Abstract
The increasing consumption of organic or ready-to-eat food may cause serious foodborne disease outbreaks. Developing microbiological culture for detection of food-borne pathogens is time-consuming, expensive, and laborious. Thus, alternative methods such as polymerase chain reaction (PCR) are usually employed for outbreaks investigation. In this work, we aimed to develop a rapid and simple protocol for the simultaneous detection of Escherichia coli (E coli), Listeria monocytogenes (L. monocytogenes), Staphylococcus aureus (S. aureus) and Salmonella enterica (S. enterica), by the combination of an enrichment step in a single culture broth and a multiplex PCR (mPCR) assay. The effectiveness of several enrichment media was assessed by culture and PCR. Buffered peptone water (BPW) was selected as the optimum one. Then, mPCR conditions were optimized and applied both to pure co-cultures and artificially inoculated food samples (organic lettuce and minced meat). In the culture medium inoculated at 100 CFU/mL, mPCR was able to detect the four microorganisms. When performed on artificially food samples, the mPCR assy was able to detect E. coli, S. enterica, and L. monocytogenes. In conclusion, BPW broth can effectively support the simultaneous growth of E. coli, S. aureus, L. monocytogenes, and S. enterica and could be, thus, used prior to a mPCR detection assay in ready-to-eat food, thereby considerably reducing the time, efforts and costs of analyzes.
1. Introduction
The development of food industrialization and the international trade in fresh and frozen foods are the two main factors that have contributed to the increasing complexity of the global food security challenge [1]. However, the resurgence of certain food trends such as massive consumption of organic food, the raw food diet, and ready-to-eat food have resulted in an alarming increase of food poisoning cases [2,3]. Despite the health benefits, the raw food diet, which is defined by the consumption of raw vegetables or animal products, when not subjected to heat treatment over 40 °C, may be the cause of serious foodborne illnesses and even disease outbreaks. Among the outbreaks reported by the Centers for Disease Control (CDC) and Prevention’s Foodborne Disease Outbreak Surveillance System (FDOSS), 18 were identified involving vegetables and dairy products from 1992 to 2014, resulting in 779 illnesses, 258 hospitalizations, and 3 deaths [4].
Among the foodborne outbreaks that represent a cause for concern, most come from animal origin food, including beef meat, poultry, eggs, raw milk and milk products [5,6,7,8], and vegetables such as radishes, cucumbers, carrots, leafy foods such as lettuce, spinach, sprouts, cabbage, or even fruits [9,10,11,12,13,14,15], which may be contaminated by multiple pathogens such as Listeria monocytogenes (L. monocytogenes), Staphylococcus aureus (S. aureus), Salmonella enterica (S. enterica), and Escherichia coli (E. coli) [10,15,16]. Those microorganisms are defined as significant foodborne pathogen due to the severity of diseases and the number of illness cases caused [15,17,18,19]. To guarantee food safety, obtain the possibility of locating potential outbreaks, and limit the spreading of infection, the screening for these pathogens needs to be carried out on raw food products prior to and throughout the complete distribution process [20,21].
The current detection methods based on microbiological cultures require pre-enrichment and selective enrichment steps, followed by isolation on several selective media and biochemical or serological tests to obtain a definitive result [7,22]. These procedures are still considered as the “gold Standard”, although they are complicated, extremely labor intensive, costly, time consuming (from days to weeks), and might be subject to handling errors. The recent development of molecular methods such as polymerase chain reaction (PCR) has shown a high potential in foodborne outbreak investigation, food analysis and food monitoring [14,23,24,25,26]. Unfortunately, the sensitivity and reliability of PCR methods depend on the number of target bacterial cells. Thus, low contamination levels in food samples make the detection of target pathogens very difficult [7]. Consequently, an enrichment step prior to DNA extraction is generally required to increase the target pathogen concentration in the sample and resuscitate the stressed or sub-lethally injured cells, which improves the detection limits of the PCR and avoids false negative results [7,11,27,28]. To simplify the work required, save time, and reduce costs and humans errors, several studies have attempted to build protocols for the simultaneous detection of multiple pathogens in a single reaction [29,30,31,32]. Most of the recent works are based on the development of a selective co-enrichment broth, to ensure a simultaneous growth of target pathogens and suppress the background microbiota, prior to the detection by multiplex PCR (mPCR). Unfortunately, the presence of several selective agents may have serious repercussions on the growth balance between the different target pathogens, which can favor one target and inhibit the growth or delay the recovery of the others at the same time [19,33]. In addition, most of those synthetic media developed to be selective, are not specific to the target pathogens [5,11,30,33,34,35].
In the present work, we aimed to develop a rapid and simple protocol for the simultaneous detection of four foodborne pathogens E. coli, L. monocytogenes, S. aureus, and S. enterica, which could be used for the analyses of ready-to-eat food, by the combination of a co-culture step in a single culture broth and mPCR detection. Buffered peptone water (BPW) broth was selected after comparing its performances with multiple selective and non-selective culture media, as well as analyzing its recovery capacities from low initial inoculums, in individual pure cultures, in co-culture, and in the presence of artificially inoculated food matrices with background microbiota. The construction of our mPCR was based on the combination of four highly selective primers, mentioned in several previous works and optimized to ensure the most sensitive detection possible. Finally, the potential application of BPW as basic co-culture broth before the mPCR, for the simultaneous detection E. coli, L. monocytogenes, S. aureus, and S. enterica from artificially inoculated raw food was evaluated.
2. Materials and Methods
2.1. Bacterial Strains
In this work, the four most relevant pathogenic bacteria in food have been studied. The strains used, E. coli CECT 101, S. enterica CECT 4266, L. monocytogenes CECT 936, and S. aureus CECT 435, were procured from the Spanish Type Culture Collection (CECT, Valencia, Spain).
For the evaluation of the mPCR specificity, strains from different origins were used: E. coli CECT 425, E. coli CECT 418, E. coli CECT 4558, Citrobacter freundii (C. freundii) CECT 401, Micrococcus luteus (M. luteus) CECT 245, Staphylococcus epidermidis (S. epidermidis) CECT 231 and 3 laboratory isolates: Listeria innocua (L. innocua), Listeria grayi (L. grayi) and Bacillus cereus (B. cereus).
Cultures started from frozen stocks preserved at −20 °C, by streaking inoculation on Plate Count Agar (PCA-Scharlau), the different bacterial strains were separately cultivated and the plates were incubated 24 h at 37 °C.
2.2. Selection of a Co-Culture Medium
To select the most suitable co-culture broth for the four target bacteria, the effect of several selective and non-selective liquid media (commercial or synthetic) on the growth of E. coli, S. enterica, L. monocytogenes, and S. aureus were compared, according to the maximum population rates obtained under the same culture conditions (24 h at 37 °C).
2.2.1. Effect of Various Culture Broths on Individual Growth
The effect of multiple commercial non-selective broths, Luria-Bertani broth (LB) (DifcoTM-BD-France), nutrient broth (NB) (Difco™-BD-France) and buffered peptone water (BPW) (Scharlau-Spain), on the individual culture of each pathogen was evaluated.
In the same way, the commercial selective media recommended by the ISO and UNE-EN ISO protocols, as specific selective enrichment for the target microorganisms were tested: Brilliant green bile lactose broth (BGBLB) (DifcoTM-BD-France) for E. coli [36], Rappaport Vassiliadis-R10 Broth (RV) (DifcoTM-BD-France) for S. enterica [37], Giolitti-Cantoni (GC) broth (Scharlau-Spain) supplemented with the 06-011-100 potassium tellurite solution 3.5% (Scharlau-Spain) for S. aureus [38] and the Fraser enrichment broth base (FB) (Scharlau-Spain) supplemented with Listeria UVMII Selective Supplement 06-111-LY01 (Scharlau-Spain) and ferric ammonium citrate supplement 06-112-LY01 (Scharlau-Spain) for L. monocytogenes [39]. Moreover, the selective synthetic broth (SSSLE broth), developed by Chen et al. (2015) [5] for the simultaneous growth of S. enterica, S. aureus, S. flexneri, L. monocytogenes and E. coli, was tested in our work.
To evaluate the growth rates of each microorganism during individual cultures in several broths, one isolated colony of each reference strains cultivated 24 h on PCA, was separately inoculated in each evaluated broth, for a final volume of 10 mL. Then, inoculated broths were incubated during 24 h, at 37 °C for BN, BPW, LB, BGBLB, GC, FB, SSSLE and at 41 °C for RV.
The maximum population rates obtained after incubation from each broth were determined by count, on the respective selective media described in the UNE-EN ISO protocols: tryptone bile glucuronic agar (TBX) (Scharlau-Spain) for E. coli, xylose-lysine-deoxycholate agar (XLD) (Scharlau-Spain) for S. enterica, Baird Parker agar base (BPA) (Scharlau-Spain) mixed with the egg yolk emulsion potassium tellurite (EYEPT) 06-026-100 (Scharlau-Spain) for S. aureus and Palcam agar base (PAL) (Scharlau-Spain), mixed with selective supplement 06-110LY01 (Scharlau-Spain) for L. monocytogenes.
All plates were incubated for 24–48 h at 37 °C. The number of colony forming units/mL (CFU/mL) was determined according to the formula mentioned in UNE-EN ISO 7218:2008 [40].
2.2.2. Evaluation of BPW Recovery Capacity
Preparation of the Inoculum
Fresh pure cultures were prepared by inoculating separately one isolated colony from each target bacteria in 10 mL BPW, then incubated 24 h at 37 °C under 150 rpm agitation. The final concentration ranging 108–109 CFU/mL was determined in accordance with UNE-EN ISO 7218:2008 [40].
Effect of BPW on Individual and Co-Culture Growth
To evaluate BPW recovery capabilities, from low initial inoculum concentrations in individual pure cultures, three initial inoculum levels were tested: 103; 102; 101 CFU/mL, in a final volume of 10 mL, for each target pathogen. The concentration of the four target bacteria was equal in each experiment: Experiment I: (103; 103; 103; 103 CFU/mL); Experiment II: (102; 102; 102; 102 CFU/mL); and Experiment III: (101; 101; 101; 101 CFU/mL).
All plates were incubated on the selective media previously mentioned for 24 h at 37 °C and the number of CFU/mL was determined according to the formula mentioned in UNE-EN ISO 7218:2008 [40]. One mL aliquots from each resulting culture were frozen at −20 °C, to be tested by PCR.
Effect of BPW on Co-Culture Growth, from Artificially Inoculated Food Matrix
To evaluate BPW recovery efficiency in co-culture, from artificially inoculated food with background microbiota, two kinds of ready-to-eat food were tested, namely eco-organic lettuce and minced meat, purchased from local stores and used fresh without any disinfectant treatment, in order to preserve the background microbiota of the foods.
Samples (lettuce and minced meat) were handled following the same protocol: 10 g of sample were artificially inoculated with the appropriate pure culture dilutions (prepared in Section 2.2.2.), to obtain 103 CFU/mL initial inoculum concentration of each pathogen, then mixed to 90 mL of sterile BPWin a stomacher bag. Mixtures were homogenized for 5 min and then incubated 24 h at 37 °C.
The number of CFU/mL was determined according to the formula mentioned in UNE-EN ISO 7218:2008 [40]. One mL aliquots from each resulting co-culture were frozen at −20 °C, to be tested by PCR.
2.3. PCR Detection
2.3.1. Preparation of DNA Template
DNA templates were extracted by using the GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MI, USA) with Lysozyme.
To evaluate the thermal lysis extraction protocol effectiveness, without lysozyme, for both Gram-positive and Gram-negative [11], aliquots collected from each individual culture were submitted to DNA extraction by both methods.
Thermal Lysis Method
One mL of the collected aliquots was subjected to 10 min centrifugation at 14,000 rpm (≈14,462× g), the pellet was resuspended in 100 µL of sterile Milli-Q water, after vortex, the suspension was boiled 10 min at 100 °C and immediately cooled on ice for 5 min. The supernatant obtained after 5 min centrifugation at 12,000 rpm (≈10,625× g) was stored at −20 °C, until further use as PCR template.
GenElute™ Bacterial Genomic DNA Kit
One mL of the collected aliquots was subjected to 2 min centrifugation at 16,000× g and the pelleted cells were used for DNA extraction following Gram-positive manufacturer’s extraction protocol of GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich), which requires the use of Lysozyme (Lysozyme BioChemica-PanReac AppliChem). The eluted DNA was stored at −20 °C to be used as PCR template.
2.3.2. Primers
Multiplex PCR (mPCR) was carried out with the combination of four specific pairs of primers in one reaction (Table 1). For the detection of E. coli, primers GADA 670-F/R targeting the glutamate decarboxylase enzyme encoding gene (gadA), were chosen from McDaniels et al. (1996) [41] study. Nuc 484-F/R primers, targeting the encoding gene of S. aureus thermostable nuclease (nuc) were selected from the work of Xu et al. (2006) [42]. LM 404-F/R primers from Wu et al. (2004) [43] targeting the listeriolysin gene (lisA), were used for the detection of L. monocytogenes and the primers SalinvA 284-139/141 from the work of Rahn et al. (1992) [44], targeting the encoding gene of the Invasion protein A (invA), were used for the detection of S. enterica. The specificity of each primer was checked in silico by using the Primer-Blast program, Basic Local Alignment Search Tool, http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 25 November 2021), to avoid nonspecific products [45]. Then, the primers were synthesized by TIB MOLBIOL (Syntheselabor GmbH-Germany).

Table 1.
Primers used.
2.3.3. Simplex PCR
Primer validation was made individually by simplex PCR, under the same conditions. These conditions were obtained after optimization tests, including an annealing temperature gradient (55; 56; 57; 58; 60 °C), for each simplex PCR.
PCRs were performed according to an initial denaturation at 94 °C for 2 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 60 s and a final extension at 72 °C for 7 min.
Reaction mixtures had similar composition: 2.9 µL Reaction Buffer 10 × NH4; 3 mM MgCl2 Solution; 0.20 mM dNTPs (dNTP Mix-BIOLINE) and 2.5 Units Taq DNA polymerase (BIOTAQ ™ DNA Polymerase-BIOLINE), mixed with 4 µL of target DNA. The amount of primers used for each simplex was 0.4 µM GADA670 for E. coli, 0.4 µM Nuc484 for S. aureus, 0.4 µM LM404 for L. monocytogenes and 0.2 µM SalinvA284 for S. enterica. Reaction mixtures with Milli-Q distilled water, instead of DNA template, were used as a negative control.
After amplification, 5 μL of each PCR products were mixed with Ready-to-Load (GeneRuler-Thermo Scientific) and submitted to electrophoresis for 80 min at 80 V on 1.5% agarose gel (Agarose D1 low EEO-CONDA/TAE buffer PanReac AppliChem, 0.05 M EDTA-Na2·2H2O, 1 M Acetic Acid glacial, 2 M Tris), then visualized under a UV transilluminator.
2.3.4. Multiplex PCR (mPCR)
mPCR was developed for the simultaneous detection of E. coli, S. aureus, L. monocytogenes and S. enterica, in a single reaction [46]. After multiplexing, several optimization tests were carried out including testing of annealing temperature gradient (58; 59; 60; 61; 62 °C), number of cycles variations (30; 25; 20 cycles), amount of MgCl2 (3.2; 3.5; 4 mM), dNTPs (0.22; 0.3; 0.4 mM), and the balance between primers.
The best conditions were: A total volume of 29 µL containing at least 100 ng of extracted DNA, 2.9 µL Reaction Buffer 10 × NH4; 3.5 mM MgCl2 Solution; 2.5 Units Taq DNA polymerase (BIOTAQ™ DNA Polymerase-BIOLINE) and 0.22 mM dNTP’s (dNTP Mix-BIOLINE). The primers concentrations were 0.2 µM GADA670, 1 µM Nuc484, 0.16 µM LM404, and 0.46 µM SalinvA284.
Reaction mixture with Milli-Q water was used as a negative control. The amplification was carried out using a Thermal Cycler (Eppendorf AG-Germany) with an initial denaturation at 94 °C for 2 min; followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 59 °C for 30 s, extension at 72 °C for 60 s, and a final extension step at 72 °C for 7 min.
After mPCR, 5 μL of PCR products were mixed with Ready-to-Load (GeneRuler-Thermo Scientific) and tested by electrophoresis during 80 min at 80 V on 1.5% agarose gel (Agarose D1 low EEO-CONDA/TAE buffer PanReac AppliChem, 0.05 M EDTA-Na2·2H2O, 1 M Acetic Acid glacial, 2 M Tris). Amplification products were visualized under a UV transilluminator.
2.3.5. Evaluation of Specificity
The specificity of the mPCR was evaluated by determining the ability of this protocol to distinguish the target bacteria from non-target ones. To check the presence of any cross-hybridization, which could lead to mis-priming between the four primers and four target DNAs, the selectivity of each primer was confirmed by mixing the four pairs of primers with several random combinations of positive control DNAs and then amplified according to the optimized PCR multiplex conditions mentioned above. Extracted DNA from pure cultures of E. coli CECT 101, S. aureus CECT 435, L. monocytogenes CECT 936, and S. enterica CECT 4266 were used as positive controls.
Then, E. coli CECT 425; E. coli CECT 418; E. coli CECT 4558, C. freundii CECT 401; M. luteus CECT 245; S. epidermidis CECT 231, and the 3 laboratory isolates L. innocua, L. grayi and B. cereus, were used for the evaluation of the mPCR specificity.
2.3.6. Evaluation of Sensitivity
The sensitivity was evaluated, firstly by using DNAs extracted separately from the four pure cultures in BPW, to determinate detection limits of each simplex and mPCR. Afterwards, tests were carried out by using DNA extracted from co-cultures made in BPW, with and without food matrix artificially inoculated, under background microbiota, as described in Section 2.2.2.
Detection Limits of Simplex and mPCR, from Individual Cultures
Detection limits of each simplex PCR were determined by multiple reactions whose composition was similar except for DNA amount, which were tested with a decreasing gradient of DNA amount in a final volume of 29 µL.
The evaluation of the mPCR detection limits was also carried out by several reactions, with the mix of the four specific primers and equal ratios of the four target DNAs, according to a decreasing gradient of DNA amount in a final volume of 29 µL. All of the PCRs were conducted following the optimized conditions mentioned previously.
Detection Limits from BPW Co-Culture Recovery
DNA extracted from 1 mL aliquots of each co-culture with the four target microorganisms in BPW broth, initially inoculated at 103; 102; 101; 100 CFU/mL, were tested by mPCR following the optimized conditions mentioned above.
Detection Limits from BPW Co-Culture Recovery, with Artificially Inoculated Food Matrices
The detection by mPCR was made by using the DNA extracted from co-culture recovery aliquots from artificially inoculated food matrices (eco-organic lettuce and minced meat) at 103 CFU/mL of each target pathogen, according to optimized conditions mentioned above.
3. Results
3.1. Selection of a Co-Culture Medium
3.1.1. Effect of Various Culture Broths on Individual Growth
One isolated colony of each reference strains cultivated 24 h on PCA was separately inoculated in each evaluated broth, for a final volume of 10 mL. Then, inoculated broths were incubated during 24 h, at 37 °C. The two non-selective media showing the best recovery performance for the four target bacteria in individual growth without any competition were NB and BPW. E. coli growth rates exceeded 108 CFU/mL in all of the non-selective media after 24 h of incubation. For S. enterica, growth ranged between 107 and 108 CFU/mL and varied between 109 and 1010 CFU/mL for S. aureus. Regarding L. monocytogenes, the growth rates recorded in all non-selective media were close, approximately 108 CFU/mL.
Maximum population rates of the selective broths recommended for the enrichment step by ISO protocols (E. coli in BGBLB, S. enterica in RV and S. aureus in GC) were much lower than growth rates obtained in the non-selective broths tested, except for FB broth, which got the highest growth rate for L. monocytogenes.
The selective co-enrichment medium SSSLE was the least effective of all evaluated media in this study, with the weakest growth rate for E. coli and no growth for S. enterica, S. aureus or L. monocytogenes (Table 2).

Table 2.
Individual culture growth (CFU/mL).
3.1.2. Effect of BPW on Individual and Co-Culture Growth
The evaluation of BPW recovery capacities from low initial inoculum, in individual pure culture, showed relatively stable and close rates, over 108 CFU/mL for E. coli, S. aureus and S. enterica, and over 107 for L. monocytogenes (Table 3).

Table 3.
Recovery rates values in CFU/mL from individual and co-culture growths in BPW.
3.1.3. Effect of BPW on Co-Culture Growth from Artificially Inoculated Food Matrix
Regarding the co-culture in BPW broth from food matrices artificially inoculated at 103 CFU/mL of each target bacteria, the recovery rates from the eco-organic lettuce sample reached 108 CFU/mL for the Gram-negative E. coli and S. enterica, and 106 CFU/mL for the Gram-positive S. aureus and L. monocytogenes.
For the artificially inoculated minced meat sample, the recovery rates recorded were lower than those obtained for the lettuce sample, about 107 CFU/mL for the Gram-negative microorganisms and S. aureus, while for L. monocytogenes it was up to 105 CFU/mL (Table 4).

Table 4.
Recovery rates values of co-culture growth in CFU/mL from BPW with and without food matrices initially inoculated at 103 CFU/mL.
3.2. PCR Detection
3.2.1. Simplex PCR
Simplex PCR detection from individual cultures was applied to check the correct performance of each primer pair and the performance of DNA extraction method. Results showed that the specific amplification of E. coli, S. aureus, L. monocytogenes, and S. enterica produced amplicons of different sizes, corresponding to 670 bp, 484 bp, 404 bp, and 284 bp, respectively, appearing as distinct bands on electrophoresis gel, without any non-specific product.
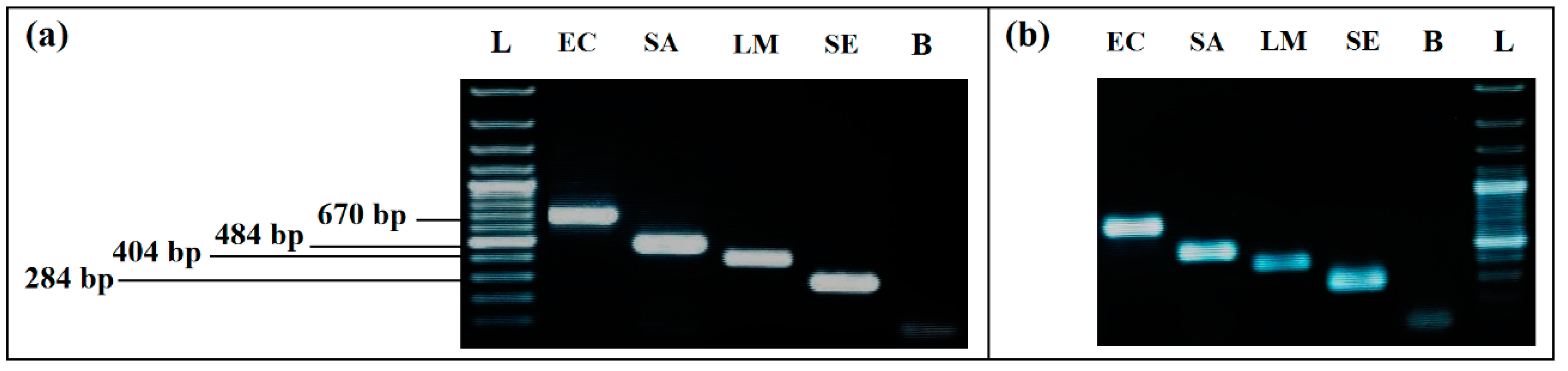
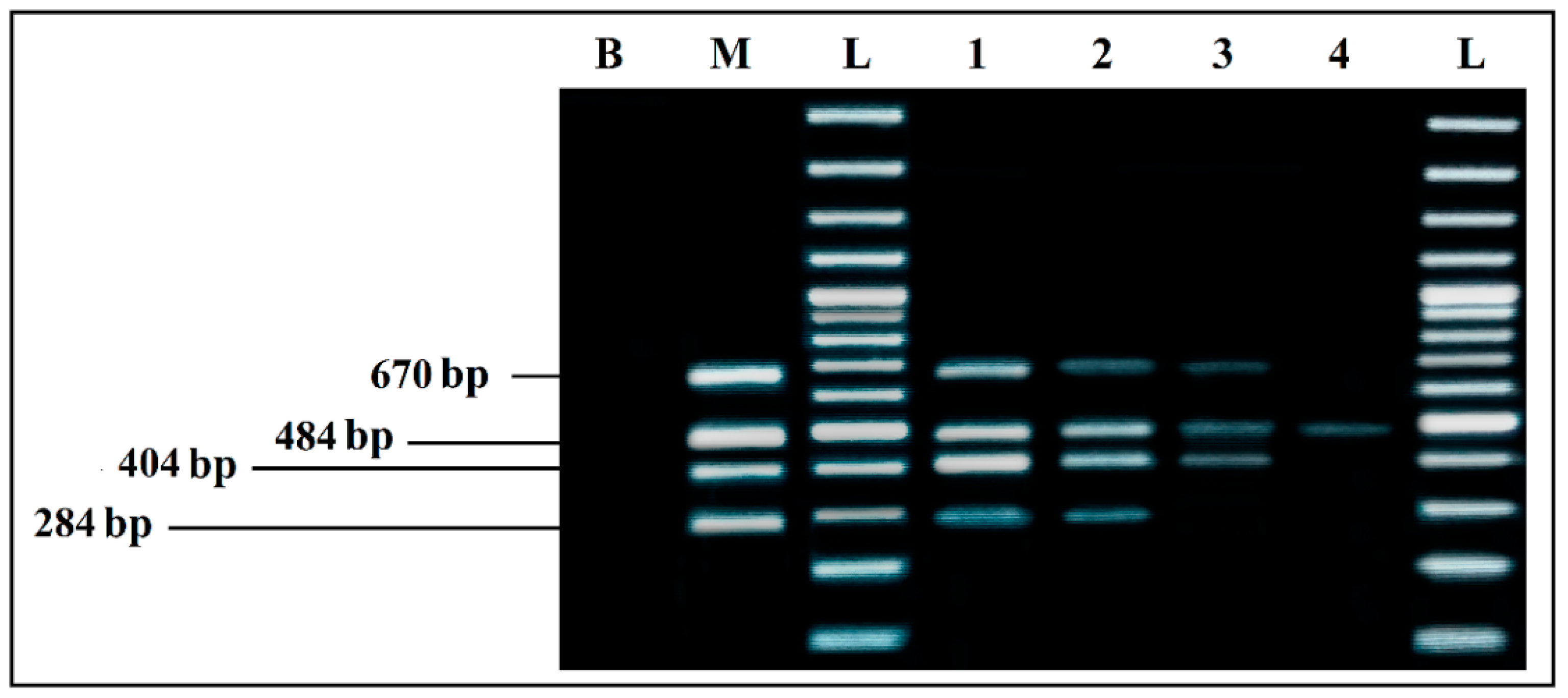
A difference in bands intensity for L. monocytogenes and S. enterica was recorded for the Thermal lysis extraction protocol. By using the GenElute ™ bacterial genomic DNA Extraction kit with lysozyme, intensity of all bands was much higher, without difference between the four bacteria (Figure 1). According to this result, the kit was used to extract the DNA of the samples for all of the remaining assays.
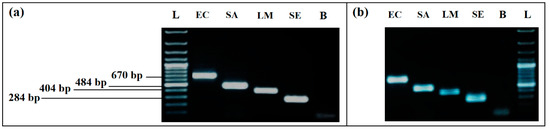
Figure 1.
Comparison of the two DNA extraction methods effect on PCR detection: (a) simplex PCR with DNA extracted by GenElute™ bacterial genomic DNA Extraction kit, with lysozyme. (b) simplex PCR with DNA extracted by thermal lysis protocol, without lysozyme. L, 100 bp DNA ladder; EC, E. coli (8.55 × 108 CFU/mL); SA, S. aureus (3.27 × 108 CFU/mL); LM, L. monocytogenes (2.1 × 107 CFU/mL); SE, S. enterica (2.2 × 108 CFU/mL); L, 100 bp DNA ladder; B, negative control.
3.2.2. mPCR
After the primers were validated in silico and by simplex PCR, multiplexing was carried out by a progressive integration of primers, in several duplex and triplex PCR reactions, then the four primers pairs were mixed in a single reaction with their respective target DNAs (quadruplex PCR). To ensure the amplification of the four target fragments and to avoid non-specific reactions, conditions such as the annealing temperature and concentration balance of the four primers in the reaction, were optimized.
The detection could be made correctly in simplex, duplex, triplex, and quadruplex PCR, without any non-specific product visible on the electrophoresis gel.
3.2.3. Evaluation of Specificity
Regarding the evaluation of multiplex detection specificity, for DNAs extracted from reference strains (E. coli 101 CECT, S. aureus 435 CECT, L. monocytogenes 936 CECT, S. enterica 4266 CECT), each primer amplified exclusively its own target gene.
On the other hand, for the reference strains C. freundii 401 CECT, M. luteus 245 CECT and S. epidermidis 231 CECT or the three laboratory isolate strains B. cereus, L. innocua and L. grayi no detection was recorded, confirming that each primers pair is exclusively selective to its target gene.
3.2.4. Evaluation of Sensitivity
Detection Limits of Simplex and mPCR, from Individual Cultures
To determine detection limits of each PCR simplex under the optimized conditions, several PCRs were carried out following a decreasing gradient of the DNA used in the reactions. DNA quantities used in each PCR mixes were calculated from a linear regression curve. This curve was established using the EXCEL software, from measurements made on concentrated dilutions of each DNA used (Figure 2, Figure 3, Figure 4 and Figure 5).
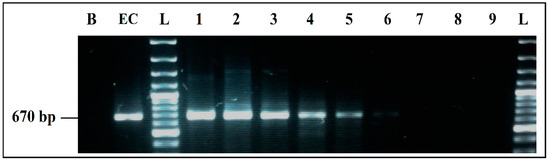
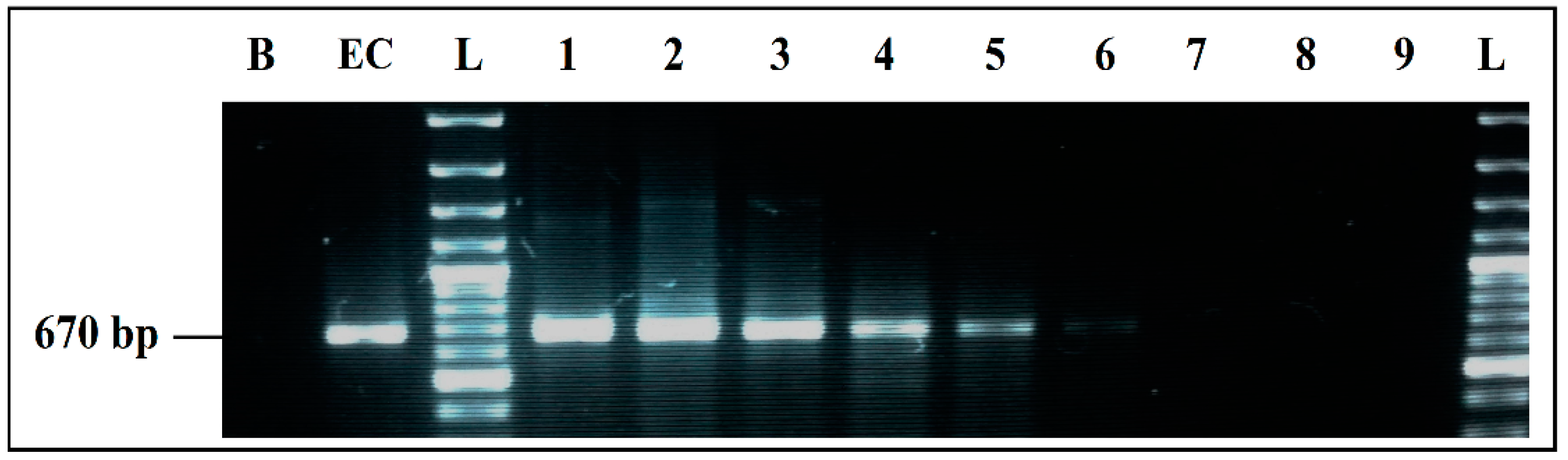
Figure 2.
Detection sensitivity of E. coli primers GADA 670 bp in simplex PCR. B, negative control; EC, positive control E. coli 101 CECT; L, 100 bp DNA ladder; lane 1 to 6, PCR results from 6078 pg/µL to 0.06 pg/µL DNA; Lane 7 to 9, PCR results from 0.006 pg/µL to 0.00006 pg/µL DNA; L, 100 bp DNA ladder.
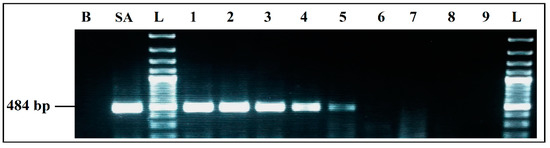
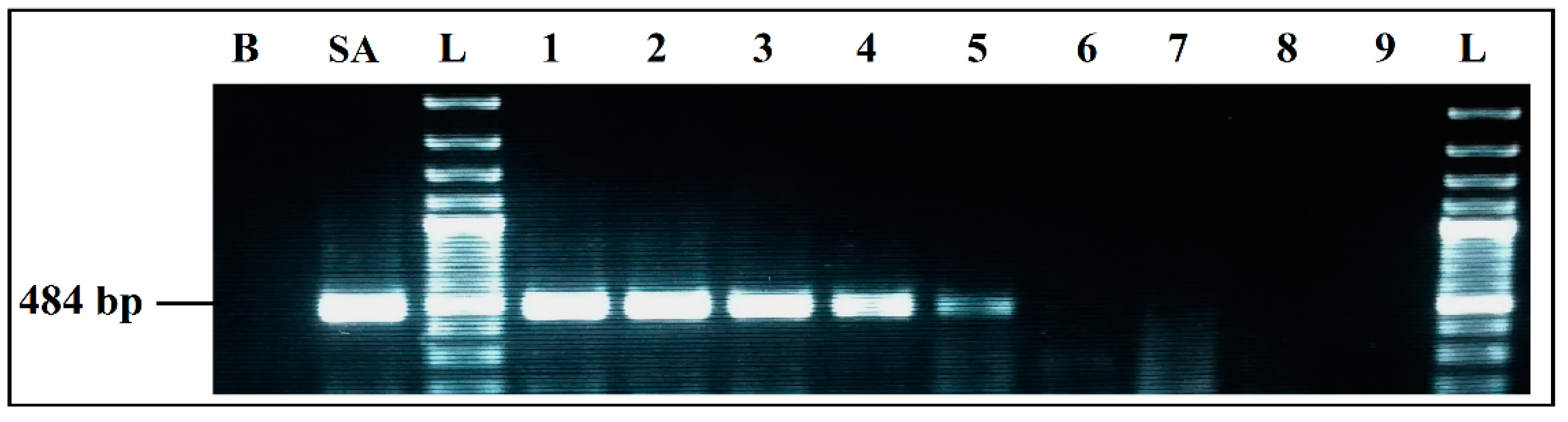
Figure 3.
Detection sensitivity of S. aureus primers Nuc 484 bp, in simplex PCR. B, negative control; SA, positive control S. aureus 435 CECT; L, 100 bp DNA ladder; lane 1 to 6, PCR results from 2457 pg/µL to 0.03 pg/µL DNA; lane 7 to 9, PCR results from 0.003 pg/µL to 0.00003 pg/µL DNA; L, 100 bp DNA ladder.
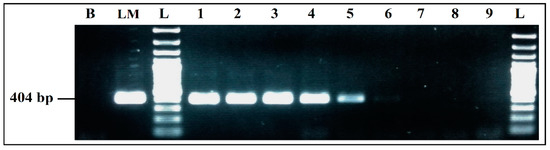
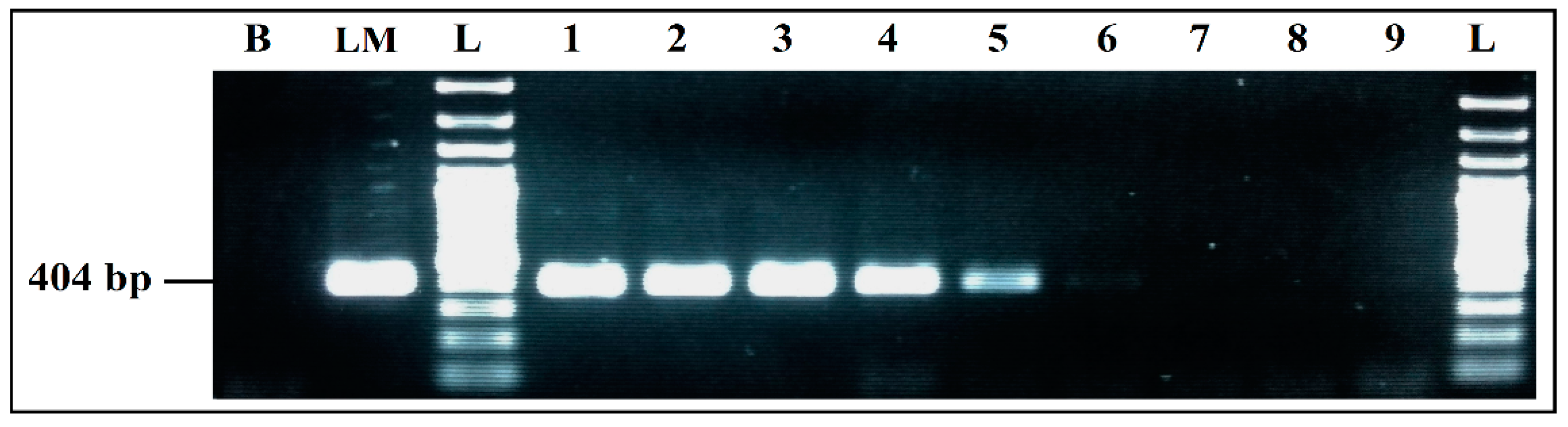
Figure 4.
Detection sensitivity of L. monocytogenes primers LM 404 bp, in Simplex PCR. B, negative control; LM, positive control L. monocytogenes 936 CECT; L, 100 bp DNA ladder; lane 1 to 6, PCR results from 359 pg/µL to 0.004 pg/µL DNA; lane 7 to 9, PCR results from 0.0004 pg/µL to 0.000004 pg/µL DNA; L, 100 bp DNA ladder.
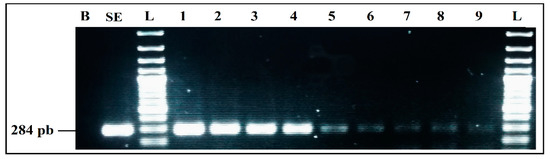
Figure 5.
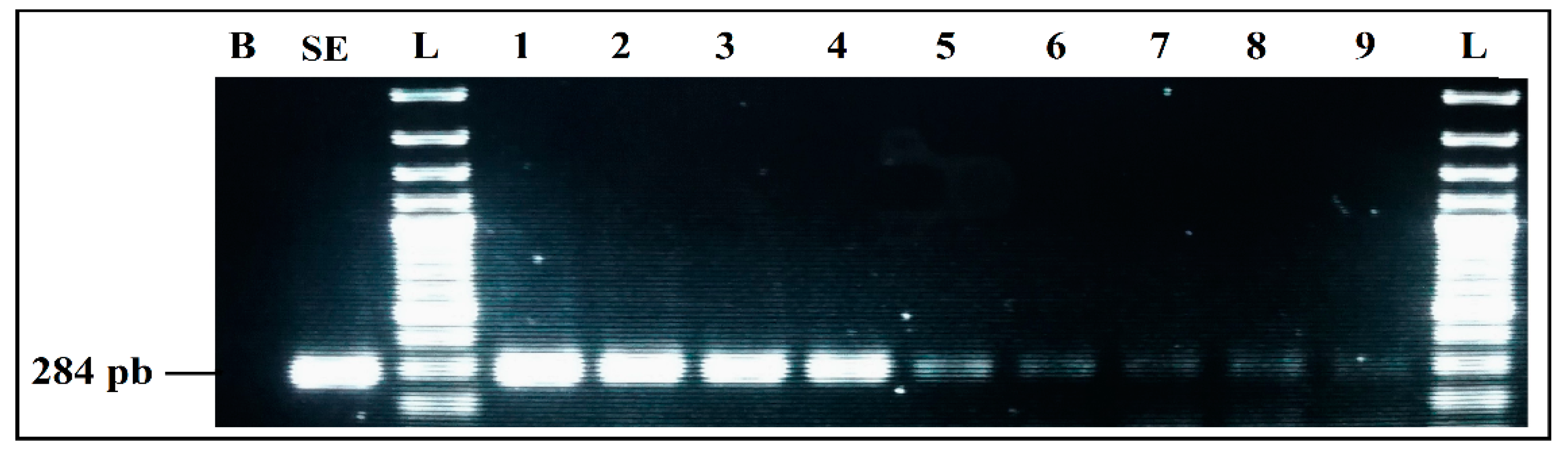
Detection sensitivity of S. enterica primers SalinA 284 bp, in Simplex PCR. B, negative control; SE, positive control S. enterica 4266 CECT; L, 100 bp DNA ladder; lane 1 to 9, PCR results from 4547 pg/µL to 0.00005 pg/µL DNA; L, 100 bp DNA ladder.
PCR detection limits, when the four primer pairs and their target DNAs were mixed in a single reaction, was evaluated by several PCR mixes containing the same components, whose DNA concentrations differed, following a decreasing gradient, with equal ratios between the four target microorganisms in the same reaction. DNA quantities used were calculated, following the same procedure previously mentioned.
The results showed a very clear detection of the four microorganisms (Figure 6), with intense bands (Lane 1). However, by reducing the DNA amount even more (Lane 2), all of the bands were still present, but intensity was lost, which means that the detection limits of the quadruplex PCR are approximately 10 pg/µL of each DNA template.
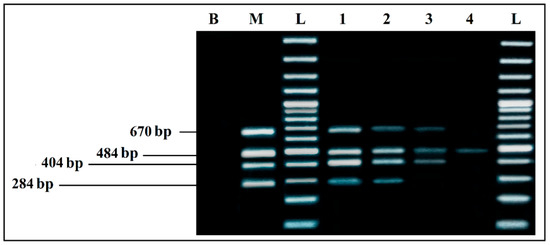
Figure 6.
Detection sensitivity of mPCR from individual cultures. B, negative control; M, mPCR positive control; L, 100 bp DNA ladder; lane 1, mPCR detection with mix DNA (100–70 pg/µL) for each target; lane 2, mPCR detection with mix DNA (10–8 pg/µL) for each target; lane 3, mPCR detection with mix DNA (5–1 pg/µL) for each target; lane 4, mPCR detection with mix DNA (0.6–0.1 pg/µL) for each target; L, 100 bp DNA ladder.
Detection Limits for Co-Culture in BPW
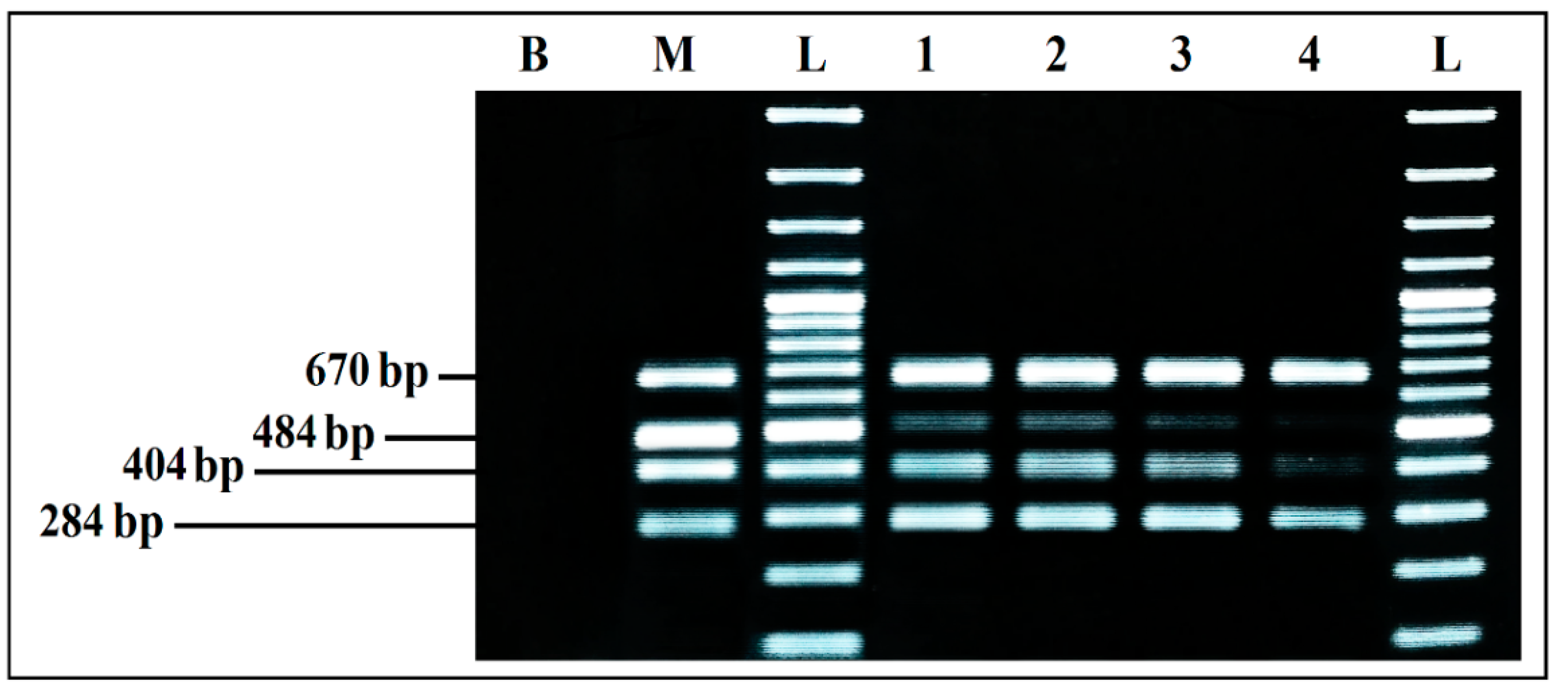
Aliquots of each co-culture were tested by mPCR, to evaluate the effect of BPW as a co-culture medium. The mPCR was able to detect simultaneously the four microorganisms, from the DNA extracted from co-cultures initially inoculated at 103, 102, 101, and 100 CFU/mL, even if the bands intensity clearly decreased for 101 CFU/mL and 100 CFU/mL initial inoculum, especially for S. aureus and L. monocytogenes. On the other hand, the intensity of E. coli and S. enterica bands was strong and invariable from 103 to 100 CFU/mL initial inoculum, with predominance of E. coli (Figure 7).
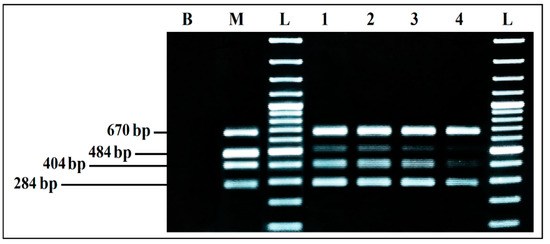
Figure 7.
mPCR detection, from co-cultures in BPW. B, negative control; M, mPCR positive control; L, 100 bp DNA ladder; lane 1, mPCR results from co-culture in BPW initially inoculated at 103 CFU/mL; lane 2, mPCR results from co-culture in BPW initially inoculated at 102 CFU/mL; lane 3, mPCR results from co-culture in BPW initially inoculated at 101 CFU/mL; lane 4, mPCR results from co-culture in BPW initially inoculated at 100 CFU/mL; L, 100 bp DNA ladder.
Detection Iimits from BPW Co-Culture from Artificially Inoculated Food Matrix
Multiplex PCR (mPCR) was applied to determine the efficiency of detection from BPW co-cultures in the presence of ready-to-eat food matrices artificially inoculated, with the presence of background microbiota.
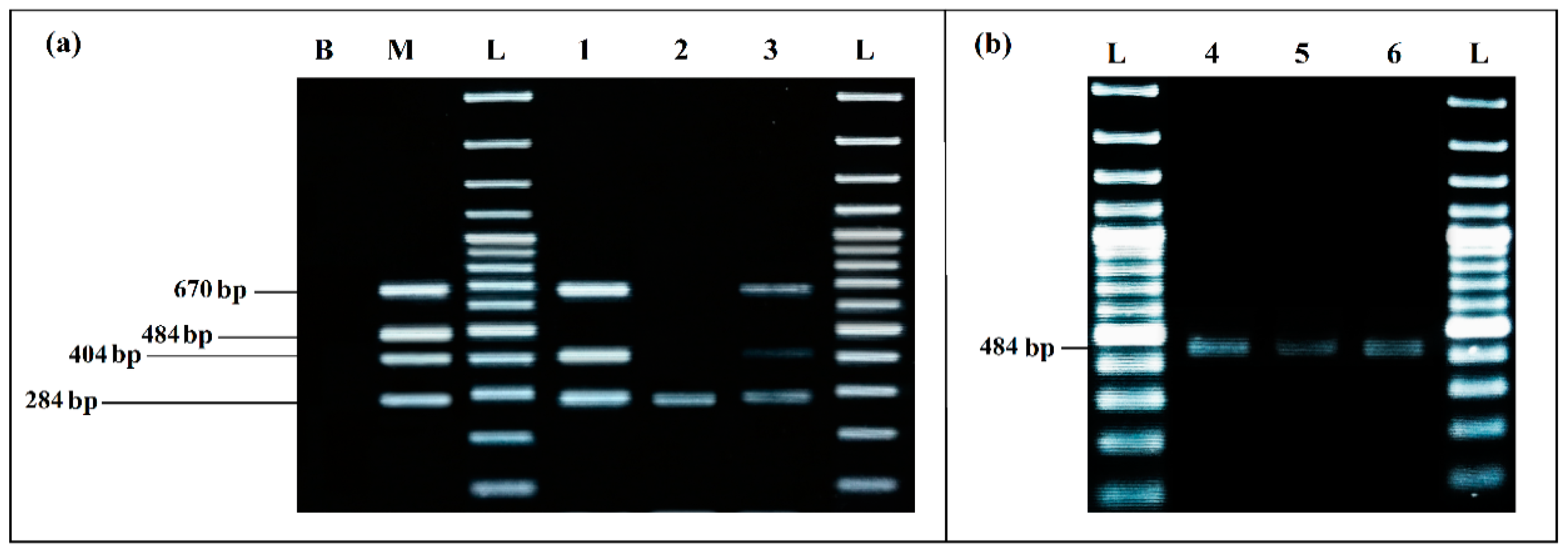
The mPCR performed with the extracted DNA of co-culture from the eco-organic lettuce artificially inoculated with 103 CFU/mL of each target bacteria was able to detect E. coli, S. enterica and L. monocytogenes very clearly, without any non-specific products (Figure 8a).
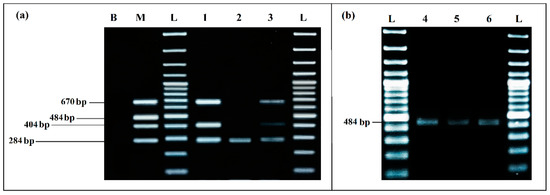
Figure 8.
PCR detection from food matrices artificially inoculated and enriched in BPW: (a) mPCR. B, negative control; M, positive control; L, 100 bp DNA ladder; lane 1, mPCR results from lettuce artificially inoculated (103 CFU/mL); lane 2, mPCR results from minced meat artificially inoculated (103 CFU/mL); lane 3, mPCR results from minced meat artificially inoculated (103 CFU/mL); L, 100 bp DNA ladder. (b) Simplex PCR for S. aureus; L, 100 bp DNA ladder; lane 4, PCR results from lettuce artificially inoculated (103 CFU/mL); lane 5, PCR results from minced meat artificially inoculated (103 CFU/mL) before incubation; lane 6, PCR results from minced meat artificially inoculated (103 CFU/mL) after incubation; L, 100 bp DNA ladder.
For the co-culture from meat sample artificially inoculated with 103 CFU/mL of each target pathogen, after incubation mPCR was able to detect E. coli, S. enterica and L. monocytogenes (Figure 8a, Lane 3).
S. aureus could not be detected by mPCR from any of the aliquots tested (Lane 1, Lane 2 and Lane 3). However, it was clearly detected by simplex PCR from the co-culture aliquots of the two artificially inoculated samples (Figure 8b): from lettuce aliquot at 8.75 × 106 CFU/mL, results presented in (Lane 4) and, more especially, from the meat samples, where a difference in the bands intensity was noticed, before incubation at 1.10 × 103 CFU/mL (Lane 5) and after incubation at 4.45 × 107 CFU/mL (Lane 6).
4. Discussion
This study started from the concept of using the simplest techniques, as well as the least expensive means possible, available in most of laboratories, to develop a rapid method for a simultaneous detection of the four microorganisms E. coli, S. aureus, L. monocytogenes, and S. enterica from raw and ready-to-eat foods. Our goal was to develop a protocol consisting of the combination of a 24 h co-culture step in a single broth combined with a standard mPCR detection.
The results obtained from efficiency comparison between BPW and the two non-selective liquid broths, LB and NB, in individual culture were globally very close, especially for BPW and NB, with a growth exceeding 9.00 log10 CFU/mL for E coli and S. aureus, and equal or above 8 log10 CFU/mL for L. monocytogenes and S. enterica. However, we noticed that the growth of the target bacteria in NB was slightly higher than the resulting from BPW, especially for S. aureus and L. monocytogenes, which may be linked to the presence of beef extract in NB composition. Abd El-Salam et al. (2010) [47] made the same observations about the growth of S. enterica in BPW and L. monocytogenes in NB broth.
Although NB showed better recovery rates for L. monocytogenes and S. aureus, the performance of both culture media was very good. Thus, we chose BPW medium since it is generally used as pre-enrichment broth in many ISO protocols, in addition to its high ability to elute the bacteria from leafy vegetables as lettuce [16]. Moreover, NB contains beef extract and some of its components, such as fats or muscle proteins, are considered as inhibitors that can affect PCR at multiple steps [48]. Wang and Suo (2011) [49] obtained very good detection results after 16 h of growth in BPW, using meat artificially contaminated by E. coli O157 and Salmonella Enteritidis, without any problematic interference due to the presence of background microbiota. The same observation was made by Alarcon et al. (2004) [50] for the simultaneous detection of Salmonella spp., S. aureus and L. monocytogenes from artificially inoculated samples.
The efficiency of BPW to support the growth of each target bacteria in individual culture was compared with other selective and non-selective media. Regarding the selective respective media (BGBLB, GC, RV, FB) used for the enrichment step in the ISO protocols of each target pathogen, BPW showed better performance for the growth of E. coli, S. aureus and S. enterica than BGBLB, GC, and RV, respectively. Similar results were observed for the growth of S. enterica serotype Enteritidis, in RV broth by Yu et al. (2010) [35].
The only selective medium that achieved better growth compared to BPW was Fraser medium, with a maximum density of L. monocytogenes at 9.94 log10 CFU/mL, compared to 8.00 log10 CFU/mL in BPW, which nevertheless remains more than satisfactory. The same observation was made in the work of Yu et al. (2010) [35] and Chen et al. (2015) [5].
Due to the promising results of the selective multi-pathogen enrichment broth SSSLE of Chen and al. (2015) [5], it was added to this study, in order to specifically enrich E. coli, S. aureus, L. monocytogenes, and S. enterica. And since this broth was formulated based on BPW composition, inhibitors for selectivity against food background microbiota and growth promoters, such as esculin for L. monocytogenes and mannitol for S. aureus, were added. Unfortunately, the only growth recorded during our test was E. coli at 5.72 log10 CFU/mL and no growth was observed for the other three target microorganisms.
To evaluate the recovering abilities, individual cultures, and co-culture in BPW broth were performed, with low initial inoculums of 103, 102 and 101 CFU/mL. In individual culture, the maximum densities obtained after 24 h of incubation in BPW E. coli yielded the highest and the most stable recovery levels, with a maximum density of 8.93–8.71 log10 CFU/mL, closely followed by S. aureus 8.51–8.12 log10 CFU/mL and S. enterica 8.34–8.03 log10 CFU/mL. L. monocytogenes obtained the lowest maximum density, ranging between 7.32–7.18 log10 CFU/mL, which nevertheless remains widely detectable by most biomolecular tools.
Regarding the 24 h co-cultures in BPW broth, we found that the growths recorded for all target microorganisms were only slightly affected by the initial inoculum levels decrease. However, the competition effect significantly seemed to affect the resulting recovery rates. The competitive effect between the four microorganisms was demonstrated by the growth rates comparison of each target bacteria, obtained from individual cultures and co-culture under the same inoculation and growth conditions: Gram-negatives seemed to be the least affected, with an average decrease between 0.09–1.10 log10 CFU/mL, while Gram-positives showed a more significant decrease in co-culture, with an average decrease of 2.03–2.39 log10 CFU/mL. This competitive effect during simultaneous growth has been previously mentioned [5,51].
E. coli was not affected by the presence of other bacteria and its growth rates remained stable, while S. enterica showed a slight but insignificant decrease. Regarding L. monocytogenes, the growth rates recorded during co-culture were lower than in individual culture, which is in accordance with Daley et al. (2014) [52], who showed that the competitive effect exerted by Enterobacteriaceae could lead to a decrease in L. monocytogenes population ranging from 1 to 4 logs during 48 h of enrichment. Kim and Bhunia (2008) [34] suggested that the fast-growing E. coli and Salmonellae probably used up most of the nutrients and depleted the culture medium, resulting in a lower growth rate for L. monocytogenes, which is known to be a slow-growing bacteria and a poor competitor.
The maximum population rate in co-culture of S. aureus was not the lowest (6.23–5.62 log10 CFU/mL), but it was the most affected pathogen by the presence of other bacteria, showing the most important density decrease in co-culture, compared to its individual growth. This inhibition decreased with reduction of E. coli inoculum. Oberhofer and Frazier (1961) [53] reported that, although E. coli did not cause any obvious inhibition of S. aureus on spot plates, it could nevertheless strongly suppress growth of S. aureus in liquid medium.
Although the recovery of the four microorganisms in BPW co-culture seems less effective compared to individual culture rates, the resulting growth remain more than sufficient, for mPCR detection.
Recovery capacity of BPW from food matrices artificially inoculated at low initial concentrations (103 CFU/mL) was evaluated, in the presence of background microbiota. Both lettuce and minced meat matrices tested were chosen as ready-to-eat food models, since they are usually consumed (lettuce) or can be consumed (minced meat) without cooking treatment. Lettuce, mainly contaminated by soil or irrigation water [16], and minced meat, mainly by handling [21], can be vectors of Shiga toxin-producing E. coli (STEC), Salmonella spp., L. monocytogenes and S. aureus [54,55].
Regarding the co-culture from lettuce, the recovery of target microorganisms from 103 CFU/mL initially inoculated was greater than 8 Log10 CFU/mL for Gram-negative, thus dominating the Gram-positive, which showed a recovery greater than 6 Log10 CFU/mL. However, compared to the co-culture in medium, a slight increase in the cell density in the presence of lettuce was recorded, around 0.92 Log10 CFU/mL for S. enterica, 0.71 Log10 CFU/mL for S. aureus, as well as 0.45 Log10 CFU/mL for L. monocytogenes, and a decrease of 0.59 Log10 CFU/mL for E. coli. The same observation was made for the co-culture from minced meat compared to the co-culture in medium, where the cell concentrations recovered were more for S. aureus (+1.42 Log10 CFU/mL) and for S. enterica (+0.54 Log10 CFU/mL). For L. monocytogenes, difference was not as significant (+0.03 Log10 CFU/mL). As for the test with lettuce, the recovery of E. coli showed a decrease of 1.17 Log10 CFU/mL compared to the co-culture only in culture medium. This means there was no significant inhibitory effect of the matrix, as well as its background microbiota, during the co-culture in BPW. Therefore, the use of selective agents was not necessary. Moreover, the presence of foods seems to have enriched the co-culture, thus stimulating the growth of target bacteria.
The design of our mPCR assay was made by the combination of four pairs of primers developed in previous research, for simultaneous detection of E. coli [41], S. aureus [42], L. monocytogenes [43], and S. enterica [44], and they have been described as very specific and reliable primers to target the bacteria studied [3,11,16,56,57,58].
The specificity of the primer sequences was checked first in silico by the Primer-BLAST software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/ access on 25 November 2021) [54], then in vitro by duplex, triplex and quadruplex PCR combinations. Multiplex PCR (mPCR) conditions such as annealing temperature, DNA template quantity and concentrations of the reaction mix components were optimized to simultaneously amplify the four target fragments, obtaining the highest detection sensitivity possible. One of the most critical points of this optimization was to find the balance between the concentrations of the four primers, to avoid the preferential amplification of certain target over another and ensure the most homogeneous amplification.
One of the essential factors affecting the PCR sensitivity is the establishment of a reliable DNA extraction procedure [20,59]. On this basis, we compared two widely used methods, namely, the extraction by Thermal lysis method (boiling method) without lysozyme, proposed by Zhang et al. (2012) [11], as well as the extraction by GenElute ™ bacterial genomic DNA extraction kit with lysozyme. After detection by simplex PCR, we were able to confirm that the extraction could be carried out correctly by thermal lysis, even without lysozyme. However, even if the boiling method costs nothing, saved time and eliminate the need of intensive labor [3,60], the DNA extraction by kit is much more efficient regarding the quantity and quality of DNA recovered, which is important during the detection of small amounts of DNA.
mPCR abilities were evaluated firstly, by the determination of its selectivity, which was confirmed by the absence of amplification for non-target bacteria tested. Then the detection sensitivity of each pair of primers was tested by simplex PCR, according to a DNA concentration gradient, from individual pure cultures in BPW. Using the same approach, the detection sensitivity of the mPCR was evaluated, by mixing the four pairs of primers with the four target DNAs, with approximately the same DNA ratio for each target bacteria, in the same reaction. The results obtained showed that reliable simultaneous amplification of the four target microorganisms (quadruplex PCR), under the optimized conditions could be observed until approximately 10 pg/µL of each DNA template in one reaction, which means that each primers pair, tested simultaneously or separately under these optimized conditions, was sensitive and specific enough to detect its target pathogen.
The mPCR detection carried out on co-cultures from the different BPW broth inoculations (103, 102, 101, 100 CFU/mL) was able to successfully amplify DNA fragments of E. coli, S. aureus, L. monocytogenes and S. enterica up to 100 CFU/mL initial inoculum, which confirms the ability of BPW as a co-culture medium for mPCR-based detection. Difference in bands intensity during mPCR detection may be related to the difference in cell concentrations recovered for each pathogen [61]. However, according to Yuan et al. (2009) [57] the traditional mPCR cannot avoid disproportionate amplification between the different primers during the whole reaction, due to the fact that each primers pair have different amplifying efficiencies. On this point, Zhang et al. (2012) [11] and Wei et al. (2018) [3] who used the same primers, for the simultaneous detection of S. aureus and Salmonella spp., noticed that there was fierce competition between the different amplification pathways.
mPCR from co-culture in the presence of food matrices artificially inoculated with 103 CFU/mL of each target microorganisms was able to detect very clearly the presence of E. coli, S. enterica and L. monocytogenes, despite the background microbiota, after incubation. The difference of bands intensity between the two food matrices could be due to the difference in recovered cell concentrations, which were more important from the lettuce, but also to a possible inhibitory effect of meat. According to Garrido et al. (2013) [62] and Rajabzadeh et al. (2018) [16], fats and glycogen are considered as inhibitors and could affect PCR at multiple steps.
Unfortunately, in the presence of food, S. aureus could be detected only by simplex PCR from co-culture. Nevertheless, we noticed an increase of detection intensity after incubation in BPW, compared to the detection from initial inoculum. We assume that the problem is related to the sensitivity of the primers used, and that they should be replaced by more efficient ones. Wei et al. (2018) [3] indicated that detection of S. aureus with Nuc484 primers was not characterized by high sensitivity compared to Salmonella spp. primers invA284 in artificially contaminated food products. Zhang et al. (2012) [11] confirmed that the biomass of S. aureus had to be greater than other bacteria for a perfect amplification of all of the target genes, due to a possible competition established by a preferential amplification. According to Elizaquível and Aznar (2008) [10], the preferential amplification of a target in mPCR can be induced by the presence of a low template concentration. In our case, the template-primer combinations of E. coli, S. enterica., and L. monocytogenes, seems to be more favored, thus affecting the amplification efficiency for S. aureus [63].
5. Conclusions
In conclusion, BPW broth can effectively support the individual and simultaneous growth of E. coli, S. aureus, L. monocytogenes, and S. enterica from low initial contamination levels in the presence of ready-to-eat food matrix such as eco-organic lettuce or minced meat, and could be, thus, used as a co-culture medium prior to mPCR detection. DNA extraction by thermal lysis method was effective on both Gram-negative and Gram-positive bacteria, even without lysozyme. However, the specific DNA extraction kit used in this work can improve the efficiency of the mPCR assay. Although mPCR could not detect S. aureus in artificially inoculated food matrices, the proposed protocol, which includes a BPW pre-enrichment step and detection by mPCR, can confirm the presence or absence of E. coli, Salmonella sp. and L. monocytogenes in foods in approximately 30 h, compared to cultural methods which require at least seven days, therefore considerably reducing the time, efforts, and costs of analyzes.
Author Contributions
Conceptualization: S.B.; methodology: S.B., A.B. and A.G.; formal analysis: S.B., M.A.F.; investigation: A.B., M.G.-F. and A.G.; supervision: S.B.; original draft preparation: A.B.; review and editing: M.A.F.; funding acquisition: M.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Ciencia e Innovación, Spain, Grant number PID2019-105691RB-I00. Miguel García-Ferrús is the recipient of a PEJ2018- 003746A Grant from Ministerio de Economía, Industria y Competitividad, Spain.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| BGBLB | Brilliant Green Bile Lactose Broth |
| BPA | Baird Parker Agar Base |
| BPW | Buffered Peptone Water |
| CECT | Spanish Type Culture Collection |
| CFU | Colony Forming Units |
| dNTPs | Deoxynucleotide triphosphates |
| F/R | Forward/Reverse |
| FB | Fraser Enrichment Broth |
| GC | Giolitti-Cantoni Broth |
| ISO | International Organization for Standardization |
| LB | Luria-Bertani Broth |
| mPCR | Multiplex PCR |
| NB | Nutrient Broth |
| PAL | Palcam Agar |
| PCR | Polymerase Chain Reaction |
| RV | Rappaport Vassiliadis Broth |
| TBX | Tryptone Bile Glucuronic Agar |
| UNE-EN | Spanish Association for Standardisation |
| XLD | Xylose-Lysine-Deoxycholate Agar |
References
- Ruppitsch, W.; Pietzka, A.; Cabal, A.; Chakeri, A.; Schmid, D.; Lakicevic, B.; Lepuschitz, S.; Allerberger, F. Advances in foodborne outbreak investigation and source tracking using whole genome sequencing. Conf. Ser. Earth Environ. Sci. 2019, 333, 012010. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- Wei, C.; Zhong, J.; Hu, T.; Xihong, Z. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.R.; Zakhour, C.M.; Gould, L.H. Foodborne Disease Outbreaks Associated with Organic Foods in the United States. J. Food Prot. 2016, 79, 1953–1958. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Bhunia, A.K.; Tang, C.; Wang, C.; Shi, H. Development of a multi-pathogen enrichment broth for simultaneous growth of five common foodborne pathogens. J. Gen. Appl. Microbiol. 2015, 61, 224–231. [Google Scholar] [CrossRef]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knøchel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela, S.; Tresse, O. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef]
- Ding, T.; Suo, Y.; Zhang, Z.; Liu, D.; Ye, X.; Chen, S.; Zhao, Y. A Multiplex RT-PCR Assay for S. aureus, L. monocytogenes, and Salmonella spp. Detection in Raw Milk with Pre-enrichment. Front. Microbiol. 2017, 8, 989. [Google Scholar] [CrossRef]
- Okafor, A.C.; Ogbo, F.C. Occurrence and enumeration of multiple bacterial pathogens in edible snails from Southeast Nigeria. J. Food Sci. Technol. 2019, 7, 23–30. [Google Scholar] [CrossRef][Green Version]
- Leclerc, V.; Dufour, B.; Lombard, B.; Gauchard, F.; Garin-Bastuji, B.; Salvat, G.; Brisabois, A.; Poumeyrol, M.; Buyser, M.L.; Besse, N.G.; et al. Pathogens in meat and milk products: Surveillance and impact on human health in France. Livest. Prod. Sci. 2002, 76, 195–202. [Google Scholar] [CrossRef]
- Elizaquível, P.; Aznar, R. A multiplex RTi-PCR reaction for simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Staphylococcus aureus on fresh, minimally processed vegetables. Food Microbiol. 2008, 25, 705–713. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Zhou, W.W.; Zhou, Y.; Wang, X.F.; Xu, J.F. Response surface methodology to design a selective co-enrichment broth of Escherichia coli, Salmonella spp. and Staphylococcus aureus for simultaneous detection by multiplex PCR. Microbiol. Res. 2012, 167, 405–412. [Google Scholar] [CrossRef]
- Harris, L.J.; Farber, J.N.; Beuchat, L.R.; Parish, M.E.; Suslow, T.V.; Garrett, E.H.; Busta, F.F. Outbreaks associated with fresh produce: Incidence, growth, and survival of pathogens in fresh and fresh cut produce. Compr. Rev. Food Sci. Food Saf. 2003, 2, 78–141. [Google Scholar] [CrossRef]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M.; Shaharoona, B. Hiding in fresh fruits and vegetables: Opportunistic pathogens may cross geographical barriers. Int. J. Microbiol. 2016, 2016, 4292417. [Google Scholar] [CrossRef]
- Zeng, D.; Chen, Z.; Jiang, Y.; Xue, F.; Li, B. Advances and challenges in viability detection of foodborne pathogens. Front. Microbiol. 2016, 7, 1833. [Google Scholar] [CrossRef]
- Kothe, C.I.; Pessoa, J.P.; Malheiros, P.S.; Tondo, E.C. Assessing the growth of Staphylococcus aureus and Escherichia coli on fruits and vegetables. J. Infect. Dev. Ctries. 2019, 13, 480–486. [Google Scholar] [CrossRef]
- Rajabzadeh, S.; Bahreini, M.; Sharifmoghadam, M.R. A rapid method for separating and concentration of food-borne pathogens using elution from ready-to-eat vegetables. Iran. J. Microbiol. 2018, 10, 385–393. [Google Scholar] [PubMed]
- Centers for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food-10 states, United States, 2005. Morb. Mortal. Wkly. Rep. 2006, 55, 392–395. [Google Scholar]
- World Health Organization. Food Safety and Foodborne Illness; World Health Organization: Geneva, Switzerland, 2007; Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 15 November 2021).
- Suo, B.; Wang, Y. Evaluation of a multiplex selective enrichment broth SEL for simultaneous detection of injured Salmonella, Escherichia coli O157:H7 and Listeria monocytogenes. Braz. J. Microbiol. 2013, 44, 737–742. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Omiccioli, E.; Amagliani, G.; Bandi, G.; Magnani, M. A new platform for real time PCR detection of Salmonella spp., L. monocytogenes and E. coli O157 in milk. Food Microbiol. 2009, 26, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Latha, C.; Anu, C.J.; Ajaykumar, V.J.; Sunil, B. Prevalence of Listeria monocytogenes, Yersinia enterocolitica, Staphylococcus aureus, and Salmonella enterica Typhimurium in meat and meat products using multiplex polymerase chain reaction. Vet. World 2017, 10, 927–931. [Google Scholar] [CrossRef][Green Version]
- Feng, K.; Hu, W.; Jiang, A.; Sarengaowa, X.Y.; Zou, Y.; Yang, L.; Wang, X. A dual filtration-based Multiplex PCR method for simultaneous detection of viable Escherichia coli O157:H7, Listeria monocytogenes, and Staphylococcus aureus on fresh-cut cantaloupe. PLoS ONE 2016, 11, e0166874. [Google Scholar] [CrossRef] [PubMed]
- Burnett, S.; Beuchat, L. Human pathogens associated with raw produce and unpasteurized juices, and difficulties in decontamination. J. Ind. Microbiol. Biotechnol. 2001, 27, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Cantoni, C.; Comi, G. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 2002, 68, 6273–6282. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Hogan, J.; Smith, K.L. Development of a PCR-based assay to detect Shiga toxin producing Escherichia coli, Listeria monocytogenes, and Salmonella in milk. Food Microbiol. 2003, 20, 345–350. [Google Scholar] [CrossRef]
- Dwivedi, H.P.; Jaykus, L.A. Detection of pathogens in foods: The current state-of-the-art and future directions. Crit. Rev. Microbiol. 2011, 37, 40–63. [Google Scholar] [CrossRef]
- Nam, H.M.; Srinivasan, V.; Gillespie, B.E.; Murinda, S.E.; Oliver, S.P. Application of SYBR green real-time PCR assay for specific detection of Salmonella spp. in dairy farm environmental samples. Int. J. Food Microbiol. 2005, 102, 161–171. [Google Scholar] [CrossRef]
- Bai, J.; Shi, X.; Nagaraja, T.G. A multiplex PCR procedure for the detection of six major virulence genes in Escherichia coli O157:H7. J. Microbiol. Methods 2010, 82, 85–89. [Google Scholar] [CrossRef]
- Qin, H.; Shi, X.; Yu, L.; Li, K.; Wang, J.; Chen, J.; Yang, F.; Xu, H.; Xu, H. Multiplex real-time PCR coupled with sodium dodecyl sulphate and propidium monoazide for the simultaneous detection of viable Listeria monocytogenes, Cronobacter sakazakii, Staphylococcus aureus and Salmonella spp. in milk. Int. Dairy J. 2020, 108, 104739. [Google Scholar] [CrossRef]
- Qu, Y.; Bai, Y.; Liu, Y.; Zhou, C.; Zhou, X.; Zhang, D.; Shi, C.; Suo, Y. SSEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Staphylococcus aureus, Escherichia coli O157: H7, and Listeria monocytogenes. J. Food Saf. 2020, 40, 12837. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Wang, Q. Sensitive and specific detection of E. coli, Listeria monocytogenes, and Salmonella enterica serovar Typhimurium in milk by microchip electrophoresis combined with multiplex PCR amplification. Microchem. J. 2020, 157, 104876. [Google Scholar] [CrossRef]
- Shi, X.; Yu, L.; Lin, C.; Li, K.; Chen, J.; Qin, H. Biotin exposure–based immunomagnetic separation coupled with sodium dodecyl sulfate, propidium monoazide, and multiplex real-time PCR for rapid detection of viable Salmonella Typhimurium, Staphylococcus aureus, and Listeria monocytogenes in milk. Int. J. Dairy Sci. 2021, 104, 6588–6597. [Google Scholar] [CrossRef]
- Jacobse, C.N. The influence of commonly used selective agents on the growth of Listeria monocytogenes. Int. J. Food Microbiol. 1999, 50, 221–226. [Google Scholar] [CrossRef]
- Kim, H.; Bhunia, A.K. SEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 4853–4866. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.G.; Wu, H.; Liu, Y.Y.; Li, S.L.; Yang, X.Q.; Xiao, X.L. A multipathogen selective enrichment broth for simultaneous growth of Salmonella enterica serovar Enteritidis, Staphylococcus aureus, and Listeria monocytogenes. Can. J. Microbiol. 2010, 56, 585–597. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization. ISO 4832 (2006); Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coliforms—Colony Count Techniqu; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2006.
- Spanish Association for Standardization. UNE-EN ISO 6579-1 (2017); Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2017.
- Spanish Association for Standardization. UNE-EN ISO 6888-3 (2003); Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coagulase-positive Staphylococci (Staphylococcus aureus and Other Species)—Part 3: Detection and MPN Technique for Low Numbers; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2003.
- Spanish Association for Standardization. UNE-EN ISO 11290-1 (2017); Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2018.
- Spanish Association for Standardization. UNE-EN ISO 7218 (2007); Microbiology of Food and Animal Feeding Stuffs—General Requirements and Guidance for Microbiological Examinations; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2008.
- McDaniels, A.E.; Rice, E.W.; Reyes, A.L.; Johnson, C.H.; Haugland, R.A.; Stelma, G.N., Jr. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β Dglucuronidase. Appl. Environ. Microbiol. 1996, 62, 3350–3354. [Google Scholar] [CrossRef]
- Xu, Y.P.; Cheng, W.; Shao, Y.C.; Chen, F.S. Detection of Salmonella spp., Escherichia coli and Staphylococcus aureus by multiplex PCR. Chin. J. Microbiol. Immunol. 2006, 33, 89–94. [Google Scholar]
- Wu, S.J.; Chan, A.; Kado, C.I. Detection of PCR amplicons from bacterial pathogens using microsphere agglutination. J. Microbiol. Methods 2004, 56, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galan, J.E.; Ginocchio, C.; Curtiss, R., III; Gyles, C.L. Amplification of an invA gene sequence of Salmonella enterica by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Goli, M.; Ezzatpanah, H.; Ghavami, M.; Chamani, M.; Aminafshar, M.; Toghiani, M.; Eghbalsaied, S. The effect of multiplex-PCR-assessed major pathogens causing subclinical mastitis on somatic cell profiles. Trop. Anim. Health Prod. 2012, 44, 1673–1680. [Google Scholar] [CrossRef]
- Abd El-Salam, A.F.; Abd El-Ghany, Z.M.; El-Tahan, M.H. The comparison between different enrichment broth media and selective solid media for growing of Salmonella typhimurium and Listeria monocytogenes. JASB 2010, 7, 351–363. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Suo, B. A new 7-plex PCR assay for simultaneous detection of shiga toxin-producing Escherichia coli O157 and Salmonella Enteritidis in meat products. JCF 2011, 6, 441–447. [Google Scholar] [CrossRef]
- Alarcoän, B.; Garciäa-Canas, V.; Cifuentes, A.; Gonzaälez, R.; Aznar, R. Simultaneous and Sensitive Detection of Three Foodborne Pathogens by Multiplex PCR, Capillary Gel Electrophoresis, and Laser-Induced Fluorescence. J. Agric. Food Chem. 2004, 52, 7180–7186. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.S.; Cox, N.A. Universal preenrichment broth for the simultaneous detection of Salmonella and Listeria in foods. J. Food Prot. 1992, 55, 256–259. [Google Scholar] [CrossRef]
- Dailey, R.C.; Martin, K.G.; Smiley, R.D. The effects of competition from non-pathogenic foodborne bacteria during the selective enrichment of Listeria monocytogenes using buffered Listeria enrichment broth. Food Microbiol. 2014, 44, 173–179. [Google Scholar] [CrossRef]
- Oberhofer, T.R.; Frazier, W.C. Competition of Staphylococcus aureus with other organisms. J. Milk Food Technol. 1961, 24, 172–175. [Google Scholar] [CrossRef]
- Herman, K.M.; Hall, A.J.; Gould, L.H. Outbreaks attributed to fresh leafy vegetables, United States, 1973–2012. Epidemiol. Infect. 2015, 143, 3011–3021. [Google Scholar] [CrossRef]
- Zilelidou, E.; Manthou, E.; Skandamis, P. Growth differences and competition between Listeria monocytogenes strains determine their predominance on ham slices and lead to bias during selective enrichment with the ISO protocol. Int. J. Food Microbiol. 2016, 235, 60–70. [Google Scholar] [CrossRef]
- Kim, J.; Demeke, T.; Clear, R.M.; Patrick, S.K. Simultaneous detection by PCR of Escherichia coli, Listeria monocytogenes and Salmonella typhimurium in artificially inoculated wheat grain. Int. J. Food Microbiol. 2006, 111, 21–25. [Google Scholar] [CrossRef]
- Germini, A.; Masola, A.; Carnevali, P.; Marchelli, R. Simultaneous detection of Escherichia coli O175: H7, Salmonella spp., and Listeria monocytogenes by multiplex PCR. Food Control 2009, 20, 733–738. [Google Scholar] [CrossRef]
- Yuan, Y.; Xu, W.; Zhai, Z.; Shi, H.; Luo, Y.; Chen, Z.; Huang, K. Universal Primer-Multiplex PCR Approach for Simultaneous Detection of Escherichia coli, Listeria monocytogenes, and Salmonella spp. in Food Samples. J. Food Sci. 2009, 74, 446–452. [Google Scholar] [CrossRef]
- Ma, K.; Deng, Y.; Bai, Y.; Xu, D.; Chen, E.; Wu, H.; Li, B.; Gao, L. Rapid and simultaneous detection of Salmonella, Shigella, and Staphylococcus aureus in fresh pork using a multiplex real-time PCR assay based on immunomagnetic separation. Food Control 2014, 42, 87–93. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, L.; Xu, H.; Aguilar, Z.P.; Liu, C.; Gan, B.; Xiong, Y.; Lai, W.; Xu, F.; Wei, H. Detection of non-emetic and emetic Bacillus cereus by propidium monoazide multiplex PCR (PMA-mPCR) with internal amplification control. Food Control 2014, 35, 401–406. [Google Scholar] [CrossRef]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: A practical approach. J. Clin. Lab. Anal. 2002, 16, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Chapela, M.J.; Román, B.; Fajardo, P.; Vieites, J.M.; Cabado, G. In-house validation of a multiplex real-time PCR method for simultaneous detection of Salmonella spp., Escherichia coli O157:H7 and Listeria monocytogenes. Int. J. Food Microbiol. 2013, 164, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Mukhopadhyay, U.K. Novel multiplex PCR approaches for the simultaneous detection of human pathogens: Escherichia coli 0157:H7 and Listeria monocytogenes. J. Microbiol. 2007, 68, 193–200. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).