | Peak Area | ||||
|---|---|---|---|---|---|---|---|
| PA (+) | PA (−) | PA (+)/(−) | PA (+) | PA (−) | PA (+)/(−) | ||
| 686.535 | 9.86 | 11.9 | 0.83 | 2037.374 | — | 42.4 | — |
| 696.46 | — | 9.35 | — | 2059.347 | 101 | 194 | 0.52 |
| 911.601 | 21.4 | 26.6 | 0.80 | 2069.348 | — | 31.2 | — |
| 984.637 | 12.3 | 12.9 | 0.95 | 2091.317 | 64 | 126 | 0.51 |
| 1105.699 | 16.8 | 22.7 | 0.74 | 2093.502 | 17.7 | 32.9 | 0.54 |
| 1110.711 | 15.6 | 21.9 | 0.71 | 2108.472 | 40.2 | 57.9 | 0.69 |
| 1122.732 | 75.2 | 83.4 | 0.90 | 2168.432 | 36.1 | 82.7 | 0.44 |
| 1144.706 | 15.3 | 23.1 | 0.66 | 2190.411 | 220 | 439 | 0.50 |
| 1146.787 | 77.1 | 71.8 | 1.07 | 2205.39 | 25.2 | 27.3 | 0.92 |
| 1322.883 | 15 | 17.8 | 0.84 | 2207.556 | 312 | 649 | 0.48 |
| 1543.022 | — | 19.5 | — | 2223.534 | 25.5 | 49.6 | 0.51 |
| 1905.241 | — | 23.9 | — | 2533.751 | 51.7 | 84.9 | 0.61 |
| 1960.269 | 21.6 | 44.9 | 0.48 | 2632.834 | 467 | 1314 | 0.36 |
| 2004.277 | — | 22.1 | — | 2648.81 | 38.7 | 80 | 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, R.; Zhang, M.; Ahmed, T.; Wu, Z.; Lv, L.; Zhou, G.; Li, B. Metabolic and Proteomic Profiles Reveal the Response of the ASD-Associated Resistant Strain 6-1 of Lactobacillus plantarum to Propionic Acid. Int. J. Environ. Res. Public Health 2022, 19, 17020. https://doi.org/10.3390/ijerph192417020
Yu R, Zhang M, Ahmed T, Wu Z, Lv L, Zhou G, Li B. Metabolic and Proteomic Profiles Reveal the Response of the ASD-Associated Resistant Strain 6-1 of Lactobacillus plantarum to Propionic Acid. International Journal of Environmental Research and Public Health. 2022; 19(24):17020. https://doi.org/10.3390/ijerph192417020
Chicago/Turabian StyleYu, Rongrong, Muchen Zhang, Temoor Ahmed, Zhifeng Wu, Luqiong Lv, Guoling Zhou, and Bin Li. 2022. "Metabolic and Proteomic Profiles Reveal the Response of the ASD-Associated Resistant Strain 6-1 of Lactobacillus plantarum to Propionic Acid" International Journal of Environmental Research and Public Health 19, no. 24: 17020. https://doi.org/10.3390/ijerph192417020
APA StyleYu, R., Zhang, M., Ahmed, T., Wu, Z., Lv, L., Zhou, G., & Li, B. (2022). Metabolic and Proteomic Profiles Reveal the Response of the ASD-Associated Resistant Strain 6-1 of Lactobacillus plantarum to Propionic Acid. International Journal of Environmental Research and Public Health, 19(24), 17020. https://doi.org/10.3390/ijerph192417020








